Abstract
Oleoylethanolamide (OEA) is an endogenous lipid mediator that decreases food intake and enhances lipid catabolism. Dietary fat stimulates OEA mobilization in the proximal small intestine, through a mechanism that requires the participation of the membrane glycoprotein CD36 (fatty acid translocase, FAT). CD36 is highly expressed in small-intestinal enterocytes and is involved in fatty acid uptake and intracellular signaling. Here, we analyze the impact of genetic CD36 deletion on OEA production in various mouse tissues under free-feeding conditions and at different times of the light/dark cycle. CD36 ablation decreases OEA levels in jejunum and plasma during the dark phase, when mice consume most of their daily food. CD36 deletion is also associated with reduced OEA levels in kidney, but not in other tissues including duodenum, stomach, adrenals, white and brown fat, heart, liver, pancreas, skeletal muscle and brain. The results underscore the important role of CD36 in jejunal OEA production linked to feeding.
Keywords: CD36, feeding, oleoylethanolamide, small intestine
1. Introduction
Oleoylethanolamide (OEA) is an endogenous fatty acid ethanolamide (FAE) that is produced on-demand by neurons and other cells by the consecutive action of two enzymes: N-acyltransferase (NAT), which generates N-oleoyl-phosphatidylethanolamine (NOPE) and N-acylphosphatidylethanolamine-specific phospholipase D (NAPE-PLD), which cleaves NOPE to release OEA [1, 2]. OEA is in turn the substrate of two distinct intracellular amidases: fatty-acid amide hydrolase (FAAH) [3, 4], and N-acylethanolamine-hydrolyzing acid amidase (NAAA) [5, 6]. In the proximal small intestine, feeding stimulates enterocytes to produce OEA [7-9], whereas fasting produces an opposite effect. Duodenal infusion experiments have shown that dietary fat, not carbohydrates or protein, selectively stimulates small-intestinal OEA generation [10]. More specifically, it has been found that infusion of monounsaturated oleic acid (C18:1Δ9) causes OEA accumulation in jejunum, whereas infusion of saturated palmitic acid (C16:0) has no such effect. Oleic acid administration increases NOPE production, stimulates NAPE-PLD activity and inhibits FAAH activity in enterocytes [10], three effects that are also elicited by feeding. In addition to these regulatory actions, biochemical studies have demonstrated that dietary oleic acid, but not plasma-derived oleic acid, serves as a metabolic precursor for OEA biosynthesis [10].
Systemic administration of OEA causes, in rodents, a dose- and time-dependent decrease in food intake. This effect is structurally and behaviorally selective, requires intact sensory vagal fibers and is accompanied by activation of the nucleus of the solitary tract in the brainstem and the paraventricular nucleus in the hypothalamus [9, 11]. The anorexic actions of OEA are mediated by the peroxisome proliferator-activated receptor-α (PPAR-α), a nuclear receptor that is involved in regulating the absorption, storage and utilization of dietary fat [12-14]. Furthermore, the hypophagic effects of OEA are abolished in PPAR-α-null mice and are mimicked by the synthetic PPAR-α agonists Wy-14643 and GW7674 [11]. Indeed, OEA binds to PPAR-α with high affinity (K0 = 40 nM) and activates it with high potency (half-maximal effective concentration, EC50, 120 nM) [11]. Importantly, during the daytime OEA reaches concentrations in the small intestine that can fully activate PPAR-α (∼300 nM). The OEA analog, palmitoylethanolamide (PEA), which is also present in the small intestine [7], also activates PPAR-α, albeit with lower potency than does OEA (EC50 = 3.1 μM).
CD36 is a glycosylated membrane protein that binds long-chain fatty acids (LCFAs) and facilitates their translocation across cell membranes [15-23]. CD36 expression along the gastrointestinal tract parallels the distribution of LCFAs absorption, being higher in duodenum and jejunum and lower in ileum, colon and stomach [24]. CD36 localizes to the apical enterocyte membrane, where it is highly concentrated [22]. CD36 is thought to play an important role in the absorption of fatty acids by the proximal small intestine [10]. Moreover, we have recently shown that CD36 may also contribute to the oleic acid-stimulated production of OEA under fasting/re-feeding conditions [10]. To further characterize the role of CD36 in OEA mobilization, in the present study we investigated the impact of deletion of the CD36 gene on OEA content in tissues of mice kept under free-feeding conditions.
2. Materials and Methods
2.1. Animals
CD36-null mice on a C57BL/6J background were generated from breeding pairs kindly provided by Dr. Maria Febbraio (Lerner Research Institute, Cleveland, OH) [19, 25-29]. Animals were maintained at 22 °C on a 12h light/dark cycle (lights on at 5:30 am) and had free access to water and standard chow pellets (Prolab RMH 2500; PMI Nutrition International, Brentwood, MO). All procedures met the National Institutes of Health guidelines for the care and use of laboratory animals, and were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine.
2.2. Plasma and tissue levels of OEA, PEA, anandamide and fatty acids
Mice were sacrificed under slight anesthesia (isoflurane, USP, Phoenix Pharmaceutical, Inc., St. Joseph, MO) at 11:30 pm or 11:30 am. The following tissues were harvested, snap-frozen in liquid nitrogen and stored at -80°C until analysis: adrenal glands, brown adipose tissue (BAT), duodenum, stomach, epididymal fat, heart, jejunum, kidney, liver, pancreas, skeletal muscle (vastus lateralis) and brain. Brains were further dissected into brainstem, cerebellum, cortex, hippocampus, hypothalamus, striatum and thalamus. Blood was also collected through a left cardioventricular puncture and centrifuged at 2000×g for 30 min to obtain plasma. OEA, PEA, anandamide and various fatty acids were extracted from tissues or plasma. Frozen tissues were weighed and homogenized in methanol (1 ml/100 mg of tissue) containing [2H4]-OEA, [2H4]-PEA, [2H4]-anandamide (prepared as described previously, [7, 10]) and heptadecanoic acid (Nu-Check Prep, Inc., Elysian, NM) as internal standards. Lipids were extracted using 2 volumes of chloroform and washed with 1 volume of water. Organic phases were collected and dried under nitrogen. Lipids were reconstituted in 60 μl of methanol and quantified by liquid chromatography mass spectrometry (LC/MS) [7, 10, 30]. For quantification purposes we monitored the [M+Na]+ ions of [2H4]-OEA (mass-to-charge ratio, m/z 352), OEA (m/z 348), [2H4]-anandamide (m/z 374), anandamide (m/z 370), [2H4]-PEA (m/z 326) and PEA (m/z 322). The following fatty acids were monitored as [M-H]+: heptadecanoic acid (C17:0, m/z 269), oleic acid (C18:1Δ9, m/z 281), erucic acid (C22:1Δ9, m/z 337), nervonic acid (C24:1Δ9, m/z 365), linoleic acid (C18:2Δ9,12, m/z 279), linolenic acid (C18:3Δ9,12,15, m/z 277), arachidonic acid (C20:4Δ5,8,11,14, m/z 303), palmitoleic acid (C16:1Δ9, m/z 253), eicosapentaenoic acid (C20:5Δ5,8,11,14,17, m/z 301), docosapentaenoic acid (C22:5Δ7,10,13,16,19, m/z 329), docosahexaenoic acid (C22:6Δ4,7,10,13,16,19, m/z 327), palmitic acid (C16:0, m/z 255) and stearic acid (C18:0, m/z 283).
