Abstract
Human immunodeficiency virus type 1 (HIV-1) integrase is one of three virally encoded enzymes essential for replication and, therefore, a rational choice as a drug target for the treatment of HIV-1 infected individuals. In 2007 raltegravir became the first integrase inhibitor approved for use in the treatment of HIV infected patients, more than a decade since the approval of the first protease inhibitor (saquinavir, Hoffman La-Roche, 1995) and two decades since the approval of the first reverse transcriptase inhibitor (retrovir, Glaxo Smithkline, 1987). The slow progress towards a clinically effective HIV-1 integrase inhibitor can at least in part be attributed to a poor structural understanding of this key viral protein.
Here we describe the development of a restrained molecular dynamics protocol that produces a more accurate model of the active site of this drug target. This model provides an advance on previously described models as it ensures that the catalytic DDE motif makes correct, monodentate, interactions with the two active site magnesium ions. Dynamic restraints applied to this coordination state create models with the correct solvation sphere for the metal ion complex and highlight the coordination sites available for metal binding ligands. Applying appropriate dynamic flexibility to the core domain allowed the inclusion of multiple conformational states in subsequent docking studies.
These models have allowed us to (1) explore the effects of key drug resistance mutations on the dynamic flexibility and conformational preferences of HIV integrase and to (2) study raltegravir binding in the context of these dynamic models of both wild type and the G140S/Q148H drug resistant enzyme.
Keywords: HIV integrase inhibitor docking, restrained Molecular Dynamics, Relaxed Complex Scheme, metalloprotein modeling, mechanisms of drug resistance
Introduction
Human immunodeficiency virus type 1 (HIV-1) encodes three essential viral enzymes: reverse transciptase, protease, and integrase. Nucleoside and nonnucleoside inhibitors of the reverse transcriptase enzyme and inhibitors of the viral protease enzyme are used for the treatment of infection in combinations known as highly active antiretroviral therapy, or HAART. Despite the undoubted beneficial impact of HAART in the treatment of HIV-1 infection, resistance to these classes of inhibitors has led to the erosion of the efficacy of these combinations, increasing the need for new classes of inhibitor.
Since HIV-1 integrase is absolutely required for viral replication, it represents a rational choice as a target for antiviral drugs. This enzyme performs two essential functions in the process of inserting the viral genome into the human host cell DNA. As part of a cytoplasmic complex known as the pre-integration complex, it first creates reactive CpA 3′-hydroxyl ends (i.e., the “Cytosine-Adenosine overhangs”) by cleaving off two nucleotides from the viral cDNA in a step known as 3′-processing. Following translocation to the nucleus, the integrase enzyme uses the hydroxyl ends in a nucleophilic attack on the host chromosomal DNA in a strand transfer reaction.
Raltegravir, the first FDA-approved HIV integrase inhibitor, blocks the strand transfer reaction.1 Inhibitors that block strand transfer bind to the complex of HIV integrase and the cleaved viral cDNA, of which there is no atomically-detailed structural data available. There are crystal structures that contain one or two of the three domains of HIV integrase, but these structures only contain a single metal ion in the active site, which is likely due to the absence of DNA. However, HIV integrase inhibitors such as the pioneering compounds L-731,988 and S-1360 contain structural features consistent with a two metal chelation motif.2,3 Subsequently, a planar two metal chelation region comprised of oxygen or nitrogen heteroatoms has become a standard feature of the inhibitors disclosed in the 130 plus patent applications on integrase inhibitors and appears to be an essential feature of highly potent inhibitors.1,4,5 We refer to this bis chelation motif, in which the three chelating heteroatoms are in the plane of the aromatic ring to which they are attached or adjacent, as “three coplanar oxygen atoms.” Clinically evaluated compounds such as raltegravir, elvitegravir and GSK364735 highlight this common bis chelation feature (see Fig. 1).
Fig. 1.
Advanced HIV integrase inhibitors raltegravir, elvitegravir, and GSK364735, illustrating coplanar heteroatom two-metal chelation motifs in red.
Although it is a very new anti-HIV drug, raltegravir-resistant mutants of HIV integrase, such as E92Q/N155H and G140S/Q148H in the catalytic core domain, have already been identified in patients.1,6,7 No structural data are available on these drug-resistant double mutants.
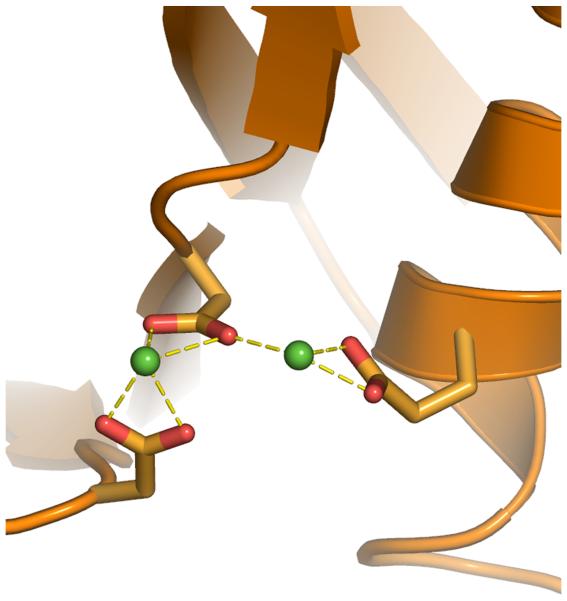
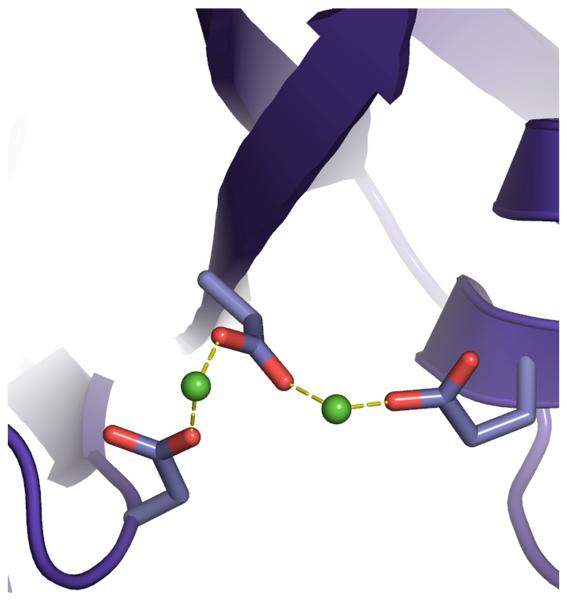
Rigorous new dynamic models of HIV integrase’s catalytic core domain from the wild type and these two raltegravir-resistant double mutants are presented. Although there are several published models that predict binding modes of HIV integrase inhibitors, none of these models contained a flexible representation of the DDE + 2 Mg motif that displayed the proper monodentate interactions with the metals.8-15 Most models have unfavorable bidentate interactions between the carboxylate groups of Asp64, Asp116, Glu152 (i.e., the DDE motif) and the two Mg’s (see Fig. 2). A detailed analysis of the Cambridge Structural Database and the Protein Data Bank showed that magnesium always displays monodentate interactions when it binds to carboxylate groups, with a strong preference for octahedral geometries, at an optimal Mg-O distance of 2.07 Angstroms.16 Instead of modeling the way that integrase dynamically interacts with the two critical magnesium ions in the active site, previous studies generally froze the magnesium-oxygen interactions into a rigid and improper geometry.
Fig. 2.
Comparing 1WKN.pdb (a), and most other published models of integrase that contain two magnesium ions in the active site, to our new models of integrase (b). Before we developed the Mg-O restraints, the coordination spheres of the magnesium ions displayed less-favorable geometries and improper bidentate interactions with the two Mg’s, similar to the flaws in the published model 1WKN.pdb8,9. Coordinations are displayed as dashed lines between a carboxylate group and a green sphere that represents a magnesium ion (at reduced size to enhance clarity). Note: the newest model from the Chimirri group contains only one unfavorable bidentate coordination.15
Improved handling of the two catalytic Mg’s was first proposed with a model that approximates proper monodentate interactions14; however, the Mg-O distances in this model were much longer than the optimal value16. Additionally, although presenting a more catalytically relevant active site, this model was not flexible or dynamic. The lack of Molecular Dynamics (MD) simulations means that only a single conformation of the target was used during these previous docking studies.
Multiple conformations were considered in early simulation studies of HIV integrase by Schames et al.17 Although this work used simulations with a single magnesium ion in the active site and other limitations, it revealed that dynamic fluctuations of the protein allow ligands to bind in more than one orientation, a finding that was influential in the development of raltegravir.18 The possibility of flipped binding modes for the metal chelating groups in several different integrase inhibitors was also predicted in recent “induced fit docking” studies by the Chimirri group, which involved docking inhibitors to a new (static) two Mg model, rearranging the rotamers for residues within 6 Å of the docked ligand, performing an energy minimization on that 6 Å zone, and Glide re-docking.15
The structural description of HIV integrase remains a key goal in HIV research. Despite the success in the development of raltegravir, it is acknowledged that the lack of accurate structural data is hampering drug discovery and development efforts for this enzyme target. We developed restraints that generate dynamic models of the core domain that are consistent with existing experimental evidence on protein-magnesium coordination. Several different types of magnesium-oxygen restraints were tested before an appropriate protocol was produced. This protocol was used to create dynamic models of the wild type, E92Q/N155H, and G140S/Q148H drug-resistant mutants of the catalytic core domain of HIV integrase. Raltegravir was then docked to ensembles that included many different backbone and side-chain conformations that this flexible drug target displayed in our MD simulations.
