Abstract
LRRK2 is a 250kDa multidomain protein, mutations in which cause familial Parkinson’s disease. Previously, we have demonstrated that the R1441C mutation in the ROC domain decreases GTPase activity. Here we show that the R1441C alters the folding properties of the ROC domain, lowering its thermodynamic stability. Similar to small GTPases, binding of different guanosine nucleotides alters the stability of the ROC domain, suggesting that there is an alteration in conformation dependent on GDP or GTP occupying the active site. GTP/GDP bound state also alters the self-interaction of the ROC domain, accentuating the impact of the R1441C mutation on this property. These data suggest a mechanism whereby the R1441C mutation can reduce the GTPase activity of LRRK2, and highlights the possibility of targeting the stability of the ROC domain as a therapeutic avenue in LRRK2 disease.
Keywords: LRRK2, ROCO protein, GTPase, Parkinson’s disease, differential scanning fluorimetry, circular dichroism
1. Introduction
Leucine rich repeat kinase 2 (LRRK2) is a member of the ROCO family of proteins, defined by the presence of a ROC (Ras of complex proteins) domain followed by a domain of unknown function termed C-terminal of ROC, or COR [1]. In LRRK2, these domains are flanked towards the C- and N- termini respectively by leucine rich repeat (LRR) and kinase domains that give this protein its name, along with a number of protein/protein interaction domains [2]. The function of LRRK2 is unknown, but several mutations in this protein have been shown to cause autosomal dominant Parkinson’s disease (PD) [3, 4]. Overall, mutations in LRRK2 are the most common genetic cause of PD [5, 6]. Due to their recent description and large size, very little is known about the biochemistry and function of the ROCO proteins. Studies focusing on the enzymatic activities of LRRK1 and LRRK2 have suggested that the GTPase activity of these proteins regulates their kinase activity, in a manner analogous to the interaction of small GTPases and associated kinases such as Ras and Raf [7–9]. Studies of mutations linked to Parkinson’s disease have shown that the kinase activity of LRRK2 is required for cytotoxicity associated with mutations [10–14]. However, while mutations in the kinase domain have a modest activating effect on kinase activity, mutations in the ROC domain of LRRK2 have not consistently been shown to affect kinase activity [15]. These same ROC domain mutations consistently disrupt the GTPase activity of the protein, which might lead to altered regulation of kinase activity, although the mechanistic basis for this is unclear [16–19]. These studies highlight the role of the enzymatic activities of LRRK2 in the disease process, but the exact functional and spatial relationship between these two domains is unknown.
We have previously demonstrated that the R1441C mutation decreases the GTPase activity of LRRK2 in the context of the full-length protein [16]. The crystal structure of LRRK2 ROC domain showed that the R1441 residue is distal to the active site of the protein and revealed that the R1441 residue sits at the interface between two constituent monomers in a dimeric structure [19]. This observation suggests that mutations at this residue could destabilize the interaction between the monomers, which might be the mechanism of enzymatic dysfunction. Consistent with this observation, we demonstrated that the ROC domain containing the R1441C mutation has a decreased ability to precipitate the full-length protein from cell lysates. However, the recent report of the crystal structure of a fragment of a ROCO protein from the prokaryote Chlorobium Tepidium consisting of the ROC and COR domains (the C. Tepidium ROCO protein does not contain a kinase domain) suggested that role of R1441 as mainly inter-domain association. This alternative model implied that that mutations at LRRK2 R1441 should not perturb the structure of the ROC GTPase domain itself, and instead acts to disrupt its interaction with the COR domain [20]. To further investigate the mechanism whereby the R1441C mutation disrupts ROC/ROC interactions and how it decreases the GTPase activity of LRRK2, we have used a variety of biophysical approaches including circular dichroism spectropolarimetry and differential scanning fluorimetry to assess the folding characteristics of the ROC domain and to test whether the R1441C mutation alters these characteristics.
2. Methods and materials
2.1 Protein production and purification
The ROC domain of LRRK2 was expressed and purified as previously reported [21]. Where only the wild type protein was used, the 6x polyhistidine tag was removed as described [21]. The R1441C mutant protein was expressed and purified as a 6xHis tagged fusion and was used to compare with the 6xHis tagged WT ROC protein. Attempts to remove the 6xhis tag from the mutant protein failed due to the drastically decreased stability of the protein (data not shown). Attempts to purify a GTP binding dead (K1347A) mutant form of the protein in a soluble form were also unsuccessful (data not shown).
