Determination of the protonation state of titratable protein residues is of critical importance for the interpretation of active site chemistry, as well as for understanding the role of electrostatic interactions in protein folding and stability.1-5 NMR spectroscopy provides the most powerful approach available for the determination of pK values for individual protein residues, particularly in combination with isotopic labeling strategies. Nevertheless, the limited pH stability range of most proteins poses a significant obstacle to obtaining reliable pK values based on the pH dependence of resonance shifts. This problem is particularly acute for lysine residues since, with a few exceptions,6 it is not possible to obtain good asymptotic shift values at high pH. However, lysine residues often play critical roles in the active site chemistry of proteins.7-9 The problem becomes more acute as more challenging and stability-challenged proteins are being investigated using this approach.
The 13C shift of lysine C∊ has often been used as a reporter group for the titration of the ∊-amino group, and several studies have utilized proteins specifically labeled with [6-13C]lysine in order to obtain lysyl pK values.6,10-12 Regardless of the labeling approach followed, analysis of the data obtained typically uses a three or four parameter fit:
where δU is the limiting shift of the unprotonated resonance, Δδ = δP - δU the total titration shift, and n is a Hill coefficient, sometimes introduced in order to account for interactions among titrating residues.11,14,15 In general, use of a four parameter fit does not work well if data at both asymptotic shift limits cannot be obtained,14 and even the three parameter fits can result in significant errors when the pH range over which data are obtained is limited. Additionally, the shift observed at high pH can be influenced by the presence of partially unfolded conformations, resulting in significant error in the calculated pK value. In the present communication, we have utilized protein labeled with L-[5-13C]- lysine, which was prepared via alkylation of the Oppolzer sultam glycinate; the synthesis is described in the Supporting Information. The use of the lysine C-5 resonance as a reporter group for the protonation state of the ∊-amino group offers significant advantages over the use of C-6 due to the greater shift dispersion that results because the C-5 carbon is positioned closer to the backbone of the protein, and due to the 5-fold greater shift response to titration of the ∊-amino group, that is, Δδ = 5.0 ppm compared with 1.0 ppm for C-6. This effect is thought to reflect the partial cancellation of several contributions to the shift of C-6.16 In general, the titration shift for C-5 is sufficiently large to dominate conformational effects, allowing analysis of a limited data set near neutral pH in combination with a two parameter fit with Δδ set equal to 5.0 ppm. Some examples are presented below.
A critical step in the base-excision repair process for DNA is the lyase reaction which removes the 5′-deoxyribose phosphate flap that is formed after damaged DNA is incised.17,18 This reaction has been shown to proceed through a β-elimination reaction that occurs after the initial formation of a Schiff base adduct between the C-1′ of the abasic deoxyribose-5′-phosphate and the active site lysine residue.17,18 Biochemically significant Schiff base reactions typically utilize lower pK α-amino groups7-9 or active site lysine residues with unusual environments that result in a lowered pK value. However, recent NMR studies of the lyase domain of DNA Pol λ demonstrated that the pK value for the active site Lys312 residue, which is important for the lyase reaction,19 is 9.6. The 1H-13C HSQC spectrum of the lyase domain of the base-excision repair enzyme DNA polymerase β grown on a medium containing [6-13C]-lysine did not provide sufficient resolution for analysis of the active site Lys72 residue (Figure 1).
Figure 1.
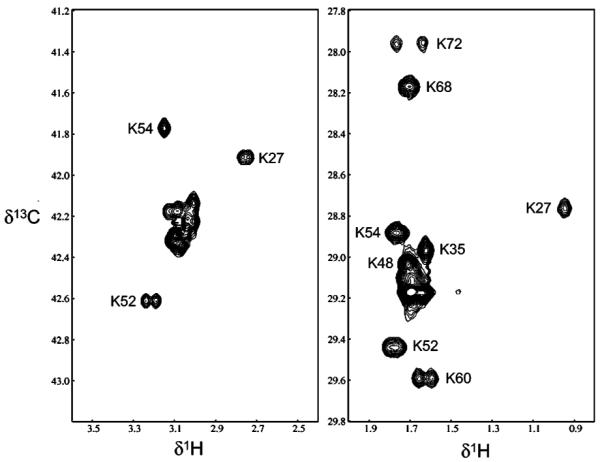
1H-13C HSQC spectra of the Pol β lyase domain labeled either with L-[6-13C]- (left panel) or L-[5-13C]lysine (right panel). 1H-13C HSQC spectra were obtained by using Varian's gChsqc sequence on a UNITY-INOVA 600 MHz spectrometer at pH 6.8, 25 °C, with 128 × 1024 complex points and acquisition time of 128 ms in both t1 and t2. Sixteen scans were acquired per increment, with a 1.0 s delay between scans.