Results
Performance of the new restraint protocol
The different types of Mg-O restraints investigated produced significantly different behavior with respect to the dynamic interactions between the catalytic domain of integrase, the two magnesium ions, and the water molecules that coordinate the magnesiums. If either the wrong type of Mg-O restraints are applied or if the right type of Mg-O restraints are initially applied and then turned off, then improper bidentate interactions quickly form and are maintained throughout MD.
These different Mg-O restraints did not deleteriously affect the overall dynamics of the core domain of HIV integrase, which lends support to the suitability of the new modeling approach. Using these different Mg-O restraints when minimizing the models and then performing 1 ns equilibration MD runs produced models of the core domain that superimposed well on each other. As expected, snapshots harvested from the different restrained and unrestrained MD simulations displayed significant differences in only the highly-flexible regions of the catalytic domain, which demonstrates that the new restraints did not significantly perturb the overall structure of the catalytic core domain in the new dynamic models.
When our best Mg-O restraints are applied during MD, the new models of HIV integrase all display and dynamically maintain the proper monodentate interactions between the DDE motif and the two Mg’s (see Fig. 2). This behavior was completely reproducible in six different, 20 nanosecond-long MD simulations. Thus, our new protocol models the way that HIV integrase interacts with these two metals in a more realistic manner, while maintaining full flexibility in the active site, which is a significant improvement in the methods used to model integrase.
Monodentate interactions between the DDE motifs and the two magnesiums in the new models cause one oxygen atom of the carboxylate groups of the peripheral D116 and E152 side-chains to be available. These exposed oxygen atoms can then interact with and alter the locations of water molecules that coordinate the Mg’s. The free carboxylate oxygen atom of E152 can also interact with and alter the location of N155’s side-chain. Since N155H is a primary mutation that confers raltegravir-resistance in clinical trials,6,7 properly modeling the dynamic interactions that control the orientation of residue 155 is very important.
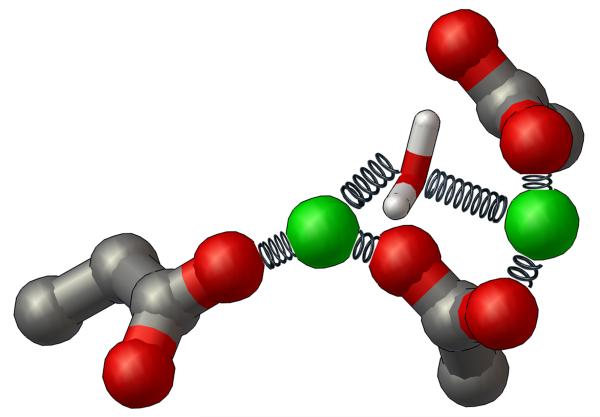
To mimic the effects of an oxygen atom that is likely provided by either a catalytic water or a phosphate group of the cleaved viral cDNA in the relevant drug-binding state,19 a protocol for keeping a specific water molecule held between the two magnesium ions during these MD simulations was also implemented (see Fig. 3). This oxygen atom shared by both Mg’s is likely displaced by the central oxyanion of the integrase inhibitor when it binds to the HIV integrase-DNA complex. The restrained water molecule’s oxygen atom (which has a partial negative charge) provides electrostatic shielding between the two positively-charged magnesium ions, which affects both the Mg-Mg interactions and the way in which the DDE motif dynamically coordinates the two Mg’s. With the new Mg-O restraints applied, seven waters coordinate the two Mg’s at all times, and they always displayed the proper octahedral coordination geometries. Without these restraints, five to seven waters can coordinate the Mg’s, and unfavorable, distorted pyramidal and trigonal biplanar coordination geometries can form. The new restraints are thus the key to obtaining both (1) reasonable locations of the water molecules that coordinate the two Mg’s and (2) proper dynamic interactions that control the structure of the surface of the drug-binding site.
Fig. 3.
Diagram of the NMR-type restraints applied to the DDE motif, the two magnesium ions, and the bridging water between the magnesiums during Molecular Dynamics. The side-chains of the DDE motif are shown as ball-and-sticks, the magnesium ions are displayed as CPK (at reduced size to enhance clarity), and the restrained water molecule is shown as thin sticks. The restraints applied to the magnesium-oxygen interactions during the energy minimization calculations and the subsequent MD simulations are depicted as metal springs. Each spring represents a separate Mg-O restraint that was applied during the creation of these dynamic models of HIV integrase.
Applying the new restrained MD protocol
During clinical studies of raltegravir, three pathways to resistance have been observed involving residues N155H, Q148H/K/R, and Y143C/R.20 E92Q has been shown to enhance the raltegravir resistance associated with N155H when both appear on the same clone.21 A similar relationship exists between G140S and mutations at Q148,22 where G140S appears to mitigate, in part, the fitness loss associated with Q148 mutations.23,24 Possibly as a consequence of this increased fitness, over longer periods of therapy, viruses containing G140S/Q148H tend to expand to replace other resistance mutations, even where the E92Q/N155H mutation has previously predominated.22,25,26 Consequently, the new modeling protocol was applied to three different variants of the catalytic core domain of HIV integrase: the wild type, the E92Q/N155H drug-resistant mutant, and the G140S/Q148H drug-resistant mutant.
The “140s” loop (i.e., Gly140 – Gly149) near the active site is known to be critical to the catalytic function of HIV integrase.27 Since one of the most raltegravir-resistant mutants involves replacing one of the flanking glycine residues with a much more constrained serine residue (i.e., G140S), it is reasonable to expect this mutant to display different structural preferences in the dynamics of its 140s loop. As illustrated in Fig. 4-5, our models predict a marked reduction in conformational flexibility for the G140S/Q148H mutant.
Fig. 4.
Significant differences were displayed in the dynamics of the critical 140s loop during the MD simulations of the wild type, the E92Q/N155H mutant, and the G140S/Q148H mutant of the catalytic domain of HIV integrase. Ribbon diagrams of the QR Factorization-derived ensembles extracted from MD simulations of these three variants at a QH2 = 0.89 are presented. The 140s loop is located at the left side of each image. 27 snapshots from the wild type’s MD simulation are displayed in panel a. 51 conformations of the E92Q/N155H mutant are displayed in panel b, and 47 target conformations of the G140S/Q148H mutant are presented in panel c. The darkest snapshot in each panel corresponds to the beginning of each MD simulation, and the ribbon diagrams get progressively lighter towards the end of each MD simulation. The snapshots were superimposed using the backbone atoms of the DDE motifs.
Fig. 5.
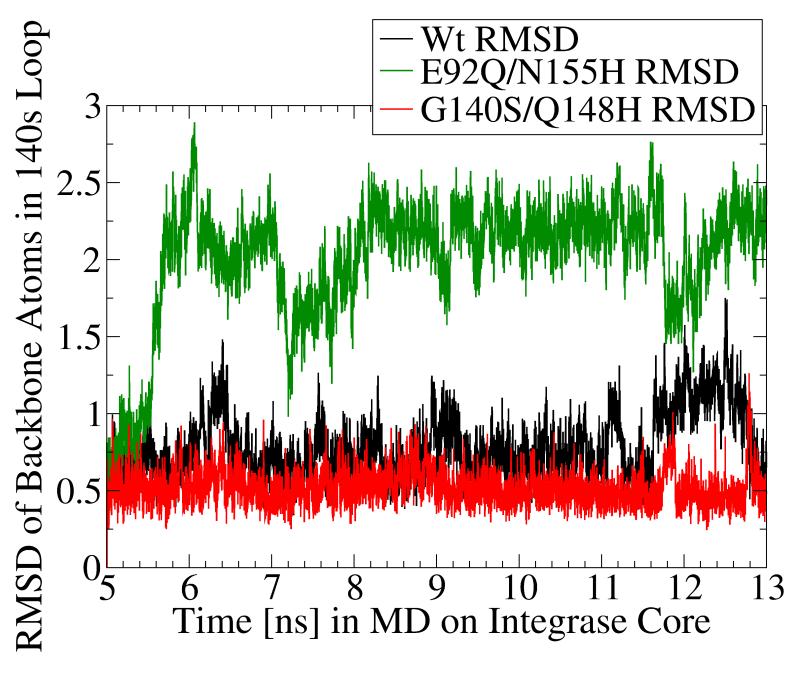
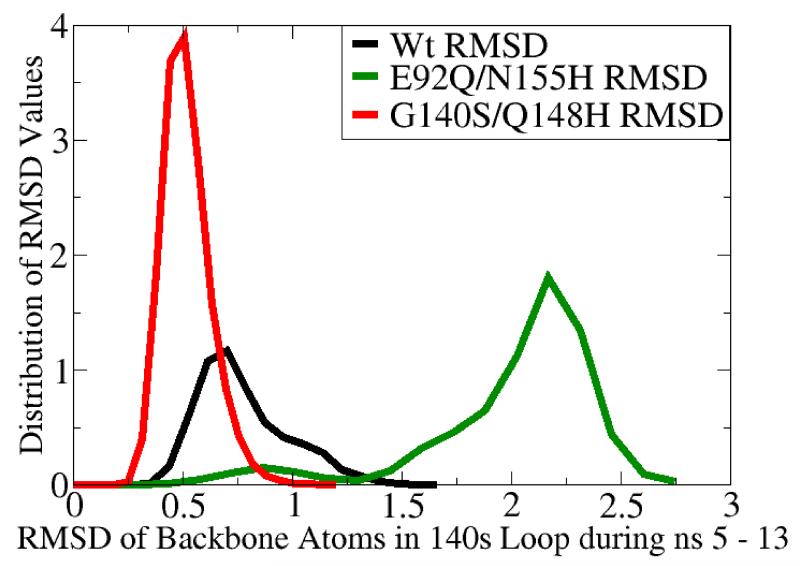
MD simulation of the G140S/Q148H raltegravir-resistant mutant displayed significantly dampened dynamics of the flexible 140s loop. In panels a and b, the Root-Mean-Square-Deviation (RMSD) displayed by the backbone atoms of the flexible 140s loop (i.e., Gly140 - Gly149) during MD simulations of the wild type (in black), the E92Q/N155H mutant (in green), and the G140S/Q148H mutant (in red) are presented. The segment of MD from the fifth to the thirteenth nanosecond is displayed, but the same trends were observed throughout the entire MD simulations. RMSD data indicate the amount of structural variation displayed in each and every conformation with respect to a reference state (i.e., to the first input structure in this segment of MD on each system, which is defined as having an RMSD = 0.0). Panel a presents the trajectory of these RMSD values, while panel b displays histograms of the relative distributions of the different RMSD values present in the 8,000 conformations corresponding to this 8 ns section of MD. The three systems displayed the following RMSD trends in ns 5-13: the wild type system displayed an average RMSD value of 0.776, with a maximum of 1.751 and a standard deviation of 0.209. The E92Q/N155H mutant system displayed an average RMSD value of 2.005, with a maximum of 2.891 and a standard deviation of 0.404, while the G140S/Q148H mutant displayed an average RMSD of 0.519, with a maximum of 1.263 and a standard deviation of 0.110.