2.2 Circular dichroism
Spectra were recorded with a Jasco J715 spectropolarimeter and measured at a protein concentration of 1 mg ml−1 with a 0.01 cm path length. The spectra are an average of 50 scans at 25°C, with background molecular ellipticity due to the buffer subtracted, in the presence of 4mM GDP. Analysis of protein stability was carried out with a protein concentration of 0.1 mg ml−1 and a 5mm path length (measurements were taken over a 10 nm bandwidth centered around 220 nm). Unfolding was carried out by melting over a temperature gradient from 25°C to 85°C, ramping by 1° every minute. The ellipticity signal (h) was converted to proportion of molecules in the native state αN, according to the relationship αN=(θ−θU)/(θN−θU), where θU and θN are the ellipticity signals for the unfolded and native states, respectively. Data were fitted to the Van’t Hoff function and the mid-point of thermal denaturation (Tm) defined as DH/DS.
2.3 Differential scanning fluorimetry
Thermal denaturation curves were also obtained using a Bio-Rad iCycler Single Wavelength Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA) and SYPRO Orange fluorescence dye (Invitrogen, Carlsbad, CA) with an excitation and emission wavelength of 490nm and 520nm, respectively. The 6xHis tagged ROC domain proteins (WT and R1441C) were subjected to a thermal gradient of 0.5°C increments from 23°–75°C. The 50µl reaction consisted of 0.25mg/ml protein in 100mM HEPES, 150mM NaCl, 20mM MgCl2, pH 7.5 and a 2x SYPRO Orange final concentration diluted from concentrated stock. Guanine nucleotide ligands (GDP, GTP and GppNP) were added at a final concentration of 2mM accordingly. All experiments were performed with five replicates. The Tm for each sample was calculated using the first derivative of the normalized fluorescence.
2.4 Pulldown Assay
Purified WT and R1441C 6xhis tagged ROC proteins were mixed with excess amounts of purified WT v5-tagged ROC. The protein mixtures were then incubated with Ni-NTA superflow resin for 1 hour at 4°C, in buffer A containing 20mM Tris, 500mM NaCl, 20mM Imidazole, 20 mM MgCl2, 10% Glycerol, pH 8.0. Different guanine nucleotide ligands (GDP, GTP and GppNP) were added to a final concentration of 2 mM accordingly. After extensive wash with buffer A, proteins were eluted from Ni-NTA. Co-purified proteins were analyzed by SDS-PAGE and western blotting with biotinylated anti-V5 antibody (1:10000). Streptavidin conjugated HRP (1:2000) were used for developing the membrane. The SDS gels were scanned by CanonPI CS-U 3.8.1× scanner and the western blot was developed and scanned on Alpha Innotech FluoChem Instrument. The intensities of the bands were integrated and recorded using the software Kodak Digital Science 1D 3.0.2 analysis package. Pulldowns were replicated 3 times.
3. Results
To examine the secondary structure and folding properties of the ROC domain, we used two approaches - circular dichroism spectropolarimetry and differential scanning fluorimetry. CD analysis showed that the ROC domain forms ordered secondary structure in solution, with an absorbance spectrum suggestive of a predominantly α-helical fold with double minima at λ208 and 222 (figure 1A). This is in agreement with data from the crystal structure of the ROC domain, which revealed 46.5% α-helix and 21.6% β-pleated sheet, and with molecular modeling studies based upon the small GTPases [21]. Using graduated thermal melting, and following the percentage of folded protein using absorbance at 220nm with CD, the ROC domain was found to unfold in a co-operative manner (figure 1B). The mid-point of thermal denaturation or Tm determined by the loss of α-helical structure was 41°C for the ROC domain in water in the absence of nucleotides. Using DSF to report the exposure of the hydrophobic core in the absence of nucleotides, the ROC domain unfolded with a very similar Tm of 39°C (figure 1C). As previously demonstrated with H-Ras, the presence of guanosine nucleotides has a major impact on the stability of the LRRK2 ROC domain (figure 1D), increasing the Tm to 52.5°C and 50.4°C in the presence of GDP and GDP respectively [22]. The R1441C mutation caused a destabilization of the ROC domain, decreasing the Tm by 4° in the presence of GDP and 5.5° in the presence of GTP as compared to the wild type domain (figure 1D and table 1).
Figure 1.