In contrast with the spectrum on the left, half of the lysine residues in the Pol β lyase domain labeled with [5-13C]lysine were resolved in the 1H-13C HSQC spectra (Figure 1). In addition, the large shift range for lysine C-5 makes it possible to observe significant shifts for the lysine residues below pH 9.0, as shown in Figure 2.
Figure 2.

1H-13C HSQC spectra of [5-13C]lysine-labeled DNA Pol β lyase domain (residues 2-87). The assignments of 10 resolved lysine residues are indicated. Superimposed spectra were obtained at pH 6.80 (black), 7.70 (red), and 8.75 (blue). Over this limited pH range, lysine residues 52, 54, and 27 show small pH-dependent shifts, Lys60 and 68 show somewhat larger shifts, and Lys72 still larger shifts. These differences are reflected in the calculated pK values (Table 1). NMR spectral parameters are the same as in the caption of Figure 1.
The pK values corresponding to the resolved lysine resonances obtained using a two parameter fit with Δδ = 5.08 ppm are summarized in Table 1.
Table 1.
Fit of Titration Data for [5-13C]- and [6-13C]Lysine-Labeled Pol β Lyase Domain
| 5-13C label |
6-13C Label |
|||||
|---|---|---|---|---|---|---|
| residue | pK(2)a | pK(3)b | Δδbppm | pK(2)a | pK(3)b | Δδbppm |
| 27 | 11.13 | 10.67 | 3.02 | 10.50 | 9.85 | 0.45 |
| 35 | 10.40 | c | c | d | d | d |
| 52 | 10.50 | 10.30 | 4.53 | 10.76 | 10.59 | 0.77 |
| 54 | 10.71 | c | c | 10.59 | c | c |
| 60 | 10.39 | 10.23 | 4.68 | d | d | d |
| 68 | 10.21 | c | c | d | d | d |
| 72 | 9.84 | c | c | d | d | d |
Two parameter fit of the 13C titration data with Δδ set equal to 1.05 ppm for C-6 or 5.08 ppm for C-5.
Three parameter fit of the 13C titration data.
Nonlinear fit did not converge.
Not resolved.
The pK value of 9.8 for the active site Lys72 residue of the domain is very similar to the value of 9.6 recently determined for the homologous residue in the lyase domain of DNA Pol λ.11 This result is consistent with a recent crystallographic study of Pol β complexed with DNA containing stable analogues of the abasic sugar,20 in which it was concluded that there was no obvious structural basis for a significant depression of the pK of Lys72. In that study, it was suggested that a simple rotation about the abasic sugar 3′-phosphate would position the abasic sugar C-1′ close to the ∊-amino group of Lys72. The C-1′ proximity and environment of Lys72 are expected to facilitate β-elimination.20
The data in Table 1, as well as data from the literature for titration of the [6-13C]lysine-labeled TEM-1 β-lactamase summarized in the Supporting Information, indicate that the use of a three parameter fit results in a systematic lowering of the pK and Δδ values. We have been able to simulate this effect by generating titration data using a Hill coefficient of 0.8—a value typical of titrating residues which exhibit negative cooperativity.15 The data so generated can be fit with a standard two, three, or four parameter Henderson-Hasselbach equation (Supporting Information). However, as titration data at the higher pH end of the curve are eliminated, the three parameter fit breaks down, leading to progressively lower pK and Δδ values. The four parameter fit of the simulated data was successful, but small deviations present in real data due to shift contributions from the titration of nearby residues or from conformational changes make such fitting procedures unstable and generally unreliable. This limitation was apparent in titration studies of the U-[13C,15N]Pol β lyase domain (Supporting Information). Alternatively, reasonable pK values are obtained using a two parameter fit of the data, even if the pH range does not reach the pK of the titratable group. For real data, the success of this two parameter fit is critically dependent on the assumption that the magnitude of the titration shift is much greater than the shift contributions arising from other effects. For this reason, the use of [5-13C]lysine with its 5-fold greater shift response to titration of the ∊-amino group is a superior approach for lysine pK determination in proteins over which the pH accessible range is limited.
Supplementary Material
Acknowledgment
This research was supported in part by the Intramural Research program of the NIH, and NIEHS, and by the National Stable Isotopes Resource (5P41 EB002166 (C.J.U. and S.N.L.)).