Measuring the changes that occurred in the critical 140s loop revealed that both of these drug-resistant double mutants displayed different dynamic behavior than was observed in the wild type’s MD simulation. The G140S/Q148H mutant displayed a much tighter distribution of the distance between the beta carbons of residues 148 and 152 than the other two systems [data not shown]. The other side of the 140s loop was also less flexible in this mutant system. Conversely, the E92Q/N155H mutant displayed much more flexibility in the 140s loop than the wild type, as reflected in the RMSD trends displayed in Fig. 5. The RMSD data agreed with the other types of measurements, which indicated the G140S/Q148H mutant displayed very little variation in the conformation of the 140s loop during MD. The wild type’s MD simulation displayed a moderate amount of flexibility in this critical loop, while the E92Q/N155H mutant displayed much more dynamic flexibility than the other two systems (see Fig. 4-5). A comparative analysis of these MD simulations suggests that two different mechanisms of drug resistance are likely utilized by these two double mutants. This hypothesis agrees well with the observation that, under raltegravir selection pressure, E92Q/N155H and G140S/Q148H are selected independently and most likely employ different, mutually exclusive mechanisms to resist raltegravir.21
Relaxed Complex calculations predict the binding modes of raltegravir
Representative ensembles of conformations of the wild type and G140S/Q148H mutant were utilized in docking experiments with AutoDock4.28,29 These Relaxed Complex calculations30,31 were performed against wild type and mutant ensembles that were extracted with the QR Factorization tool in VMD.32,33 The predicted binding modes for raltegravir (see Fig. 6) are consistent with the main Structure-Activity Relationships (SAR) trend governing the potency of HIV integrase inhibitors.1,5 The relative frequency of integrase conformations that displayed these consistent binding modes with raltegravir was then characterized (see Fig. 7).
Fig. 6.
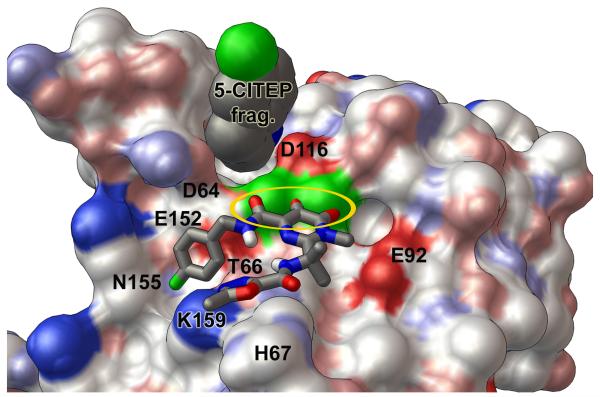
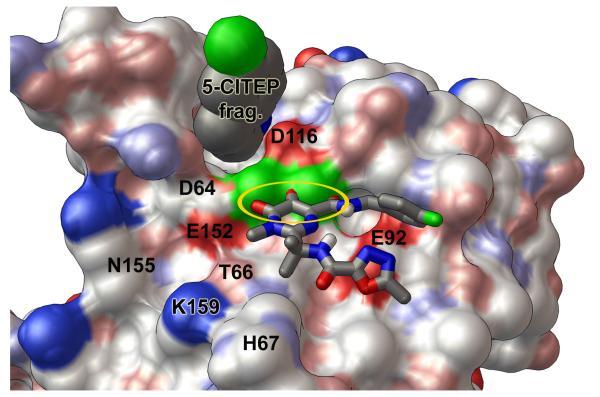
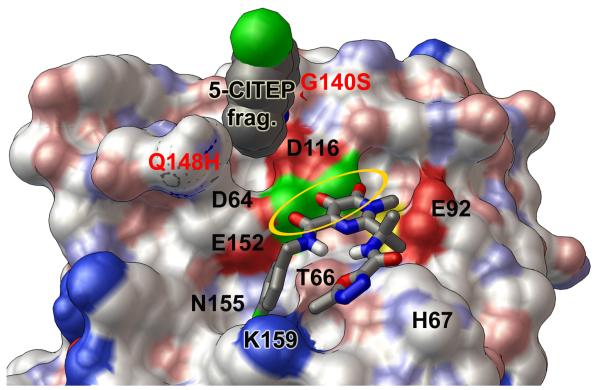
Predicted binding modes of raltegravir against the wild type and the G140S/Q148H mutant of the catalytic core domain. A solvent-accessible surface, colored by the “David Goodsell” convention, is shown with labels for key residues. Panel a presents the “primary binding mode” of raltegravir with the wild type HIV integrase. In this binding mode, “three coplanar oxygen atoms” of raltegravir exhibit four optimal Mg-O distance values (highlighted by the golden oval), and the non-coordinating end of raltegravir has two oxygen atoms forming favorable electrostatic interactions with the side-chain of the critical H67 residue. Other important interactions in this binding mode involve T66, K159, and N155. Panel b displays the “flipped mode” of raltegravir with the wild type catalytic domain, which involves two optimal and two acceptable Mg-O distance values as well as favorable electrostatic interactions with E92. Panel c presents the predicted binding mode of raltegravir with the G140S/Q148H mutant. This binding mode is similar to the primary mode presented in panel a, but raltegravir does not interact as strongly with His67 in the G140S/Q148H mutant. The wild type integrase conformation shown above displayed the following Mg-O distance values between the DDE motif and the two Mg’s: 1.82, 1.82, 1.82, and 1.87 Å. The Mg-Mg distance in this wild type target = 3.51 Å. The G140S/Q148H mutant shown in panel c contained the corresponding Mg-O distance values: 1.93, 1.77, 1.89, and 1.87 Å, with an Mg-Mg distance of 3.75 Å.
Fig. 7.
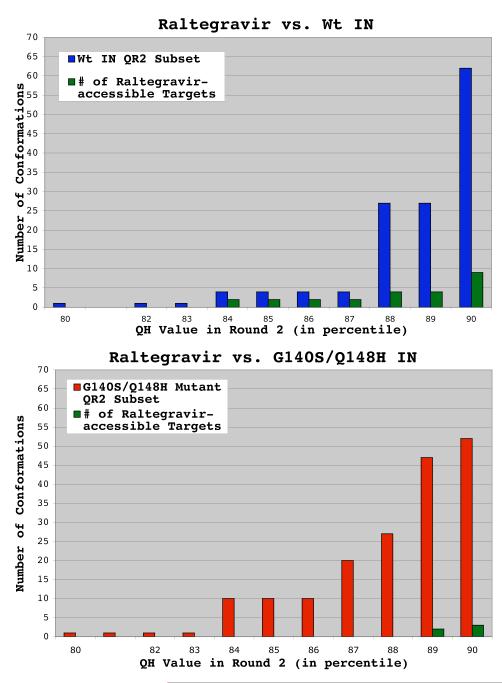
Wild type ensemble displayed a higher frequency of conformations against which raltegravir binds well. The QR Factorization tool in VMD was used to cluster and characterize snapshots harvested from MD simulations of the wild type and G140S/Q148H drug-resistant mutant of integrase. The results for wild type HIV integrase are presented in panel a, while the results for the G140S/Q148H mutant are presented in panel b. The number of conformations that passed through the structural diversity filter in the second round of QR factorization is plotted on the y-axis, while the QH value that extracted these results in the second round of filtering is plotted on the x-axis. When raltegravir was docked against the wild type ensemble of conformations extracted at a QH = 0.90, 9 of the 62 conformations produced binding modes in which raltegravir’s “three coplanar oxygen atoms” were between 1.7 and 2.3 Å from the magnesium ions. I.e., the w.t. ensemble contained 9 raltegravir-accessible targets. Using the same metrics, the G140S/Q148H mutant’s ensemble contained only 3 raltegravir-accessible targets.