2.3. Jejunal levels of NOPE and N-palmitoyl-phosphatidylethanolamine (NPPE)
Frozen tissues were weighed and homogenized in methanol (1 ml/100 mg of tissue) containing 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-heptadecanoyl as an internal standard (prepared as described previously [7, 10]). Lipids were extracted in chloroform and washed with water. Organic phases were collected and dried under nitrogen. Lipids were reconstituted in chloroform/methanol (1:3, vol/vol). NAPE species were measured by LC/MSn as described previously [7, 10, 30]. For quantification purposes, we monitored 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine-N-oleoyl (m/z 1030.8 > 744.8), 1-stearoyl-2-arachidonoyl-sn-glycero-3-phosphoethanolamine-N-palmitoyl (m/z 1004.8 > 718.8) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-heptadecanoyl (m/z 942.8 > 704.8).
2.4. Jejunal NAPE-PLD activity
Tissues were homogenized in ice-cold Tris-HCl (50 mM, pH 7.4, 10 vol) containing 0.32 M sucrose. Homogenates were centrifuged at 1,000×g for 10 min. NAPE-PLD activity was measured at 37°C for 30 min in Tris-HCl buffer (50 mM, pH 7.4) containing 0.1% Triton X-100, phenylmethylsulphonylfluoride (1 mM), protein (100 μg) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-heptadecenoyl (100 μM) as substrate. The reactions were stopped by adding chloroform/methanol (1:1, vol/vol) containing 2[H4]-heptadecenoylethanolamide as internal standard. After centrifugation at 1,500×g at 4°C for 5 min, the organic layers were collected and dried under nitrogen. The residues were suspended in 50 μl of methanol and analyzed by LC/MS. For quantification purposes, we monitored the [M+Na]+ ions of 2[H4]-heptadecenoylethanolamide (m/z, 338) and N-heptadecanoylethanolamide (m/z, 334).
2.5. Jejunal FAAH activity
Tissues were homogenized in ice-cold Tris-HCl (50 mM, pH 7.5, 10 vol) containing 0.32 M sucrose. Homogenates were centrifuged at 1,000×g for 10 min, and the supernatants were kept. Reactions were conducted at 37°C for 30 min in Tris-HCl buffer (50 mM, pH 7.5) containing fatty acid-free bovine serum albumin (0.05%), protein (50 μg) and [3H-ethanolamine]anandamide (10,000 dpm, specific activity 20 Ci/mmol). After stopping the reactions with chloroform/methanol (1:1, vol/vol, 1 ml), radioactivity was measured in the aqueous layers by liquid scintillation counting.
2.6. Statistical Analysis
Results are expressed as mean ± standard error of the mean (S.E.M.) and the significance of differences was determined using the two-way analysis of variance (ANOVA) followed by Student-Newman-Keuls multiple comparison test (SNK) as post hoc. Differences were consider significant if P < 0.05. Statistical analyses were conducted using Sigma Stat 32 Version 2.0.
3. Results
3.1. Effects of CD36 deletion on jejunal fatty acid levels
We found an overall effect of genotype on oleic acid content in the jejunum of free-feeding mice (P = 0.044). Thus, CD36 ablation resulted in a significant decrease in oleic acid levels during the dark phase (P = 0.003), but not the light phase (P = 0.982) (Table 1). Although no significant overall effect of time (P = 0.053) was observed, wild-type mice showed lower levels of oleic acid during the light phase compared with the dark phase (P = 0.027), whereas no differences with time were found in null mice. The interaction between both variables was statistically significant (P = 0.041). No such difference was observed with other fatty acids of the ω-9 series, including erucic acid (genotype: P = 0.455; time: P = 0.838; interaction: P = 0.423) and nervonic acid (genotype: P = 0.238; time: P = 0.953; interaction: P = 0.613). Within the polyunsaturated ω-6 series, no overall effect of genotype on jejunal levels of linoleic acid (P = 0.684) or a significant interaction between genotype and time (P = 0.220) was found, but an overall effect of time was observed (P = 0.012). Wild-type mice had lower levels of linoleic acid during the light phase (P = 0.023), while in null mice no differences were observed between the dark and the light phase (P = 0.537). Genotype had no an overall effect on linolenic acid in jejunum (P = 0.537) but an overall effect of time (P = 0.001) and a significant interaction between both variables (P = 0.033) was observed. Wild-type mice showed lower levels of this fatty acid in the light phase (P < 0.001), while null mice had comparable levels of linolenic acid during the dark and the light phase (P = 0.275). We found an overall effect of genotype on arachidonic acid levels on jejunum (P = 0.045), but no effect of time (P = 0.940) or significant interaction between genotype and time (P = 0.808) was observed. However, although arachidonic acid content trended to be higher in null mice than in wild-type mice in both dark and light phase, no statistical differences were observed (P = 0.201 and P = 0.096, respectively). Moreover, the total levels of ω-6 fatty acids was not significantly altered by CD36 deletion (genotype: P = 0.899), but by time (P = 0.032), although comparison between light/dark phases within genotype did not show any statistical difference. No interaction between these variables was observed (P = 0.121). Within the ω-3 series, no overall effect of genotype and time or interaction between both variables was observed for palmitoleic acid (genotype: P = 0.251; time: P = 0.156; interaction P = 0.101), docosapentaenoic acid (genotype: P = 0.075; time: P = 0.415; interaction P = 0.334) and docosahexaenoic acid (genotype: P = 0.132; time: P = 0.292; interaction P = 0.289). Eicosapentaenoic acid levels were not altered by genotype (P = 0.321) but time had an overall effect (P = 0.002), while no significant interaction was found (P = 0.052). Eicosapentaenoic acid was higher in wild-type mice during the dark phase compared to the light phase (P = 0.002), while CD36-null mice had comparable levels at both phases (P = 0.353). The total levels of ω-3 fatty acids was not affected by genotype (P = 0.540) or time (P = 0.817) and no interaction was observed between both variables (P = 0.235). Within the saturated fatty acids, palmitic acid showed no statistical differences (genotype: P = 0.381; time: P = 0.922; interaction: P = 0.423), whereas stearic acid were affected by genotype (P = 0.027) but not by time (P = 0.287) and no interaction was found between both variables (P = 0.293). CD36-null mice showed lower levels of stearic acid than wild-type mice during the dark phase (P = 0.028) but similar levels during the light phase (P = 0.338). No changes were found in total saturated fatty acids (genotype: P = 0.083; time: 0.503; interaction: P = 0.261). Finally, the ratio of ω-6/ω-3 fatty acids was not changed in CD36-null mice compared to wild-type mice (genotype: P = 0.663), but was affected by time (P = 0.011), although the post hoc test showed no statistical differences in any of the genotypes with time. No interaction was observed between genotype and time (P = 0.984). The results confirm and extend previous findings indicating that CD36 plays a key role in the small intestinal uptake of oleic acid [21].