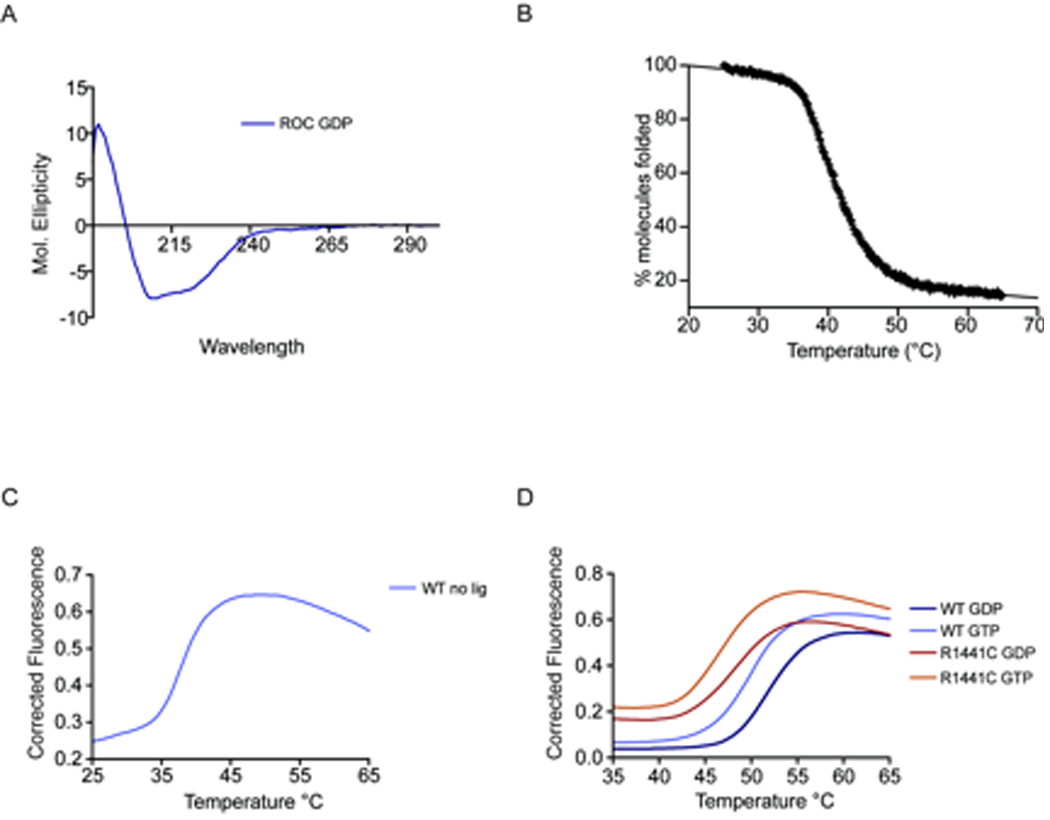
A) CD analysis of the wild type ROC domain, displaying absorption spectra typical of a predominantly α-helical fold B, C) Thermal melt data from the ROC domain in the absence of ligand using CD (B) and DSF (C) to follow loss of structure D) DSF measurement of protein stability in the presence of GDP and GTP for wild type and R1441C ROC domain
Table 1.
Tm values for ROC +/− R1441C and GDP/GTP. Differences in stability due to presence of GDP or GTP for WT (P<0.005) and R1441C (P<0.005) were significant as assessed by Welch’s T-Test, with differences due to the presence of mutation (i.e. WT vs R1441C) in the case GDP (P<0.0001) or GTP (P<0.000005) also being significant.
| Wild Type | R1441C | |
|---|---|---|
| GDP | 52.5±0.8 | 47.8±0.3 |
| GTP | 50.4±0.2 | 46.6±0.4 |
| GppNp | 40.1±0.3 | N/A |
| No ligand | 37.7±0.1 | N/A |
Based upon previous work examining LRRK2 self interaction using LRRK2 and fragments expressed in mammalian cell culture [23], we assessed ROC-ROC domain interactions between WT homodimers and WT/R1441C heterodimers with purified protein in solution, mimicking the situation in humans with heterozygous mutations. Using ROC domains with different epitope tags (6xHistag and V5), a significant decrease was observed in the interaction between mutant and WT domains (figure 2). The interaction between WT and R1441C was lower still in the presence of GTP.
Figure 2.