The “primary mode” that raltegravir produced when binding to the wild type catalytic domain is displayed in Fig. 6a. When the docking results were clustered with an RMSD tolerance of 2.0 Å, this mode was the best-ranked member of the cluster that displayed the best binding energy. The estimated free energy of binding for this mode was −8.63 kcal/mol (from AutoDock4.2’s scoring function). This cluster had 9 members (i.e., 9/100 independent AutoDock runs against this target produced this mode). Raltegravir displayed four optimal Mg-O interactions between its “three coplanar oxygen atoms” and the two magnesiums (with distances of 2.05, 2.11, 1.74, and 1.75 Å). The non-coordinating end of raltegravir contains two oxygen atoms that formed favorable electrostatic interactions with the NH atom in the side-chain of His67, with O-NH distances of 4.3 and 5.4 Å. Thus, this predicted binding mode allows raltegravir to both (1) interact strongly with the two Mg’s and to also (2) impede the chemistry that the side-chain of His67 likely performs during the catalytic cycle of integrase.34 This binding mode involves interactions with T66, N155, and K159, which are all known to be important for the function of HIV integrase.1,7,10,35
The “QH” value controls the stringency of the structural diversity filter used in the QR Factorization method to cluster the snapshots.32,33 Lower QH values will extract smaller numbers of conformations that encompass the structural diversity displayed within an ensemble of protein structures, while a QH = 1.0 causes no filtering to occur. In the wild type’s ensemble of conformations, the primary binding mode was very accessible. Of the four conformations that best represent the structural diversity displayed during the wild type’s 20 ns-long MD (i.e., the snapshots extracted with a QH = 0.87), 50% of these targets produced the primary binding mode (see Fig. 7). In the full ensemble of wild type conformations targeted, 9 of the 62 conformations (i.e., 15%) produced the primary binding mode.
Raltegravir’s “flipped mode” was only displayed against the wild type ensemble; see Fig. 6b. This flipped mode was present in the third largest cluster of the docking results, which contained 12 members. Although this cluster was slightly larger than the cluster containing the primary binding mode, this flipped mode had a lower estimated free energy of binding of −6.82 kcal/mol. The less-frequent occurrence of the flipped mode against the wild type ensemble and its less-favorable binding energy are the reasons why the other mode was ranked superior. This flipped mode also displayed four Mg-O interactions between its “three coplanar oxygens” and the two magnesiums (at distances of 2.22, 1.60, 2.31, and 1.72 Å). The flipped mode still interacted fairly well with the DDE + 2 Mg motif, but the distances displayed were not as favorable as those found in the primary binding mode. The flipped mode involved several favorable electrostatic interactions with the key residue E92. E92Q is associated with resistance to both raltegravir and elvitegravir.7
Raltegravir’s predicted binding mode against the G140S/Q148H mutant is displayed in Fig. 6c. It is important to note that the G140S and Q148H mutations are present in the 140s loop, unlike the E92Q and N155H mutations, which are part of the predicted drug-binding site. Similar to the primary mode observed against the wild type, raltegravir’s binding mode against this mutant involved interactions with T66, N155, and K159. This mode was also the best-ranked member of the cluster that displayed the best binding energy, but only five members were in this cluster. The estimated free energy of binding was −8.04 kcal/mol. Raltegravir’s “three coplanar oxygen atoms” formed four interactions with the two Mg’s (with distances of 2.01, 1.97, 2.10, and 1.75 Å). However, when compared to the wild type system, this binding mode against the mutant appears less able to impede the putative catalytic activity of H67. In the wild type’s conformations, both the NH and the N atoms in His67’s side-chain are close to two oxygen atoms of raltegravir, which will allow raltegravir to form favorable electrostatic interactions with H67 and shift its pKa. Since this side-chain has flipped in the G140S/Q148H mutant system, only the N atom of H67’s side-chain is near raltegravir (see Fig. 6). In addition, the 5-membered ring at the non-coordinating end of raltegravir has also flipped. This binding mode against the mutant displayed O-NH distances of 7.16 and 5.57 Å. The 2nd best-ranked member of this cluster, which had an estimated free energy of binding of −7.99 kcal/mol, had the same orientation of this 5-membered ring in raltegravir as the primary mode observed against the wild type, but it only formed three strong coordinating bonds to the two Mg’s.
The binding mode raltegravir displayed against the G140S/Q148H mutant is similar to the primary mode that it displayed against the wild type, but this mode was produced by a much smaller percentage of the mutant’s conformations. When the same stringent structural diversity filter was used on this mutant’s ensemble (i.e., a QH value = 0.87), the resulting subset of 20 conformations contained no targets that produced the predicted primary binding mode (i.e., 0%, instead of 50% for the wild type). In the full ensemble of G140S/Q148H mutant targets, only 3 of the 52 mutant conformations (i.e., 6%) produced the primary binding mode. The different rotameric sampling behavior of the key residue H67 amongst these systems (see Fig. 8) likely affected the observed differences in raltegravir accessibility between the wild type and G140S/Q148H mutant.
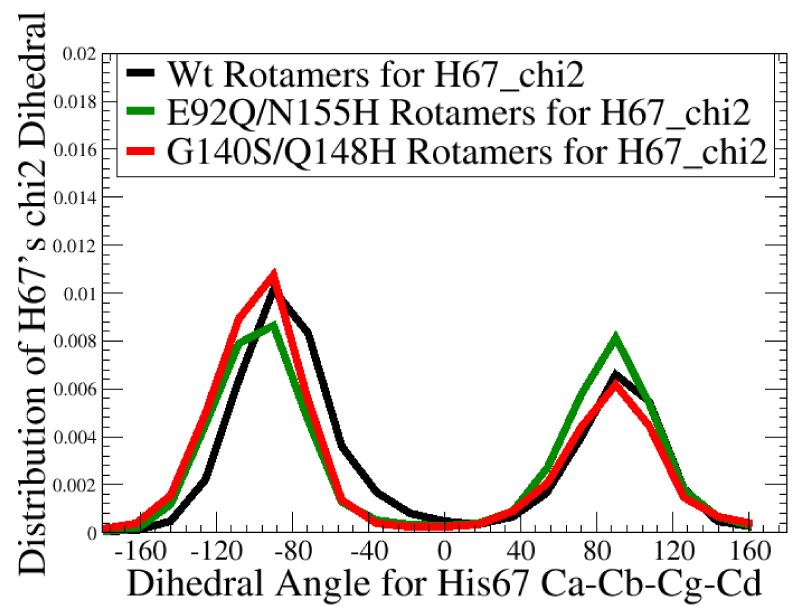
Fig. 8.
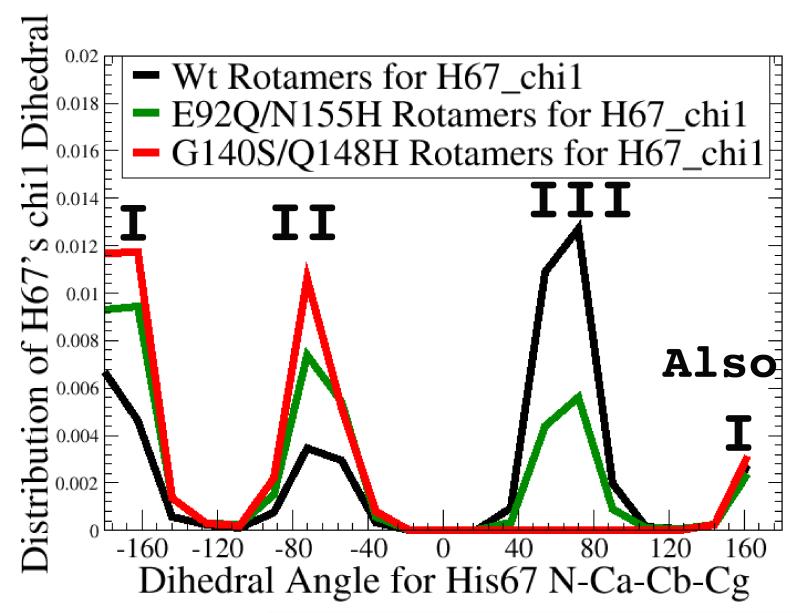
Differences in His67’s dynamic display pattern could affect whether raltegravir can bind well. The chi 1 dihedral angle (in panel a) measures the direction H67 points along the surface of integrase, while the chi 2 dihedral (in panel b) measures the librational motion (or “wobbling”) of H67’s side-chain. H67 is a critical residue: H67 mutants are noninfectious.34 Although no significant differences were observed in the chi 2 dihedral values amongst the wild type’s MD, the  , and the
, and the  , substantial differences were displayed in the chi 1 values. The 3 conformations of the G140S/Q148H mutant against which raltegravir docks well were all from chi 1’s population I. Of the 9 w.t. conformations against which raltegravir docks well, 6 were also from chi 1’s population I, but 3 were from population III, which contains rotamers of H67 that were never sampled during 20 ns of MD on the G140S/Q148H mutant.