Table 1.
Jejunal levels of fatty acids in wild-type and CD36-null mice during the dark phase (11:30 pm) and the light phase (11:30 am).
| Dark phase | Light phase | |||
|---|---|---|---|---|
| +/+ | -/- | +/+ | -/- | |
| ω-9 series | ||||
| 18:1 (nmol/g) | 20.17 ± 2.18 | 14.91 ± 1.01*** | 15.07 ± 2.06# | 15.13 ± 1.62 |
| 22:1 (pmol/g) | 594.28 ± 89.72 | 585.99 ± 179.69 | 505.75 ± 111.86 | 734.42 ± 181.62 |
| 24:1 (pmol/g) | 399.9 ± 49.72 | 287.5 ± 87.32 | 318.2 ± 71.55 | 370.9 ± 44.71 |
| ω-6 series | ||||
| 18:2 (nmol/g) | 18.90 ± 2.58 | 15.89 ± 1.33 | 12.71 ± 1.34# | 14.25 ± 1.29 |
| 18:3 (nmol/g) | 3.44 ± 0.42 | 2.48 ± 0.22 | 1.55 ± 0.23### | 1.99 ± 0.24 |
| 20:4 (nmol/g) | 4.41 ± 0.27 | 5.58 ± 0.72 | 4.31 ± 0.24 | 5.77 ± 0.91 |
| Σ ω-6 (nmol/g) | 24.98 ± 2.34 | 21.46 ± 1.95 | 17.02 ± 1.47 | 20.02 ± 1.20 |
| ω-3 series | ||||
| 16:1 (pmol/g) | 2.31 ± 0.18 | 2.06 ± 0.44 | 2.20 ± 0.511 | 3.51 ± 0.59 |
| 20:5 (pmol/g) | 1391.35 ± 140.48 | 1032.78 ± 85.66 | 733.22 ± 52.70### | 857.73 ± 139.40 |
| 22:5 (pmol/g) | 45.48 ± 5.41 | 52.46 ± 10.01 | 44.26 ± 5.78 | 66.52 ± 9.16 |
| 22:6 (nmol/g) | 2.52 ± 0.19 | 2.65 ± 0.35 | 2.52 ± 0.19 | 3.21 ± 0.29 |
| Σ ω-3 (nmol/g) | 51.71 ± 5.74 | 58.21 ± 10.81 | 49.71 ± 6.34 | 73.39 ± 9.80 |
| Saturated | ||||
| C16:0 (nmol/g) | 11.28 ± 0.89 | 9.71 ± 0.83 | 10.44 ± 0.96 | 10.37 ± 0.91 |
| C18:0 (nmol/g) | 11.66 ± 0.83 | 9.36 ± 0.75* | 10.24 ± 0.26 | 9.36 ± 0.61 |
| Σ saturated (nmol/g) | 23.21 ± 1.56 | 19.07 ± 1.49 | 20.68 ± 0.97 | 19.73 ± 1.38 |
| Ratios | ||||
| 16:1/16:0 | 5.08 ± 0.43 | 5.12 ± 0.65 | 5.40 ± 0.70 | 3.41 ± 0.60 |
| 18:1/18:0 | 1.83 ± 0.12 | 1.60 ± 0.07 | 1.48 ± 0.21 | 1.64 ± 0.20 |
| 18:2/18:3 | 6.01 ± 0.18 | 6.42 ± 0.19 | 8.46 ± 0.45 | 7.34 ± 0.47 |
| 20:4/22:6 | 1.77 ± 0.10 | 2.13 ± 0.16 | 1.72 ± 0.07 | 1.77 ± 0.19 |
| 22:1/24:1 | 1.47 ± 0.11 | 1.58 ± 0.31 | 1.60 ± 0.06 | 1.51 ± 0.06 |
| ω-6/ω-3 | 0.49 ± 0.03 | 0.40 ± 0.05 | 0.36 ± 0.03 | 0.28 ± 0.02 |
Data are expressed as mean ± S.E.M of n = 4-5. +/+: wild-type; -/-: CD36-null mice. ω-9 series: 18:1, oleic acid; 22:1, erucic acid and 24:1, nervonic acid; ω-6 series: 18:2, linoleic acid; 18:3, linolenic acid and 20:4, arachidonic acid. ω-3 series: 16:1, palmitoleic acid; 20:5, eicosapentaenoic acid; 22:5, docosapentaenoic acid and 22:6, docosahexaenoic acid; Saturated: C16:0, palmitic acid and C18:0, stearic acid.
P < 0.05 and
P < 0.005 vs. wild-type within the same phase of the light/dark cycle;
P < 0.05 and
P < 0.005 vs. dark phase within the same genotype (Two-way ANOVA followed by SNK).