Footnotes
Supporting Information Available Synthetic method for the synthesis of [5-13C]lysine, two and three parameter fits of TEM-1 β-lactamase [6-13C]lysine titration data from refs 10 and 12, analysis of titration data for the U-[13C,15N] Pol β lyase domain, and various simulations. This material is available free charge via the Internet at http://pubs.acs.org.
References
- (1).Antosiewicz J, McCammon JA, Gilson MK. Biochemistry. 1996;35(24):7819–33. doi: 10.1021/bi9601565. [DOI] [PubMed] [Google Scholar]
- (2).Loewenthal R, Sancho J, Fersht AR. J. Mol. Biol. 1992;224(3):759–70. doi: 10.1016/0022-2836(92)90560-7. [DOI] [PubMed] [Google Scholar]
- (3).Oliveberg M, Fersht AR. Biochemistry. 1996;35(8):2726–37. doi: 10.1021/bi9509661. [DOI] [PubMed] [Google Scholar]
- (4).Sancho J, Serrano L, Fersht AR. Biochemistry. 1992;31(8):2253–8. doi: 10.1021/bi00123a006. [DOI] [PubMed] [Google Scholar]
- (5).Shoemaker KR, Kim PS, Brems DN, Marqusee S, York EJ, Chaiken IM, Baldwin RL. Proc. Natl. Acad. Sci. U.S.A. 1985;82:2349–53. doi: 10.1073/pnas.82.8.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kesvatera T, Jonsson B, Thulin E, Linse S. J. Mol. Biol. 1996;259(4):828–39. doi: 10.1006/jmbi.1996.0361. [DOI] [PubMed] [Google Scholar]
- (7).Barbas CF, III, Heine A, Zhong G, Hoffmann T, Gramatikova S, Bjornestedt R, List B, Anderson J, Stura EA, Wilson IA, Lerner RA. Science. 1997;278(5346):2085–92. doi: 10.1126/science.278.5346.2085. [DOI] [PubMed] [Google Scholar]
- (8).Harris TK, Turner GJ. IUBMB Life. 2002;53(2):85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- (9).Highbarger LA, Gerlt JA, Kenyon GL. Biochemistry. 1996;35(1):41–6. doi: 10.1021/bi9518306. [DOI] [PubMed] [Google Scholar]
- (10).Damblon C, Raquet X, Lian LY, Lamotte-Brasseur J, Fonze E, Charlier P, Roberts GC, Frere JM. Proc. Natl. Acad. Sci. U.S.A. 1996;93(5):1747–52. doi: 10.1073/pnas.93.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gao G, Derose EF, Kirby TW, London RE. Biochemistry. 2006;45(6):1785–94. doi: 10.1021/bi051856p. [DOI] [PubMed] [Google Scholar]
- (12).Golemi-Kotra D, Meroueh SO, Kim C, Vakulenko SB, Bulychev A, Stemmler AJ, Stemmler TL, Mobashery S. J. Biol. Chem. 2004;279(33):34665–73. doi: 10.1074/jbc.M313143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Logan TM, Olejniczak ET, Xu RX, Fesik SW. J. Biomol. NMR. 1993;3(2):225–31. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- (14).Laurents DV, Huyghues-Despointes BM, Bruix M, Thurlkill RL, Schell D, Newsom S, Grimsley GR, Shaw KL, Trevino S, Rico M, Briggs JM, Antosiewicz JM, Scholtz JM, Pace CN. J. Mol. Biol. 2003;325(5):1077–92. doi: 10.1016/s0022-2836(02)01273-1. [DOI] [PubMed] [Google Scholar]
- (15).Song J, Laskowski M, Jr., Qasim MA, Markley JL. Biochemistry. 2003;42(10):2847–56. doi: 10.1021/bi0269512. [DOI] [PubMed] [Google Scholar]
- (16).Batchelor JG, Feeney J, Roberts GCK. J. Magn. Reson. 1975;20(1):19–38. [Google Scholar]
- (17).Deterding LJ, Prasad R, Mullen GP, Wilson SH, Tomer KB. J. Biol. Chem. 2000;275(14):10463–71. doi: 10.1074/jbc.275.14.10463. [DOI] [PubMed] [Google Scholar]
- (18).Matsumoto Y, Kim K. Science. 1995;269(5224):699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- (19).Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. J. Biol. Chem. 2001;276(37):34659–63. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- (20).Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, Wilson SH. DNA Repair (Amst) 2005;4(12):1347–57. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