, substantial differences were displayed in the chi 1 values. The 3 conformations of the G140S/Q148H mutant against which raltegravir docks well were all from chi 1’s population I. Of the 9 w.t. conformations against which raltegravir docks well, 6 were also from chi 1’s population I, but 3 were from population III, which contains rotamers of H67 that were never sampled during 20 ns of MD on the G140S/Q148H mutant.
Discussion
Several different rounds of docking experiments were performed, in which the charges on the DDE + 2 Mg motif, the charge on the central oxyanion of raltegravir, and the location and identity of the “steric wall” mimicking the viral cDNA’s cytosine-adenosine (CA) overhang were modified (see Materials and Methods). In all of these different rounds of docking experiments, the same conclusions were obtained: raltegravir displayed both the “primary mode” and the “flipped mode” against only the wild type ensemble of conformations of the catalytic core domain. The primary mode was much less accessible in the G140S/Q148H mutant’s ensemble of conformations. When the primary mode can be formed against this mutant, raltegravir seems less able to interfere with the putative catalytic role of H67, since the side-chain of H67 has flipped in the mutant, which caused its NH atom to be much farther away from the oxygen atoms of Raltegravir.
In the active site of integrase, either Y143 or H67 have the required properties to activate a catalytic water capable of hydrolyzing the phosphate backbone of DNA bound in the active site. In patients failing therapy with either raltegravir or elvitegravir, the mutations Y143R/C/H have been detected, but no mutations of H67 have been observed in patients.36,37 In addition, mutant integrase enzymes with Y143F are competent at replication, and mutants with Y143N are viable but have a delayed replication phenotype.38,39 The Y143G mutant is also infective, but H67E and H67Q/K71E are not infective, with the latter double mutant being completely defective.34 Although Y143 has been shown to cross-link to DNA, Y143 does not play a significant role in the ability of HIV to replicate.34 In an in vitro study using site-directed mutagenesis, Y143F was more than twice as active as the H67F mutant.12 However, that assay only detected ligation of target and donor DNA. Since gene expression was not part of the assay, it did not differentiate between productive and defective integration events, which can reduce the clinical relevance of that result. In another study the H67S mutant displayed integrase activity similar to a F185K “wild type model,” but this assay was performed with manganese instead of magnesium, which is known to significantly affect the sequence specificity of the interactions between HIV integrase and the viral cDNA, at least.40,41 In addition, that result may well be a consequence of serine’s ability to act as a nucleophile in a manner similar to Y or H. Since the Y143R/G/F mutants of integrase are viable and infective, and since no mutants of H67 have yet been encountered in patients, the sum of these data suggests that H67 is more likely to play a catalytic role than Y143. This notion underscores the significance of the presented observations regarding the dynamic display patterns of H67 and the ability of specific rotamers of H67 to interact strongly with raltegravir.
While the predicted binding modes of raltegravir presented herein are consistent with the main SAR trend governing the potency of advanced HIV integrase inhibitors,1,5 they are significantly different than the binding modes predicted in a previously-published model by Chen and co-workers.12 This difference may be a consequence of their published model containing improper bidentate interactions between D116 and a Mg.12 In addition, when generating the coordinates for the 140s loop that were missing in their starting crystal structure, they used a loop-building tool which constructed a model with an open conformation of the 140s loop. A bacterial transposase:DNA complex was then used as the source for the position of the DNA in their HIV integrase complex. In Chen’s model the backbone of the integrase and the entire DNA molecule were treated as rigid during the initial energy minimization calculations, which could have trapped the system in an artificial energy well. This led to a fixed open conformation of the 140s loop, when the closed conformation is more likely to be the active, DNA-bound conformation. In our approach we spliced in the coordinates of the closed 140s loop from another crystal structure of HIV integrase when we created our models. MD simulations were then used to generate many different open and closed conformations of the 140s loop, which were included in our docking studies against targets that all displayed the proper coordination geometry between the DDE motif and the two magnesium ions. The aforementioned flaws in the approach described by Chen et al. may explain their surprising conclusion that HIV integrase inhibitors only interact strongly with a single magnesium ion in the active site,12 which is at odds with the widely-known SAR trends discussed previously.
In our presented models, the wild type system displayed oscillations between open and closed states of the 140s loop throughout the entire MD simulation. The E92Q/N155H mutant’s MD exhibited a higher amplitude and frequency of oscillations between open and closed states. The G140S/Q148H mutant’s MD showed more restricted motion around a distorted, closed conformation of the 140s loop. However, these observed differences in dynamic behavior should be validated with NMR or other experimental techniques. These differences in conformational preferences and dynamic flexibility displayed by the 140s loop, combined with the significant differences in the dynamic display pattern of the critical H67 residue, contribute to the fact that the G140S/Q148H mutant’s ensemble contained many fewer conformations against which raltegravir could dock well, relative to the wild type. The G140S/Q148H mutant’s ensemble of snapshots was much less accessible to the predicted primary binding mode of raltegravir, and the flipped binding mode was never observed against this mutant. The trend in accessibility indicates that “kinetic gating” could contribute to the drug-resistance profile of the G140S/Q148H mutant. In addition, if raltegravir induces any significant structural changes in the catalytic domain to achieve its high affinity and inhibitory activity, then its binding would likely pay a larger enthalpic penalty to induce such changes in this more rigid G140S/Q148H mutant. Thus, the results presented indicate that kinetic gating and/or induced fit effects are plausible mechanisms for raltegravir resistance of the G140S/Q148H mutant.
Should this hypothesis of the mechanisms of drug-resistance for the G140S/Q148H mutant be correct, then the following strategy would be useful in guiding the design and evaluation of integrase inhibitors with resistance profiles superior to raltegravir: create fairly rigid compounds with structures that are pre-optimized to interact well with the closed conformations of this double mutant and the wild type integrase. Differences in the dynamic display pattern of His67 must also be taken into account when optimizing inhibitors against this mutant.
The single mutation N155H is a primary/signature mutation that confers raltegravir-resistance in the clinic.6 E92Q is linked with N155H to make a double mutant that is far more raltegravir-resistant than either single mutant.21 In the primary binding mode of raltegravir against the wild type catalytic domain, the fluoro-benzyl group of raltegravir forms a favorable electrostatic interaction with N155. This binding mode has a much more favorable estimated free energy of binding than the “flipped” mode, which interacts well with E92. The fact that the primary mode interacts well with N155 and displays a better binding energy than the flipped mode is in good agreement with the known trends in resistance profiles for the N155H and E92Q single mutants. Additional docking studies need to be performed before predicting raltegravir’s binding mode against this double mutant. But the two binding modes that raltegravir is predicted to display against the wild type appear to explain why the E92Q/N155H double mutant is highly raltegravir-resistant.
If the current preliminary hypothesis of the mechanism of drug resistance for the E92Q/N155H mutant is correct, then a very different strategy should be useful when designing inhibitors with enhanced efficacy against this double mutant. Finding a new class of inhibitors that prevents this mutant from sampling the active conformations of the 140s loop could be quite useful. To defeat a mechanism of drug resistance that involves enhanced flexibility of the critical 140s loop and changes to the surface structure of the active site that affect both binding modes that raltegravir displayed against the wild type, future studies will also involve searching for an allosteric binding site where inhibitors can potentially stabilize the inactive conformations of this critical loop near the active site.
Materials and Methods
Creating our initial model of the catalytic core domain
The crystal structure 1QS442, chain B (of the catalytic core domain of HIV integrase bound to one Mg and to the early Shionogi inhibitor “5-CITEP”) was used as the source for most of the starting coordinates in our model. Since this crystal structure lacks coordinates for most of the 140s loop, these missing coordinates were spliced into the model, using the crystal structure 1BL343, chain C (of the catalytic core domain of HIV integrase bound to one Mg) as the source. Similar to the approach used by both the Wang and Savarino groups11,14, the crystal structure of Avian Sarcoma Virus integrase bound to two zinc ions (in 1VSH44) was used to guide the initial placement of the two magnesium ions in the presented model. The “extract” and “merge” commands in SYBYL7.245 were used to perform the splices. This initial model with the 140s loop and the two magnesium ions spliced into it was the starting structure for the new simulations. When the Biopolymer Module in SYBYL7.245 was used to substitute specific residues to create the two double mutants, these mutations were applied to this initial spliced model. The hydrogen atoms were then added to the models with the MolProbity server,46 which accounts for pKa shifts in titratable residues. The MolProbity server also flips the side-chains of Asn, Gln, and His if doing so would provide a more favorable energy. All protocols were applied equally and consistently to all three variants of HIV integrase.
NMR-type restraint protocol on Mg-O interactions
The best restraint protocol was developed for AMBER9,47 with the parm99SB force field set,48 and with “TIP3P” water molecules.49 For the Mg-O interactions, the force constant k2 = 40 and k3= 20 kcal/mol/Å2 were used in the NMR-type restraints. A separate restraint was created for each integrase-Mg coordination, see Fig. 3. The radii values used were r1= 2.00, r2= 2.04, r3= 2.10, and r4= 2.60 Å.