3.2. Effects of CD36 deletion on jejunal FAE levels
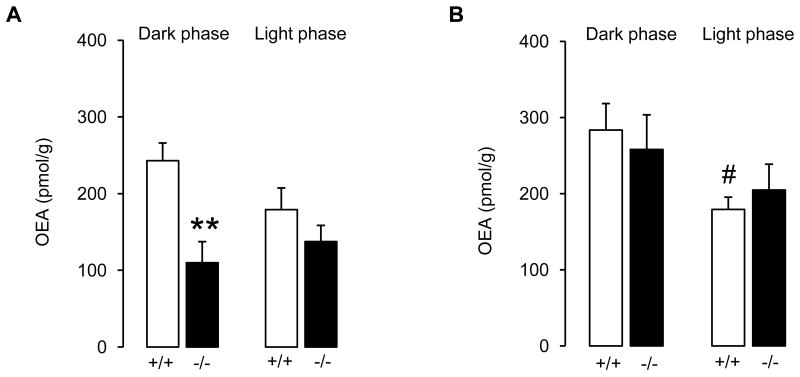
Genotype had an overall effect on jejunal OEA levels (P = 0.047), while no overall effect of time (P = 0.607) or statistical interaction genotype × time were found (P = 0.182). Deletion of CD36 resulted in a significant decrease in OEA levels in jejunum during the dark phase (P = 0.007, Figure 1A), but had only marginal effects on OEA levels during the light phase (P = 0.270, Figure 1A). Within the gastrointestinal tract, changes in OEA levels were specific for the jejunum, as no differences were found between wild-type and CD36-null mice in duodenum (Figure 1B; genotype: P = 0.998; time: P = 0.031; interaction: P = 0.454) or stomach (Table 3, genotype: P = 0.625; time: P = 0.095; interaction: P = 0.209). However, in wild-type mice, duodenal OEA levels were lower in the light phase compared to the dark phase (P = 0.037), while no differences between both phases were found in CD36-null mice (P = 0.285).
Figure 1.
CD36 deletion is associated with reduced OEA content in jejunum (A), but not duodenum (B). The effect is limited to the dark phase of the light/dark cycle, when mice consume most of their daily meals. +/+: wild-type; -/-: CD36-null mice. Results are expressed as mean ± S.E.M. of n = 4–5. **P < 0.01 vs. wild-type within the same phase of the light/dark cycle; #P < 0.05 vs. dark phase within the same genotype (two-way ANOVA followed by SNK).
Table 3.
Levels of OEA in various peripheral tissues and brain regions of wild-type and CD36-null mice during the dark phase (11:30 pm) and the light phase (11:30 am).
| Dark phase | Light phase | |||
|---|---|---|---|---|
| +/+ | -/- | +/+ | -/- | |
| Peripheral tissues | ||||
| Adrenal gland | 206.17 ± 46.04 | 172.97 ± 27.17 | 245.23 ± 32.75 | 192.37 ± 22.30 |
| BAT | 274.48 ± 43.97 | 264.76 ± 26.98 | 449.97 ± 67.79# | 451.71 ± 38.30# |
| Duodenum | 283.48 ± 34.89 | 258.17 ± 45.44 | 179.31 ± 16.11# | 204.81 ± 33.89 |
| Epididymal fat | 150.72 ± 23.02 | 121.62 ± 16.70 | 346.89 ± 57.58### | 378.70 ± 68.41### |
| Heart | 96.22 ± 15.66 | 103.42 ± 7.56 | 165.28 ± 25.90 | 160.29 ± 38.13 |
| Kidney | 296.00 ± 49.59 | 96.17 ± 13.13* | 259.3 ± 19.02 | 100.80 ± 9.72*** |
| Liver | 27.42 ± 2.48 | 31.12 ± 3.68 | 33.85 ± 5.24 | 38.96 ± 5.33 |
| Pancreas | 203.14 ± 45.44 | 97.83 ± 20.70 | 151.40 ± 50.97 | 298.29 ± 64.93# |
| Plasma | 10.60 ± 1.33 | 6.09 ± 0.78* | 14.89 ± 1.59 | 10.25 ± 2.55 |
| Stomach | 281.4 ± 19.50 | 250.9 ± 17.80 | 299.0 ± 25.62 | 366.7 ± 60.99 |
| Vastus lateralis | 92.72 ± 11.81 | 141.66 ± 21.12 | 182.82 ± 24.57# | 194.99 ± 35.25 |
| Brain regions | ||||
| Brainstem | 251.52 ± 10.18 | 267.54 ± 9.24 | 235.28 ± 10.97 | 255.62 ± 12.08 |
| Cerebellum | 236.06 ± 18.00 | 246.38 ± 10.17 | 233.32 ± 10.76 | 261.26 ± 14.21 |
| Cortex | 206.83 ± 18.21 | 199.43 ± 7.27 | 203.98 ± 10.11 | 243.00 ± 19.11 |
| Hippocampus | 135.57 ± 10.96 | 118.61 ± 5.00 | 118.01 ± 10.36 | 123.30 ± 7.69 |
| Hypothalamus | 105.93 ± 9.80 | 118.91 ± 13.57 | 100.46 ± 19.80 | 92.66 ± 17.72 |
| Striatum | 216.04 ± 11.41 | 199.64 ± 3.22 | 209.13 ± 9.58 | 222.28 ± 8.99 |
| Thalamus | 302.17 ± 24.26 | 310.18 ± 25.81 | 274.32 ± 5.17 | 302.64 ± 14.18 |
Data are expressed as mean ± S.E.M of n = 4-6. +/+: wild-type; -/-: CD36-null mice. OEA levels are expressed as pmol/g tissue.
P < 0.05 and
P < 0.005 vs. wild-type within the same phase of the light/dark cycle;
P < 0.05 and
P < 0.005 vs. dark phase within the same genotype (Two-way ANOVA followed by SNK).