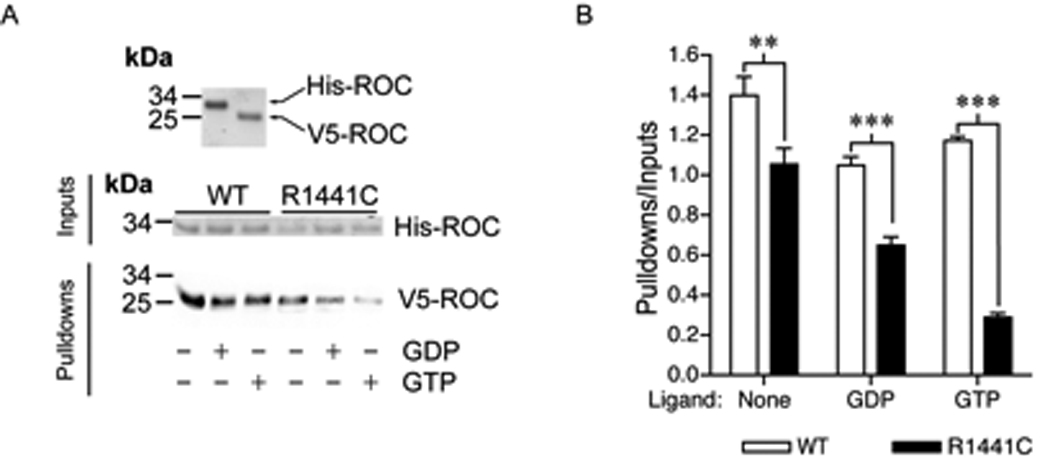
Pulldown analysis of the affinity of wild type and R1441C ROC domain for each other showing a significant decrease in the ability of ROC domain containing the R1441C mutation to precipitate the wild type domain. Analysis was carried out by two-way ANOVA with variant and ligand as separate variables, using a Bonferronni post hoc test. Indicated P values are **<0.01 and ***<0.001.
4. Discussion
We have previously shown that the R1441C mutation in the LRRK2 ROC domain results in a decrease in GTPase activity. Based on the model of a dimeric ROC-ROC structure, we predicted that the mutated residue sits at the interface in a dimeric structure for the ROC domain. In the current study, we sought to test the hypothesis that the R1441C mutation decreases the thermodynamic stability of the ROC domain and to investigate the impact of guanosine nucleotides on the folding, stability and self-interaction of the ROC domain.
We assessed how the R1441C mutation and binding of guanosine nucleotides influence the folding properties of the ROC domain in solution. Using CD to assess secondary structure, we found that the ROC domain of LRRK2 forms a predominantly α-helical secondary structure in solution, agreeing with data from crystallographic studies. Although of low resolution and describing only secondary structure, this is the first report of any structural information for LRRK2 in solution. Using molecular ellipticity as a marker for percent folded protein in solution, we carried out thermal denaturation of the ROC domain, yielding a calculated Tm of 39°C. Using DSF to measure unfolded protein, we examined the impact of ligand binding and the R1441C mutation on the stability of the ROC domain. As previously shown with single domain GTPases, MgCl2 and guanosine nucleotides substantially increase the stability of the ROC domain, with GDP having a more pronounced impact than GTP [22]. This suggests, indirectly, an alteration in the three dimensional structural organization of the domain between the GDP and GTP bound states, and supports the thesis that the ROC domain is acting as a molecular switch [9]. This finding requires further investigation using high-resolution structural approaches. Intriguingly, the R1441C mutation consistently and significantly lowered the stability of the ROC in the presence of either GTP or GDP, suggesting that this mutation acts to destabilize the ROC domain. These data suggest that R1441 plays an important role in the structural integrity of the GTPase domain itself, which is consistent with the crystal structural data of LRRK2 ROC dimer [21]. Due to the fact that the R1441 residue is not conserved between the ROCO proteins and the small GTPases such as a Ras and Rho, it is not possible to study or compare the impact of alterations at this residue in these proteins. We went on to examine ROC domain self-interaction in the light of data showing the impact of GDP and GTP on the stability of the ROC domain using differentially tagged ROC domain constructs. Results from these experiments demonstrate that the strength of interaction between differentially tagged ROC domain monomers is altered in the presence of GDP or GTP, that these have additive effects on stability of the ROC domain in the presence of the R1441C mutation. These data are consistent with structural information indicating that the R1441C mutation is distal from the GTP binding site and suggests that there are separate effects of guanine nucleotides and the R1441C mutation. These results also support previous data from our group looking at the interaction between full length LRRK2 from mammalian cell lysates and the ROC domain in the presence and absence of the mutation [21]. Both the stability data and the pulldown data suggest that there is an alteration in the three dimensional structure of the ROC domain based upon whether GDP or GTP is bound in the active site, again consistent with studies carried out on the small GTPases [24]. We are currently invesstigating this further using a variety of techniques. It is important to note that these studies need extending to larger, multi-domain fragments of LRRK2 in order to shed light on the nature and location of the interactions between the components of the LRRK2 dimer. To date, we have not been able to purify fragments larger than the ROC domain that are stable in solution, and so such further studies wait upon overcoming this technical hurdle.