One water molecule was restrained between the two Mg’s, with the force constant k2 = 40 and k3= 20 kcal/mol/Å2 (see Fig. 3). One restraint was created between a particular water molecule’s oxygen atom and the Mg in site I, and a separate restraint was applied between that oxygen atom and the Mg in site II. The radii values used in both of these Mg-water restraints are r1= 2.00, r2= 2.04, r3= 2.10, and r4= 2.60 Å. To prevent stretching the restrained bridging water between the two Mg’s, each of its hydrogen atoms was restrained to the central oxygen atom of this water. The force constant k2 = 50 and k3= 50 kcal/mol/Å2 were used. One restraint was applied between the “H1” atom and the “O” atom of this water, and a second restraint was created between the “H2” atom and the “O”. The radii values used in both of these restraints are r1= 0.92, r2= 0.95, r3= 0.97, and r4= 1.00 Å.
Energy minimizations with implicit solvent
For these minimizations and for the subsequent MD simulations, AMBER9, the “parm99SB” force-field set, and “TIP3P” water molecules were used.47,48,49 The magnesium ions had a charge of +2.0.
The best NMR-type restraint protocol was applied during the initial implicitly-solvated energy minimization calculations (i.e., only one water molecule was initially present-the water bridging the two magnesiums) and during all subsequent stages of the modeling protocol. In this stage of minimization, the rest of the solvent was mimicked with the “Generalized Born” approximation. A cut-off value of 16.0 Angstroms was applied to the Lennard-Jones interactions.
The first phase of implicitly-solvated minimization consisted of 500 steps of “steepest descent”, followed by 500 steps of “conjugate gradient” minimization. In addition to applying the NMR-type restraints on Mg-O interactions, the standard method of using harmonic restraints on all of the protein’s atoms and the two Mg’s during the early stages of simulation was also implemented, with a restraint weight of 100.0 kcal/mol/Å2. The first phase of implicitly-solvated minimization corrected the unfavorable bond lengths that resulted from the splicing process that generated the starting structure.
In the second phase the weight of the restraints on all of the protein’s atoms and the two Mg’s was decreased to 10.0 kcal/mol/Å2. 200 steps of steepest descent were followed by 300 steps of conjugate gradient minimization. In the third phase of this minimization, the harmonic restraints on all of the protein’s atoms and the two Mg’s were turned off, but the NMR-type restraints on Mg-O interactions were retained during this and all subsequent phases. Minimization using 500 steps of steepest descent were followed by 2,500 steps of conjugate gradient. The fourth phase used 1,000 steps of steepest descent, followed by 2,000 steps of the conjugate gradient method. The fifth phase used 2,000 steps of steepest descent, followed by 1,000 steps with the conjugate gradient method. In addition, the convergence criteria was increased to 0.010 (i.e., drms=0.010).
Energy minimizations with explicit solvent
The output from the previous rounds of energy minimizations was used as the input for the next stage. Each system was solvated with a pre-equilibrated box of TIP3P waters49, with a buffer distance of 10.0 Angstroms around each side of the protein. Approximately 8,000 water molecules were added to each system. Explicit chloride counter-ions were added with the AMBER suite’s “addIons2” method, which generates a coarse electrostatic grid in order to place the counter-ions in energetically favorable locations.47 The older “addIons” method ignores all of the explicit water molecules when calculating where to place the ion; if the ion that was added overlaps a water molecule, then that water is deleted and replaced. Since the older “addIons” method caused the chloride ions to be placed near the magnesium ions in the active site, the newer “addIons2” method, which treats solvent molecules the same as solute atoms, was employed. The addIons2 protocol placed the counter-ions far from the protein’s surface.
The NMR-type restraints were retained during the subsequent rounds of energy minimization with explicit solvent. A 9.0 Angstrom cut-off was applied to the Lennard-Jones interactions, and the Particle Mesh Ewald method was used to calculate the long-range electrostatic interactions.50,51
The first phase of the explicitly-solvated minimization used 500 steps of steepest descent, followed by 5,500 steps of conjugate gradient. The conventional type of harmonic restraints was applied to all of the protein’s heavy atoms and to the two Mg’s, with a restraint weight of 100.0 kcal/mol/Å2, but the protein’s hydrogen atoms and the thousands of explicit water molecules were allowed to move freely (except for the restrained water bridging the two magnesium ions). The second phase used 500 steps of steepest descent, followed by 500 steps of conjugate gradient. The harmonic restraints were still applied to all of the protein’s heavy atoms and to the two Mg’s, but the weight on the restraints was decreased to 10.0 kcal/mol/Å2.
In the third and fourth phases, the harmonic restraints were turned off. Thus, only the Mg-O distances and the water bridging the two Mg’s were restrained. The third phase used 500 steps of steepest descent, followed by 500 steps of conjugate gradient. The fourth phase used 500 steps of steepest descent, followed by 10,000 steps of conjugate gradient, and the convergence criteria was increased to 0.010.
Details of the Molecular Dynamics simulations
The NMR-type restraints were applied during the Equilibration phase of MD and during the entire production phase of MD for all three variants of integrase. Periodic boundary conditions were applied, and the “vlimit” was reduced from the default value of 20 to 15 (i.e., any component of the velocity that exceeds the absolute value of the vlimit is decreased to 15 (while preserving the sign)).
Equilibration MD (i.e., EqMD) gently heats the system to 300 K, while slowly decreasing the weight of the restraints on the protein during each subsequent phase. During all phases of EqMD, the SHAKE algorithm52 was utilized to constrain any bonds that contain a hydrogen atom. The SHAKE algorithm allowed the use of a 2 femtosecond step-size. A snapshot of the current conformation was recorded every 500 steps (i.e., every ps). During all phases of EqMD and during the production phase of MD, a 9.0 Angstrom cut-off was applied to the Lennard-Jones interactions, and the Particle Mesh Ewald method was used to calculate the long-range electrostatic interactions.50,51
The first phase of EqMD had a constant volume and used 100,000 steps. All of the protein’s atoms and the two Mg’s were restrained with conventional harmonic restraints, with a restraint weight of 10.0 kcal/mol/Å2. The starting temperature was 0 K, the final temperature was 300 K, and a time-step of 0.5 psec was used to couple the heat bath to the system.
The second phase of EqMD (and all subsequent phases) ran at a constant pressure, with isotropic position scaling, at a temperature of 300 K. 100,000 steps were involved. All of the protein’s atoms and the two Mg’s were restrained with conventional harmonic restraints, with a restraint weight of 5.0 kcal/mol/Å2. The time-step used to couple the heat bath to the system was increased to 1.0 psec. For the third phase of EqMD (and for all subsequent phases), the conventional harmonic restraints on the protein were turned off. Only the NMR-type restraints on Mg-O interactions and on the water bridging the two Mg’s were applied. This stage also involved 100,000 steps. The time-step used to couple the heat bath to the system was increased to 2.0 psec.
The fourth phase of EqMD was similar to the third phase, but the time-step used to couple the heat bath to the system was increased to 4.0 psec. In addition, the fourth phase involved 200,000 steps. The production phase of MD utilized the same settings as the fourth phase of EqMD. The ptraj module of AMBER9 was used to process and analyze the outputs of EqMD and MD.
Description of the QR Factorization approach
The QR Factorization tool in VMD32,33 was used to cluster the conformations generated in the three, 20 nanosecond-long MD simulations (i.e., to extract and characterize diverse, non-redundant ensembles of conformations). See Fig. 7. By adjusting the stringency of the structural diversity filter (i.e., the “QH value”), one can obtain different numbers of conformations that best represent the structural diversity displayed in an ensemble of protein structures.
In its original form, the “QR Factorization” approach in VMD (also known as the Structure QR Method) was designed as a homology-modeling tool.32,33 It is based on the equation for the “Q value” from energy landscape theory, which uses the changes in the pair-wise distributions of all alpha carbon-alpha carbon distances to describe the physics that controls protein folding/unfolding.53 In a pioneering study, Dr. Rommie Amaro et al. applied the QR Factorization method to increase the efficiency of Relaxed Complex applications by 10 to 100-fold (i.e., to cluster and filter the conformations harvested from MD simulations, in order to extract a small subset of conformations that, when used in docking experiments, still reproduces the overall binding spectrum that was produced by docking inhibitors to the full ensemble of thousands of snapshots from MD).31 The Amaro protocol involves loading a few hundred snapshots at a time into the QR Factorization tool in VMD (~ 100 to 400 snapshots are the limit the program can handle, depending on the number of residues in the system). By using every 10th picosecond snapshot, 200 snapshots corresponds to 2 nanoseconds of MD. Each set of 200 snapshots from the many-nanosecond-long MD simulation was analyzed independently by the QR Factorization tool, to extract a small subset of structurally-diverse, non-redundant conformations. The QH value of 0.90 was used as the cut-off for the structural diversity filter when analyzing each set of snapshots (i.e., each segment of the MD simulation), and all of the resulting subsets were then combined to produce an ensemble of conformations of the drug target against which to dock flexible ligands.31
Motivated by the possible redundancy from many separate QR factorizations, we extended this protocol to enhance the QR Factorization approach’s utility for clustering, extracting, and also characterizing structurally-diverse, non-redundant sets of conformations from MD simulations. To obtain a truly non-redundant, diverse set of conformations for subsequent docking studies, the protocol was extended. Following the Amaro protocol, sets of 200 snapshots (i.e., every 10th ps snapshot from 2 ns of MD) were loaded into the QR Factorization tool, and a QH value of 0.90 was used to filter each set of snapshots. All of the resulting QR subsets were then pooled together to form an ensemble of targets. That combined, QR-selected ensemble of targets was then used as the input for a second round of filtering with the QR Factorization tool. During this second round of filtering, the QH value was systematically modified in order to characterize the number of conformations that were extracted at a particular QH2 value (i.e., QH value in the second round of QR factorization). The QH2 value was incrementally increased from the value that produced a single snapshot in the QR2 results to the value near one that produced a QR2 subset which contained all of the non-redundant input conformations from the first round of QR factorization. The QR2 subsets extracted with a QH2 = 0.90 were targeted in the Relaxed Complex experiments presented.