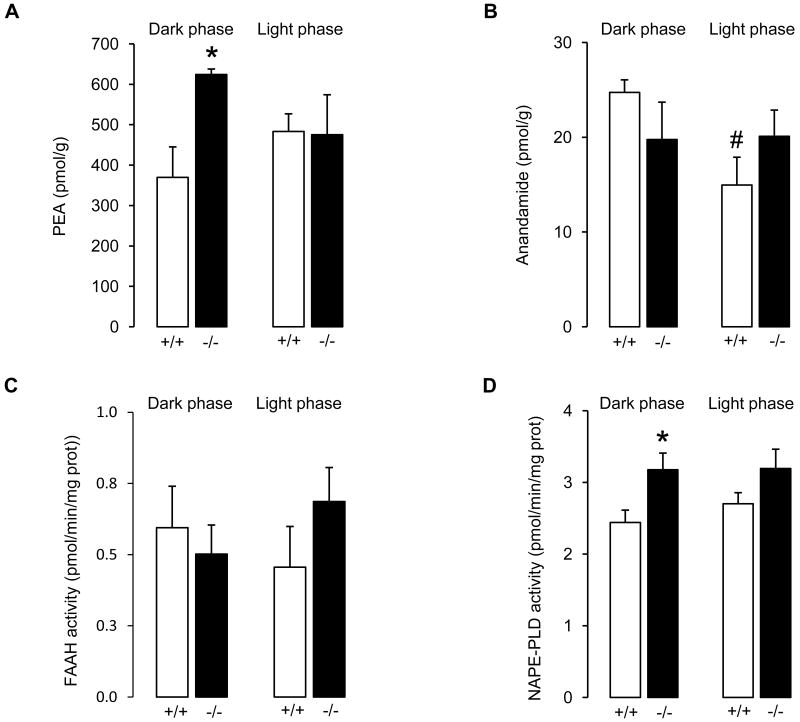
The decrease in OEA levels observed in the jejunum of CD36-null mice was accompanied by an increase in PEA concentrations in the same tissue (Figure 2A), which was also restricted to the dark phase (dark phase: P = 0.026, light phase P = 0.934) although the two-way ANOVA analysis revealed no overall effect of genotype or time and no interaction between both variables (genotype: P = 0.102; interaction: P = 0.083). By contrast, no differences in jejunal anandamide content were observed between the two genotypes (Figure 2B, genotype: P = 0.976; time: P = 0.112; interaction: P = 0.090). However, in wild-type mice anandamide levels were significantly lower during the light phase compared with the dark phase (P = 0.022), a difference not observed in null mice (P = 0.933).
Figure 2.
CD36 deletion elevates PEA levels (A) in jejunum during the dark phase, without changing significantly the levels of anandamide (B). FAAH activity is not altered (C), whereas NAPE-PLD activity is increased in CD36-null mice during the dark phase (D). +/+: wild-type; -/-: CD36-null mice. Results are expressed as means±S.E.M. of n = 4–6. *P < 0.05 vs. wild-type within the same phase of the light/dark cycle; #P < 0.05 vs. dark phase within the same genotype (two-way ANOVA followed by SNK).
3.3. Effects of CD36 deletion on the formation and degradation of OEA and PEA in jejunum
To examine the biochemical mechanisms underlying the observed changes in OEA and PEA levels, we measured the activities of FAAH and NAPE-PLD as well as the levels of NOPE and NPPE in jejunal tissue. The results show that FAAH activity was similar in wild-type and CD36-null mice at both the dark phase and the light phases (genotype: P = 0.601; time: P = 0.858; interaction: P = 0.218, Figure 2C). However, genotype had an overall effect on NAPE-PLD activity (P = 0.010). Thus, a significant increase in NAPE-PLD activity (Figure 2D) was observed in CD36-null mice during the dark phase (P = 0.034) without differences during the light phase (P = 0.146). No overall effect of time (P = 0.514) or interaction between genotype and time was observed (P = 0. 575). Jejunal NOPE (genotype: P = 0.510; time: P = 0.762; interaction: P = 0.922) and NPPE (genotype: P = 0.729; time: P = 0.146; interaction: P = 0.166) levels were comparable across genotypes and time of the day (Table 2).
Table 2.
Jejunal levels of NOPE and NPPE in wild-type and CD36-null mice during the dark phase (11:30 pm) and the light phase (11:30 am).
| Dark phase | Light phase | |||
|---|---|---|---|---|
| +/+ | -/- | +/+ | -/- | |
| NOPE | 365.84 ± 91.05 | 307.46 ± 82.59 | 381.62 ± 74.74 | 338.19 ± 44.04 |
| NPPE | 238.01 ± 50.73 | 324.72 ± 66.15 | 381.31 ± 46.93 | 328.39 ± 26.72 |
Data are expressed as mean ± S.E.M of n = 4-5. +/+: wild-type; -/-: CD36-null mice. NOPE: N-oleoyl-phosphatidylethanolamine, NPPE: N-palmytoyl-phosphatidylethanolamine. NOPE and NPPE levels are expressed as pmol/g tissue.
3.4. Effects of CD36 deletion on OEA levels in plasma, brain and other tissues
As shown in Table 3, plasma OEA levels was affected by both genotype (P = 0.018) and time (P = 0.027) but no interaction between them was observed (P = 0.972). During the dark phase, OEA was significantly lower in CD36-null mice (P = 0.015), whereas no statistical differences were observed during the light phase (P = 0.213). Among the different peripheral tissues analyzed, CD36 deletion only influenced OEA levels in kidney (genotype: P = 0.013), where it produced a marked reduction in OEA content during both dark (P = 0.010) and light phases (P < 0.001). No effect of time (P = 0.216) or interaction between both variables was observed (P = 0.230). No changes were observed on OEA levels in adrenal glands (genotype: P = 0.222; time P = 0.400; interaction: P = 0.774). In BAT no differences were found between wild-type and null mice (P = 0.936) but time had a significant effect (P = 0.002). Thus, in both wild-type and CD36-null mice OEA levels were significantly higher during the light phase (P = 0.020 and P = 0.019, respectively). No interaction was observed between genotype and time (P = 0.908). Similar results were observed in epididymal fat where genotype had no overall effect (P = 0.764), but time had a significant effect (P < 0.001) on OEA content, which was higher during the light phase in both wild-type and null mice (P = 0.004 and P < 0.001, respectively). No interaction between both variables was found (P = 0.464). In heart, genotype had no overall effect on OEA content (P = 0.967) and no interaction between genotype and time was observed (P = 0.818), while time affected significantly OEA levels (P = 0.029). However, the post hoc test revealed no significant differences between dark and light phases in either wild-type (P = 0.073) or null mice (P = 0.155). Hepatic OEA levels were not affected by genotype or time and no interaction between both variables was found (genotype: P = 0.306; time: P = 0.118; interaction: P = 0.953). In pancreas, no overall effect of genotype or time on OEA levels was observed (P = 0.689 and P = 0.165, respectively), but a significant interaction between both variables was found (P = 0.026). However, in CD36-null mice OEA levels during the light phase were significantly higher than during the light phase (P = 0.016) while no significant differences were observed between both phases in wild-type mice (P = 0.470). In skeletal muscle (vastus lateralis) genotype did not affect OEA content (P = 0.250), whereas time had an overall effect (P = 0.014), but there was not interaction between both variables (P = 0.482). In wild-type mice OEA levels were higher during the light phase (P = 0.025) whereas in CD36-null mice no differences were observed between both phases of the light/dark cycle (P = 0.160). Within the brain, no differences in OEA levels between wild-type and CD36-null mice and no effect of time or interaction between genotype and time were found in any of the regions examined (Table 3): brainstem (genotype: P = 0.119; time: P = 0.219; interaction: P = 0.847), cerebellum (genotype: P = 0.196; time: P = 0.673; interaction P = 0.542), cortex (genotype: P = 0.319; time: P = 0.204; interaction: P = 0.151), hippocampus (genotype: P =0.533; time: P = 0.492; interaction: P = 0.243), hypothalamus (genotype: P = 0.878; time: P = 0.355; interaction: P = 0.541), striatum (genotype: P = 0.864; time: P = 0.414; interaction: P = 0.135) and thalamus (genotype: P = 0.310; time: P = 0.322; interaction: P = 0.565). Plasma oleic acid concentrations (data not shown) were significantly affected by genotype (P < 0.005), being higher in CD36-null mice compared with wild-type animals, in both dark and light phases (P = 0.002 and P = 0.023, respectively), with a 3-fold increase in the dark phase and a 1.5-fold increase in the light phase. Time had an overall effect on oleic acid levels (P < 0.005), which were significantly higher in null mice during the dark phase compared to the light phase (P = 0.047). No changes were found in plasma palmitic acid levels (genotype: P = 0.480; time: P = 0.372; interaction: P = 0.046; data not shown).