In summary, our investigations into the folding, stability and self interaction properties of the ROC domain suggests a potential mechanism for dysfunction and loss of GTPase activity of the ROC domain based upon a decrease in stability associated with the R1441C mutation found in familial Parkinson’s disease. They represent the first description of a folding analysis of any of the domains of LRRK2. Future experiments dissecting the relationship between stability and function may shed light on the underlying cause of LRRK2 dysfunction linked to ROC domain mutations, and it will be important to extend these studies using larger fragments of LRRK2 and other mutations. These data also highlight the possibility of targeting the stability of the LRRK2 ROC domain as a potential therapeutic avenue to correcting the dysfunction of this domain, artificially reverting the destabilizing impact of the R1441C mutation.
Acknowledgements
This work was funded in part by the Intramural Research Program of the National Institute on Aging, NIH (NS062287), Oklahoma Agricultural Experiment Station at Oklahoma State University, the Parkinson’s Disease Society (grant #K-0812) and the Medical Research Council. P.A.L. is funded by a senior research fellowship from the Brain Research Trust. The authors would like to thank Dr. Udaya Desilva for his kind support with RT-PCR instrumentation, and declare no competing interest.
Abbreviations
- DSF
differential scanning fluorimetry
- CD
circular dichroism
- GDP
guanosine diphosphate
- GTP
guanosine triphosphate
- LRRK2
leucine rich repeat kinase 2
- ROC
Ras of complex proteins
- PD
Parkinson’s disease
References
- 1.Bosgraaf L, Van Haastert PJ. Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Marin I. The Parkinson disease gene LRRK2: evolutionary and structural insights. Molecular biology and evolution. 2006;23:2423–2433. doi: 10.1093/molbev/msl114. [DOI] [PubMed] [Google Scholar]
- 3.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Clark LN, Wang Y, Karlins E, Saito L, Mejia-Santana H, Harris J, Louis ED, Cote LJ, Andrews H, Fahn S, Waters C, Ford B, Frucht S, Ottman R, Marder K. Frequency of LRRK2 mutations in early- and late-onset Parkinson disease. Neurology. 2006;67:1786–1791. doi: 10.1212/01.wnl.0000244345.49809.36. [DOI] [PubMed] [Google Scholar]
- 6.Paisan-Ruiz C. LRRK2 gene variation and its contribution to Parkinson disease. Hum Mutat. 2009;30:1153–1160. doi: 10.1002/humu.21038. [DOI] [PubMed] [Google Scholar]
- 7.Korr D, Toschi L, Donner P, Pohlenz HD, Kreft B, Weiss B. LRRK1 protein kinase activity is stimulated upon binding of GTP to its Roc domain. Cell Signal. 2006;18:910–920. doi: 10.1016/j.cellsig.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Ito G, Okai T, Fujino G, Takeda K, Ichijo H, Katada T, Iwatsubo T. GTP Binding Is Essential to the Protein Kinase Activity of LRRK2, a Causative Gene Product for Familial Parkinson's Disease. Biochemistry. 2007;46:1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 9.Weiss B. ROCO kinase activity is controlled by internal GTPase function. Science signaling. 2008;1:pe27. doi: 10.1126/scisignal.123pe27. [DOI] [PubMed] [Google Scholar]
- 10.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O'Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 12.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 14.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at Thr558; characterisation of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007 doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greggio E, Cookson MR. Leucine Rich Repeat Kinase 2 mutations and Parkinson's disease: Three Questions. ASN Neuro. 2009 doi: 10.1042/AN20090007. doi:10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem Biophys Res Commun. 2007;357:668–671. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J Neurochem. 2007 doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotthardt K, Weyand M, Kortholt A, Van Haastert PJ, Wittinghofer A. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. Embo J. 2008;27:2352. doi: 10.1038/emboj.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng J, Lewis PA, Greggio E, Sluch E, Beilina A, Cookson MR. Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc Natl Acad Sci U S A. 2008;105:1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Matthews CR. Ligand binding is the principal determinant of stability for the p21(H)-ras protein. Biochemistry. 1998;37:14881–14890. doi: 10.1021/bi9811157. [DOI] [PubMed] [Google Scholar]
- 23.Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniels V, Lewis P, Jain S, Ding J, Syed A, Thomas KJ, Baekelandt V, Cookson MR. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]