Description of the docking experiments with AutoDock4.0 and AutoDock4.2
Before starting the docking calculations, a model of adenosine (i.e., the base and the sugar, without the phosphate group) was added to each snapshot harvested from MD to mimic the steric wall provided by the cleaved viral cDNA in the active site. The relevant fragment from 5-CITEP in 1QS4.pdb was extracted, and superimpositions of each snapshot with the 1QS4 reference were used to place either a model of adenosine or that 5-CITEP fragment into each active site (see Fig. 6). The strategy of using the early Shionogi inhibitor “5-CITEP” in 1QS4.pdb as a surrogate for the CA overhang has been used by other labs.13,14 This strategy is supported by experimental data on the kinetic properties of strand transfer inhibitors of HIV integrase, which indicated that adenosine (in the “CA overhang” generated after cleavage of the viral cDNA) acts as a “shield” or wall that impedes the rate of association of inhibitors with the active site.54
AutoDock4.028,29 was initially used when docking raltegravir against the snapshots harvested from MD. 100 independent AutoDock runs were performed against each integrase conformation targeted, with the grid centered on a magnesium ion, a grid spacing of 0.375 Å, and a grid size = 70 × 76 × 76 points. The other run parameters used in AutoDock4.0 were as follows: the number of copies of raltegravir used in each generation (the “ga_pop_size”) was set to 200, the number of energy evaluations (the “ga_num_evals”) equaled 100,000,000, the number of generations within each run (the “ga_num_generations”) was set to 15,000, and the probability of performing a local search (the “ls_search_freq”) was set to 0.07. Default values were used for the other run parameters.
Gasteiger-Marsili (G.M.) charges55 were used for all atoms in the integrase targets and in raltegravir (except for the DDE motif and the two magnesiums). During the first round of experiments with AutoDock4.0 that involved the four wild type targets and the ten targets of the G140S/Q148H mutant present in the QR2 subsets with a QH2 = 0.84, the charges on the magnesium ions and on the oxygen atoms of the DDE motif that coordinate them were derived from Quantum Mechanical simulations (QM) with Gaussian03.56 On the wild type model’s output from equilibration MD, QM calculations were performed on the DDE motif and the two magnesium ions, with and without the presence of the water molecules that coordinate the magnesiums, to investigate the amount of charge that is transferred between these atoms. These QM-calculated charges were then converted to RESP charges, to give these results: both OD atoms of Asp64 and Asp116 = −0.679, both OE atoms of Glu152 = −0.641, and the magnesium ions had a charge of +1.515. AutoDockTools 4 was used to visually inspect all of the binding modes that were produced against each and every conformation targeted.29
The next round of docking experiments with AutoDock4.0 utilized all of the targets in the QR2 subsets with a QH2 = 0.90 (i.e., 62 wild type targets and 52 mutant targets). These targets contained the adenosine model as the steric wall. In this round, the normal G.M. charges were used for the atoms in the DDE motif (OD atoms of D64 = −0.524, OD atoms of D116 = −.520, and OE atoms of E152 = −0.490), while the magnesium ions had a charge of +1.520.
The “raltegravir-accessible targets” were defined according to the results of this round of docking experiments as follows: conformations that produced binding modes in which (1) the “three co-planar oxygen atoms” in raltegravir chelated both magnesium ions of the core domain and in which (2) one or two oxygen atoms of raltegravir were near His67. Chelating a magnesium ion was defined as having a docked conformation in which the Mg-O distances were between 1.7 and 2.3 Angstroms. A docked conformation was defined as interacting strongly with H67 if two oxygen atoms of the ligand were within 5.5 Å of the NH atom in H67’s side-chain.
The final round of experiments used AutoDock4.2 to dock raltegravir against the wild type target and the mutant target that displayed the best binding mode (i.e., most consistent with the known SAR trends) and clustering properties from the previous rounds. Instead of using a single placement of the rough adenosine wall based on the structure of 5-CITEP in 1QS4, two new locations of the adenosine wall and two locations for the corresponding ring of 5-CITEP were created and manually adjusted. The identity of the adenosine wall in these new locations was also modified as follows: one model contained a methyl group “cap” that replaced the oxygen atom that would normally be attached to the phosphate group, and the other wall had all of its atom types changed to carbon. Thus, several different placements and compositions of this wall were investigated in independent docking experiments on the best wild type and mutant targets. This round used the normal G.M. charges for the DDE motifs (see above) and the rest of the targets, but the magnesium ions had a charge of +2.0, and the central enolate oxygen atom of raltegravir was given a charge of −0.500. Although the binding modes produced in this round were very similar to those from previous rounds, the clustering properties improved significantly in this final round of retesting. These results are displayed in Fig. 6.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Catherine “Kate” Burt, whose keen insights and perseverance resulted in the studies described herein. The authors thank Prof. Lou Noodleman of TSRI for suggesting excellent references on metalloproteins that contain magnesium. We thank Dr. Qing Zhang (formerly of TSRI) for advice on modeling protein-metal interactions. We thank John Eargle of the Luthey-Schulten Lab at UIUC for instruction on the use of the QR Factorization tool in VMD. We thank Dr. Maria L. Barreca of the Chimirri group for providing pdb files of their newest models of the integrase-raltegravir complex as a reference.15 Molecular images were created with the freely-available Python Molecular Viewer (PMV).29,57 Support for this TSRI-Pfizer collaboration was provided by SFP-1758 from the Anti-Virals Research Unit at Pfizer Global Research and Development, Sandwich, Kent, UK. Additional support was provided by the National Biomedical Computational Resource (NBCR), NSF, NIH (American Recovery and Reinvestment Act of 2009, grant 3P01GM083658-02S1), the Center for Theoretical Biological Physics (CTBP), and the Howard Hughes Medical Institute. This research was performed, in part, to extend the scope of the virtual screens performed in the FightAIDS@Home project, which is part of IBM’s “World Community Grid.”