4. Discussion
In the present study we show that genetic deletion of the fatty acid transporter CD36 decreases oleic acid levels in jejunual tissue of free-feeding mice during the night, when the animals consume the majority of their daily meals. This result is in line with the study by Schwartz et al. [10], which reported a reduction in the jejunal content of oleic acid in CD36-null mice after food deprivation/re-feeding. We report here that CD36 ablation does no modify the jejunal levels of other ω-9 monounsaturated fatty acids, such as erucic acid and nervonic acid, or the levels of polyunsaturated ω-3 and ω-6 fatty acids. These results suggest that CD36 plays a specific role in oleic acid uptake and metabolism by jejunal enterocytes, at least under free-feeding conditions. This interpretation is consistent with prior work showing that the absorption of free fatty acids by the intestinal lumen is similar in CD36-null mice and wild-type following intragastric infusion of 14C-labeled palmitic acid, 3H-labeled triolein [31] or synthetic fatty acid analogues [32]. Furthermore, enrichment of oleate in proximal intestine mucosa is reduced by 50% in CD36-null mice 90 min (active absorption period) after intragastric gavage of olive oil, while linoleate enrichment is increased [21]. Alternatively, regulatory mechanisms might compensate for CD36 deletion in mutant mice. One such mechanism might be the expression of fatty acid transport protein 4 (FATP4), which is present in the small intestine and is localized to the apical brush border of the epithelial cells, and is both necessary and sufficient for efficient uptake of LCFAs [33, 34]. The reduction in oleic acid levels induced by CD36 ablation under free-feeding conditions seems to be specific to the jejunum, as no such changes were noted in other sections of the gastrointestinal tract.
Another finding of the present study is that jejunal OEA levels are reduced in free-feeding CD36-null mice during the night. This observation is in agreement with previous work indicating an important role for CD36 in feeding-stimulated OEA production in the small intestine [10]. This work has shown that CD36-null mice lack the ability to absorb 10 Z-heptadecenoic acid (an analog of oleic acid that is not naturally present in mammalian tissues) and to convert this fatty acid into heptadecenoylethanolamide (an analogue of OEA) [10]. This result suggests that dietary oleic acid serves as a metabolic precursor for OEA biosynthesis in enterocytes [10]. It is likely, therefore, that reduced absorption of oleic acid in the jejunum of CD36-null mice results in lower production of OEA. Interestingly, this effect was accompanied by a marked increase in jejunal PEA levels, which could result from enhanced NAPE-PLD expression. This increase in PEA levels in jejunum of CD36-null mice does not seem to require an elevation in its fatty acid precursor, palmitic acid, since it takes place without changes in jejunal palmitic acid content, suggesting that OEA and PEA synthesis are regulated by different mechanisms. This hypothesis is in accordance with the fact that intraduodenal infusion of palmitic acid does not change PEA levels [10]. The elevation in jejunal PEA might represent a mechanism aimed at compensating the loss of OEA signaling. This idea is supported by PEA's ability to activate PPAR-α and reduce food intake when administered as a drug [35, 36].
A third noteworthy result of our experiments study was the marked reduction in OEA levels in the kidney elicited by CD36 deletion, which occurred irrespectively of the time of the day. This decrease was not accompanied by a reduction in oleic acid content, suggesting that CD36 could play different roles in OEA formation in jejunum and kidney. The biochemical mechanism and the physiological significance of this reduction in renal OEA caused by CD36 deletion are unknown, but it is tempting to speculate the existence of a link with the regulation of blood pressure. Several findings suggest that renal CD36 is involved in the homeostatic control of blood pressure: (i) the renal expression of CD36 correlates inversely with both systolic and diastolic blood pressure [37]; (ii) deficient renal expression of CD36 has been proposed as a determinant of increased blood pressure in spontaneously hypertensive rats (SHR) [37]; (iii) the blood pressure of Japanese individuals with CD36 deficiency is greater than in age-matched controls [38]; and (iv) genetic deletion of CD36 can cause hypertension [39]. Moreover, PPAR-α agonists prevent heart failure in animals [40], as OEA does [41], and decrease blood pressure in a mouse model of hypertension [42].
In conclusion, the present results suggest that, under free-feeding conditions, CD36 serves a key function in the production of OEA, in particular during the dark phase, when the mice eat the majority of their meals. These results, together with those obtained by Yang et al. [43], which showed that OEA increases CD36 mRNA expression and fatty acid uptake in enterocytes, suggest that OEA and CD36 might work in a coordinated fashion to face the metabolic challenge represented by the arrival of fat into the small intestine.
Acknowledgments
We thank Dr. Maria Febbraio for kindly providing us with CD36 null mice, and Azar Ghomian and Franzisca Pahlisch for helping with experiments. The contribution of the Agilent Technologies/UCI Analytical Discovery Facility, Center for Drug Discovery is gratefully acknowledged. This work was supported by grants from NIH grant DK073955 and the Agilent Foundation.
Abbreviations
- FAAH
fatty-acid amide hydrolase
- FAE
fatty acid ethanolamide
- LCFAs
long-chain fatty acids
- LC/MS
isotope dilution liquid chromatography mass spectrometry
- NOPE
N-oleoyl-phosphatidylethanolamine
- NAPE-PLD
N-acylphosphatidylethanolamine-specific phospholipase D
- NPPE
N-palmitoyl-phosphatidylethanolamine
- OEA
oleoylethanolamide
- PEA
palmitoylethanolamide
- PPAR-α
peroxisome proliferator-activated receptor-α
Footnotes
5. Conflict of Interest: There are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Okamoto Y, Morishita J, Tsuboi K, Tonai T, Ueda N. Molecular characterization of a phospholipase D generating anandamide and its congeners. J Biol Chem. 2004;279:5298–305. doi: 10.1074/jbc.M306642200. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Okamoto Y, Morishita J, Tsuboi K, Miyatake A, Ueda N. Functional analysis of the purified anandamide-generating phospholipase D as a member of the metallo-beta-lactamase family. J Biol Chem. 2006;281:12325–35. doi: 10.1074/jbc.M512359200. [DOI] [PubMed] [Google Scholar]
- 3.Ahn K, McKinney MK, Cravatt BF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem Rev. 2008;108:1687–707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–7. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 5.Tsuboi K, Takezaki N, Ueda N. The N-acylethanolamine-hydrolyzing acid amidase (NAAA) Chem Biodivers. 2007;4:1914–25. doi: 10.1002/cbdv.200790159. [DOI] [PubMed] [Google Scholar]
- 6.Ueda N, Yamanaka K, Yamamoto S. Purification and characterization of an acid amidase selective for N-palmitoylethanolamine, a putative endogenous anti-inflammatory substance. J Biol Chem. 2001;276:35552–7. doi: 10.1074/jbc.M106261200. [DOI] [PubMed] [Google Scholar]
- 7.Fu J, Astarita G, Gaetani S, Kim J, Cravatt BF, Mackie K, Piomelli D. Food intake regulates oleoylethanolamide formation and degradation in the proximal small intestine. J Biol Chem. 2007;282:1518–28. doi: 10.1074/jbc.M607809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu J, Kim J, Oveisi F, Astarita G, Piomelli D. Targeted enhancement of oleoylethanolamide production in proximal small intestine induces across-meal satiety in rats. Am J Physiol Regul Integr Comp Physiol. 2008;295:R45–50. doi: 10.1152/ajpregu.00126.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez de Fonseca F, Navarro M, Gomez R, Escuredo L, Nava F, Fu J, Murillo-Rodriguez E, Giuffrida A, LoVerme J, Gaetani S, Kathuria S, Gall C, Piomelli D. An anorexic lipid mediator regulated by feeding. Nature. 2001;414:209–12. doi: 10.1038/35102582. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz GJ, Fu J, Astarita G, Li X, Gaetani S, Campolongo P, Cuomo V, Piomelli D. The lipid messenger OEA links dietary fat intake to satiety. Cell Metab. 2008;8:281–8. doi: 10.1016/j.cmet.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, Rodriguez De Fonseca F, Rosengarth A, Luecke H, Di Giacomo B, Tarzia G, Piomelli D. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature. 2003;425:90–3. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- 12.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–99. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–61. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 14.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–80. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J Biol Chem. 1993;268:17665–8. [PubMed] [Google Scholar]
- 16.Bonen A, Campbell SE, Benton CR, Chabowski A, Coort SL, Han XX, Koonen DP, Glatz JF, Luiken JJ. Regulation of fatty acid transport by fatty acid translocase/CD36. Proc Nutr Soc. 2004;63:245–9. doi: 10.1079/PNS2004331. [DOI] [PubMed] [Google Scholar]
- 17.Brinkmann JF, Pelsers MM, van Nieuwenhoven FA, Tandon NN, van der Vusse GJ, Glatz JF. Purification, immunochemical quantification and localization in rat heart of putative fatty acid translocase (FAT/CD36) Mol Cell Biochem. 2006;284:127–34. doi: 10.1007/s11010-005-9033-2. [DOI] [PubMed] [Google Scholar]
- 18.Ehehalt R, Fullekrug J, Pohl J, Ring A, Herrmann T, Stremmel W. Translocation of long chain fatty acids across the plasma membrane--lipid rafts and fatty acid transport proteins. Mol Cell Biochem. 2006;284:135–40. doi: 10.1007/s11010-005-9034-1. [DOI] [PubMed] [Google Scholar]
- 19.Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SF, Silverstein RL. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem. 1999;274:19055–62. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 20.Koonen DP, Glatz JF, Bonen A, Luiken JJ. Long-chain fatty acid uptake and FAT/CD36 translocation in heart and skeletal muscle. Biochim Biophys Acta. 2005;1736:163–80. doi: 10.1016/j.bbalip.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Nassir F, Wilson B, Han X, Gross RW, Abumrad NA. CD36 is important for fatty acid and cholesterol uptake by the proximal but not distal intestine. J Biol Chem. 2007;282:19493–501. doi: 10.1074/jbc.M703330200. [DOI] [PubMed] [Google Scholar]
- 22.Poirier H, Degrace P, Niot I, Bernard A, Besnard P. Localization and regulation of the putative membrane fatty-acid transporter (FAT) in the small intestine. Comparison with fatty acid-binding proteins (FABP) Eur J Biochem. 1996;238:368–73. doi: 10.1111/j.1432-1033.1996.0368z.x. [DOI] [PubMed] [Google Scholar]