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Rowley M, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 2008;51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 2.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Miller MD, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287(5453):646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 3.Fikkert V, Van Maele B, Vercammen J, Hantson A, Van Remoortel B, Witvrouw M, et al. Development of resistance against diketo derivatives of human immunodeficiency virus type 1 by progressive accumulation of integrase mutations. J. Virol. 2003;77(21):11459–11470. doi: 10.1128/JVI.77.21.11459-11470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johns BA, Svolto AC. Advances in two-metal chelation inhibitors of HIV integrase. Expert Opin. Ther. Patents. 2008;18(11):1225–1237. [Google Scholar]
- 5.Langford HM, Williams PD, Homnick CF, Vacca JP, Felock PJ, Schleif WA, et al. Design and synthesis of substitute 4-oxo-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazine-2-carboxamides, novel HIV-1 integrase inhibitors. Bioorg. Med. Chem. Lett. 2008;18:721–725. doi: 10.1016/j.bmcl.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 6.Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Nguyen BY, et al. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 2008;359(4):355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 7.Myers RE, Pillay D. Analysis of natural sequence variation and covariation in Human Immunodeficiency Virus type 1 integrase. J. Virol. 2008;82(18):9228–9235. doi: 10.1128/JVI.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Luca L, Pedretti A, Vistoli G, Barreca ML, Villa L, Monforte P, Chimirri A. Analysis of full-length integrase-DNA complex by a modified approach for DNA docking. Biochem. Biophys. Res. Commun. 2003;310:1083–1088. doi: 10.1016/j.bbrc.2003.09.120. [DOI] [PubMed] [Google Scholar]
- 9.De Luca L, Vistoli G, Pedretti A, Barreca ML, Chimirri A. Molecular Dynamics studies of the full-length integrase-DNA complex. Biochem. Biophys. Res. Commun. 2005;336:1010–1016. doi: 10.1016/j.bbrc.2005.08.211. [DOI] [PubMed] [Google Scholar]
- 10.Ferro S, De Luca L, Barreca ML, Iraci N, De Grazia S, Chimirri A, et al. Docking studies on a new Human Immunodeficiency Virus integrase-Mg-DNA complex: phenyl ring exploration and synthesis of 1H-benzylindole derivatives through fluorine substitutions. J. Med. Chem. 2009;52(2):569–573. doi: 10.1021/jm8009266. [DOI] [PubMed] [Google Scholar]
- 11.Wang LD, Liu CL, Chen WZ, Wang CX. Constructing HIV-1 integrase tetramer and exploring influences of metal ions on forming integrase—DNA complex. Biochem. & Biophys. Res. Commun. 2005;337:313–319. doi: 10.1016/j.bbrc.2005.08.274. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Tsiang M, Yu F, Hung M, Jones GS, Chen JM, et al. Modeling, analysis, and validation of a novel HIV integrase structure provide insights into the binding modes of potent integrase inhibitors. J. Mol. Biol. 2008;380:504–519. doi: 10.1016/j.jmb.2008.04.054. [DOI] [PubMed] [Google Scholar]
- 13.Loizidou EK, Kousiappa I, Zeinalipour-Yazdi CD, Van de Vijver DAMC, Kostrikis LG. Implications of HIV-1 M group polymorphisms on integrase inhibitor efficacy and resistance: genetic and structural in silico analyses. Biochem. 2009;48:4–6. doi: 10.1021/bi8019349. [DOI] [PubMed] [Google Scholar]
- 14.Savarino A. In-silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate. Retrovirol. 2007;4:21–35. doi: 10.1186/1742-4690-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barreca ML, Iraci N, De Luca L, Chimirri A. Induced-Fit Docking approach provides insight into the binding mode and mechanism of action of HIV-1 integrase inhibitors. ChemMedChem. 2009;4:1446–1456. doi: 10.1002/cmdc.200900166. [DOI] [PubMed] [Google Scholar]
- 16.Bock CW, Katz AK, Markham GD, Glusker JP. Manganese as a replacement for magnesium and zinc: Functional comparison of the divalent ions. J. Am. Chem. Soc. 1999;121(32):7360–7372. [Google Scholar]
- 17.Schames JR, Henchman RH, Siegel JS, Sotriffer CA, Ni H, McCammon JA. Discovery of a novel binding trench in HIV integrase. J. Med. Chem. 2004;47:1879–1881. doi: 10.1021/jm0341913. [DOI] [PubMed] [Google Scholar]
- 18.Hazuda DJ, Anthony NJ, Gomez RP, Jolly SM, Wai JS, Young SD, et al. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl. Acad. Sci. USA. 2004;101(31):11233–11238. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courcot B, Firley D, Fraisse B, Becker P, Gillet JM, Ghermani NE, et al. Crystal and electronic structures of magnesium(II), copper(II), and mixed magnesium(II)-copper(II) complexes of quinoline half of styrylquinoline-type HIV-1 integrase inhibitors. J. Phys. Chem. B. 2007;111:6042–6050. doi: 10.1021/jp0687551. [DOI] [PubMed] [Google Scholar]
- 20.Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Nguyen BY, et al. BENCHMRK Study Teams: Subgroup and Resistance Analyses of Raltegravir for Resistant HIV-1 Infection. N. Engl. J. Med. 2008;359(4):355–65. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 21.Fransen S, Gupta S, Danovich R, Hazuda D, Miller M, Huang W, et al. Loss of raltegravir susceptibility in treated patients is conferred by multiple non-overlapping genetic pathways. Antivir. Ther. 2008;13(Suppl 3):A9. [Google Scholar]
- 22.Charpentier C, Karmochkine M, Laureillard D, Tisserand P, Bélec L, Piketty C, et al. Drug resistance profiles for the HIV integrase gene in patients failing raltegravir salvage therapy. HIV Med. 2008;9(9):765–70. doi: 10.1111/j.1468-1293.2008.00628.x. [DOI] [PubMed] [Google Scholar]
- 23.Delelis O, Malet I, Na L, Tchertanov L, Calvez V, Mouscadet JF, et al. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 2009;37:1193–1201. doi: 10.1093/nar/gkn1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakahara K, Wakasa-Morimoto C, Kobayashi M, Miki S, Noshi T, Fujiwara T, et al. Secondary mutations in viruses resistant to HIV-1 integrase inhibitors that restore viral infectivity and replication kinetics. Antiviral Res. 2009;81:141–146. doi: 10.1016/j.antiviral.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Miller MD, Danovich RM, Ke Y, Witmer M, Zhao J, Harvey C, Nguyen BY, Hazuda D, on behalf of the P005 Study Team Longitudinal analysis of resistance to the HIV-1 integrase inhibitor raltegravir: results from P005 a phase II study in treatment-experienced patients. Antivir. Ther. 2008;13(Suppl 3):A8. [Google Scholar]
- 26.Fransen S, Karmochkine M, Huang W, Weiss L, Petropoulos CJ, Charpentier C. Longitudinal analysis of raltegravir susceptibility and integrase replication capacity of HIV-1 during virological failure. Antimicrobial Agents and Chemo. 2009 doi: 10.1128/AAC.00651-09. [In Press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenwald J, Le V, Butler SL, Bushman FD, Choe S. The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochem. 1999;38:8892–8898. doi: 10.1021/bi9907173. [DOI] [PubMed] [Google Scholar]
- 28.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007;28(6):1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 29.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009 doi: 10.1002/jcc.21256. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility - the Relaxed Complex scheme. J. Am. Chem. Soc. 2002;124(20):5632–5633. doi: 10.1021/ja0260162. (Communication) [DOI] [PubMed] [Google Scholar]
- 31.Amaro RE, Baron R, McCammon JA. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J. Comput. Aided Mol. Des. 2008;22(9):693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Donoghue P, Luthey-Schulten Z. Evolutionary profiles derived from the QR factorization of multiple structural alignments gives an economy of information. J. Mol. Biol. 2005;346:875–894. doi: 10.1016/j.jmb.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 33.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu R, Limon A, Ghory HZ, Engelman A. Genetic analyses of DNA-binding mutants in the catalytic core domain of Human Immunodeficiency Virus type I integrase. J. Virol. 2005;79(4):2493–2505. doi: 10.1128/JVI.79.4.2493-2505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenkins TM, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA indentified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delelis O, Thierry S, Subra F, Simon F, Malet I, Mouscadet JF, et al. Impact of Y143 HIV-1 integrase mutations on resistance to raltegravir in vitro and in vivo. Antimicrobial Agents and Chemo. 2010;54(1):491–501. doi: 10.1128/AAC.01075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ceccherini-Silberstein F, Malet I, D’Arrigo R, Antinori A, Marcelin AG, Perno CF. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 2009;11:17–29. [PubMed] [Google Scholar]
- 38.Wiskerchen M, Muesing MA. Human Immunodeficiency Virus type I integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 1995;69(1):376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin CG, Taddeo B, Haseltine WA, Farnet CM. Genetic analysis of the Human Immunodeficiency Virus type I integrase protein. J. Virol. 1994;68(3):1633–1642. doi: 10.1128/jvi.68.3.1633-1642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerton JL, Ohgi S, Olsen M, Derisi J, Brown PO. Effects of mutations in residues near the active site of Human Immunodeficiency Virus type I integrase on specific enzyme-substrate interactions. J. Virol. 1998;72(6):5046–5055. doi: 10.1128/jvi.72.6.5046-5055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. The EMBO J. 1998;17(19):5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldgur Y, Craigie R, Cohen GH, Fujiwara T, Yoshinaga T, Davies DR, et al. Structure of the HIV-1 integrase catalytic domain complexed with an inhbitor: A platform for antiviral drug design. Proc. Natl. Acad. Sci. USA. 1999;96:13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maignan S, Guilloteau JP, Zhou-Liu Q, Clement-Mella C, Mikol V. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: High level of similarity of the active site with other viral integrases. J. Mol. Biol. 1998;282:359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- 44.Bujacz G, Alexandratos J, Wlodawer A, Merkel G, Andrake M, Katz RA, Skalka AM. Binding of different divalent cations to the active site of Avian Sarcoma Virus integrase and their effects on enzymatic activity. J. Biol. Chem. 1997;272:18161–18168. doi: 10.1074/jbc.272.29.18161. [DOI] [PubMed] [Google Scholar]
- 45.The Sybyl software. version 7.2 Tripos, Inc.; St. Louis, MO: 2006. [Google Scholar]
- 46.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Richardson DC, et al. Structure validation by C-alpha geometry: phi, psi, and C-beta deviation. Proteins: Str., Function, & Gen. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 47.Case DA, Darden TE, Cheatham TE, Simmerling CL, Ross WS, Kollman PA, et al. AMBER 9. University of California; San Francisco: 2006. [Google Scholar]
- 48.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple AMBER force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 50.Darden T, York D, Pedersen L. Particle mesh Ewald-an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 51.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 52.Miyamoto S, Kollman PA. SETTLE: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 53.Eastwood MP, Hardin C, Luthey-Schulten Z, Wolynes PG. Evaluating protein structure-prediction schemes using energy landscape theory. IBM J. Res. Dev. 2001;45:475–497. [Google Scholar]
- 54.Langley DR, Samanta HK, Lin Z, Walker MA, Krystal MR, Dicker IB. The terminal (catalytic) adenosine of the HIV LTR controls the kinetics of binding and dissociation of HIV integrase strand transfer inhibitors. Biochem. 2008;47(51):13481–13488. doi: 10.1021/bi801372d. [DOI] [PubMed] [Google Scholar]
- 55.Gasteiger M, Marsili M. Iterative partial equalization of orbital electronegativity. Tetrahedron. 1980;36:3219. [Google Scholar]
- 56.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Pople JA, et al. GAUSSIAN 03. Gaussian, Inc.; Wallingford, CT: 2004. [Google Scholar]
- 57.Sanner MF. Python: A programming language for software integration and development. J. Mol. Graphics. Mod. 1999;17:57–61. (i.e., PMV) [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.