- 23.Van Nieuwenhoven FA, Verstijnen CP, Abumrad NA, Willemsen PH, Van Eys GJ, Van der Vusse GJ, Glatz JF. Putative membrane fatty acid translocase and cytoplasmic fatty acid-binding protein are co-expressed in rat heart and skeletal muscles. Biochem Biophys Res Commun. 1995;207:747–52. doi: 10.1006/bbrc.1995.1250. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Yang Y, Braunstein E, Georgeson KE, Harmon CM. Gut expression and regulation of FAT/CD36: possible role in fatty acid transport in rat enterocytes. Am J Physiol Endocrinol Metab. 2001;281:E916–23. doi: 10.1152/ajpendo.2001.281.5.E916. [DOI] [PubMed] [Google Scholar]
- 25.Coburn CT, Hajri T, Ibrahimi A, Abumrad NA. Role of CD36 in membrane transport and utilization of long-chain fatty acids by different tissues. J Mol Neurosci. 2001;16:117–21. 151–7. doi: 10.1385/JMN:16:2-3:117. discussion. [DOI] [PubMed] [Google Scholar]
- 26.Coburn CT, Knapp FF, Jr, Febbraio M, Beets AL, Silverstein RL, Abumrad NA. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J Biol Chem. 2000;275:32523–9. doi: 10.1074/jbc.M003826200. [DOI] [PubMed] [Google Scholar]
- 27.Goudriaan JR, Dahlmans VE, Teusink B, Ouwens DM, Febbraio M, Maassen JA, Romijn JA, Havekes LM, Voshol PJ. CD36 deficiency increases insulin sensitivity in muscle, but induces insulin resistance in the liver in mice. J Lipid Res. 2003;44:2270–7. doi: 10.1194/jlr.M300143-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Hajri T, Han XX, Bonen A, Abumrad NA. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J Clin Invest. 2002;109:1381–9. doi: 10.1172/JCI14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajri T, Ibrahimi A, Coburn CT, Knapp FF, Jr, Kurtz T, Pravenec M, Abumrad NA. Defective fatty acid uptake in the spontaneously hypertensive rat is a primary determinant of altered glucose metabolism, hyperinsulinemia, and myocardial hypertrophy. J Biol Chem. 2001;276:23661–6. doi: 10.1074/jbc.M100942200. [DOI] [PubMed] [Google Scholar]
- 30.Giuffrida A, Rodriguez de Fonseca F, Piomelli D. Quantification of bioactive acylethanolamides in rat plasma by electrospray mass spectrometry. Anal Biochem. 2000;280:87–93. doi: 10.1006/abio.2000.4509. [DOI] [PubMed] [Google Scholar]
- 31.Goudriaan JR, Dahlmans VE, Febbraio M, Teusink B, Romijn JA, Havekes LM, Voshol PJ. Intestinal lipid absorption is not affected in CD36 deficient mice. Mol Cell Biochem. 2002;239:199–202. [PubMed] [Google Scholar]
- 32.Drover VA, Ajmal M, Nassir F, Davidson NO, Nauli AM, Sahoo D, Tso P, Abumrad NA. CD36 deficiency impairs intestinal lipid secretion and clearance of chylomicrons from the blood. J Clin Invest. 2005;115:1290–7. doi: 10.1172/JCI21514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gimeno RE, Hirsch DJ, Punreddy S, Sun Y, Ortegon AM, Wu H, Daniels T, Stricker-Krongrad A, Lodish HF, Stahl A. Targeted deletion of fatty acid transport protein-4 results in early embryonic lethality. J Biol Chem. 2003;278:49512–6. doi: 10.1074/jbc.M309759200. [DOI] [PubMed] [Google Scholar]
- 34.Stahl A, Hirsch DJ, Gimeno RE, Punreddy S, Ge P, Watson N, Patel S, Kotler M, Raimondi A, Tartaglia LA, Lodish HF. Identification of the major intestinal fatty acid transport protein. Mol Cell. 1999;4:299–308. doi: 10.1016/s1097-2765(00)80332-9. [DOI] [PubMed] [Google Scholar]
- 35.Borrelli Fand Izzo AA. Role of acylethanolamides in the gastrointestinal tract with special reference to food intake and energy balance. Best Pract Res Clin Endocrinol Metab. 2009;23:33–49. doi: 10.1016/j.beem.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Hansen HS, Diep TA. N-acylethanolamines, anandamide and food intake. Biochem Pharmacol. 2009;78:553–60. doi: 10.1016/j.bcp.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 37.Pravenec M, Churchill PC, Churchill MC, Viklicky O, Kazdova L, Aitman TJ, Petretto E, Hubner N, Wallace CA, Zimdahl H, Zidek V, Landa V, Dunbar J, Bidani A, Griffin K, Qi N, Maxova M, Kren V, Mlejnek P, Wang J, Kurtz TW. Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat Genet. 2008;40:952–4. doi: 10.1038/ng.164. [DOI] [PubMed] [Google Scholar]
- 38.Hirano K, Kuwasako T, Nakagawa-Toyama Y, Janabi M, Yamashita S, Matsuzawa Y. Pathophysiology of human genetic CD36 deficiency. Trends Cardiovasc Med. 2003;13:136–41. doi: 10.1016/s1050-1738(03)00026-4. [DOI] [PubMed] [Google Scholar]
- 39.Koonen DP, Febbraio M, Bonnet S, Nagendran J, Young ME, Michelakis ED, Dyck JR. CD36 expression contributes to age-induced cardiomyopathy in mice. Circulation. 2007;116:2139–47. doi: 10.1161/CIRCULATIONAHA.107.712901. [DOI] [PubMed] [Google Scholar]
- 40.Linz W, Wohlfart P, Baader M, Breitschopf K, Falk E, Schafer HL, Gerl M, Kramer W, Rutten H. The peroxisome proliferator-activated receptor-alpha (PPAR-alpha) agonist, AVE8134, attenuates the progression of heart failure and increases survival in rats. Acta Pharmacol Sin. 2009 doi: 10.1038/aps.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su HF, Samsamshariat A, Fu J, Shan YX, Chen YH, Piomelli D, Wang PH. Oleylethanolamide activates Ras-Erk pathway and improves myocardial function in doxorubicin-induced heart failure. Endocrinology. 2006;147:827–34. doi: 10.1210/en.2005-1098. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y, Luo P, Chang HH, Huang H, Yang T, Dong Z, Wang CY, Wang MH. Colfibrate attenuates blood pressure and sodium retention in DOCA-salt hypertension. Kidney Int. 2008;74:1040–8. doi: 10.1038/ki.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Chen M, Georgeson KE, Harmon CM. Mechanism of oleoylethanolamide on fatty acid uptake in small intestine after food intake and body weight reduction. Am J Physiol Regul Integr Comp Physiol. 2007;292:R235–41. doi: 10.1152/ajpregu.00270.2006. [DOI] [PubMed] [Google Scholar]