Abstract
Background
Members of the nucleotidyltransferase superfamily known as DNA and RNA ligases carry out the enzymatic process of polynucleotide ligation. These guardians of genomic integrity share a three-step ligation mechanism, as well as common core structural elements. Both DNA and RNA ligases have experienced a surge of recent interest as chemotherapeutic targets for the treatment of a range of diseases, including bacterial infection, cancer, and the diseases caused by the protozoan parasites known as trypanosomes.
Objective
In this review, we will focus on efforts targeting pathogenic microorganisms; specifically, bacterial NAD+-dependent DNA ligases, which are promising broad-spectrum antibiotic targets, and ATP-dependent RNA editing ligases from Trypanosoma brucei, the species responsible for the devastating neurodegenerative disease, African sleeping sickness.
Conclusion
High quality crystal structures of both NAD+-dependent DNA ligase and the Trypanosoma brucei RNA editing ligase have facilitated the development of a number of promising leads. For both targets, further progress will require surmounting permeability issues and improving selectivity and affinity.
Keywords: nucleotidyltransferase superfamily, DNA ligase, RNA ligase, RNA editing ligase 1, NAD+ dependent DNA ligase, antibiotics, anti-trypanosomal therapeutics, African sleeping sickness
1. Introduction
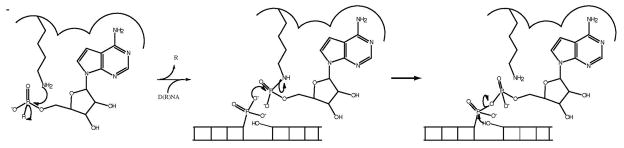
The enzymatic process of polynucleotide ligation is carried out by RNA and DNA ligases, structurally and evolutionarily related members of the nucleotidyltransferase (NTR) superfamily 1, which also includes eukaryotic GTP-dependent mRNA capping enzymes 1. Ligase superfamily members share a conserved chemical mechanism (Fig. 1), which utilizes a nucleotide cofactor, either ATP or NAD+, to furnish an AMP moiety. In the first reaction step, attack of a conserved lysine nucleophile displaces either pyrophosphate (PPi) or nicotinamide mononucleotide (NMN), and forms a phosphoramidate bond between the lysine ε-nitrogen and the AMP moiety. In the second step, a nicked double-stranded polynucleotide binds, and the AMP is transferred to the 5′PO4 end of the nick where it is connected via a phosphoanhydride linkage. The reaction is completed when the nicked 3′OH end attacks the phosphoanhydride, displaces AMP and joins the nicked-polynucleotide ends. In addition to mechanistic conservation, superfamily members share recognizably conserved structural elements 1. Prominent among these is a nucleotide-binding domain (NTBD), which comprises the fundamental catalytic unit of the superfamily and may be an example of a stand-alone ancestral enzyme from which contemporary superfamily members evolved 1, 2. Domain modules, which impart polynucleotide specificity, are covalently linked to both the N- and C-terminals of the NTBD, in a clamp-like arrangement, by peptide tethers. By virtue of these tethers, the clamp-like architecture is quite flexible 3, 4 5, 6, 7, and progression through the catalytic cycle is facilitated by clamp opening and closing, which allows substrate ingress and product release 3, 5, 6. One notable exception is the ATP-dependent RNA editing ligase from the pathogenic organism Trypanosoma brucei. In this organism, which will be discussed in greater detail in subsequent sections, modular domains associate non-covalently to the NTBD 8. Despite this difference, movement through the catalytic cycle is also thought to coincide with domain opening and closing 8.
Figure 1. Three-step nick joining mechanism by DNA and RNA ligases.
In step one, the active-site lysine attacks the alpha phosphate of NAD+ or ATP, displacing R (where R=PPi in ATP-dependent DNA and RNA ligases and R=NMN in NAD+-dependent DNA ligases) and forming an enzyme-AMP intermediate. Following polynucleotide binding, the nicked 5′PO42− attacks the enzyme-AMP intermediate, displacing the active-site lysine, forming a D(R)NA-AMP intermediate in step two. In step three, the nicked 3′OH attacks the D(R)NA-AMP intermediate, displacing AMP and joining the nicked polynucleotide strand.
In the repertoire of biochemical reactions, polynucleotide ligation, catalyzed by members of the NTR superfamily, is vitally important in a wide spectrum of cellular processes. For example, the 5′ to 3′ directionality of DNA replication results in a lagging strand whose synthesis is fragmented into discontinuous blocks, called Okazaki fragments. Lagging strand synthesis is completed when a DNA ligase joins the 5′PO4 and 3′OH ends of apposing fragments. In addition to normal cellular process like DNA replication, the integrity of DNA may be interrupted by various exogenous assaults that result in oxidative damage 9, as well as alkylation 10 and deamination 11. In order to preserve genomic integrity in the face of these assaults, cells have evolved various repair strategies, like long and short patch base excision repair 12, 13, and nonhomologous end joining 14, 15. Among other factors, the efficacy of these repair mechanisms is critically dependent upon nicked polynucleotide joining, which is generally the last step in these pathways 16–18. Polynucleotide ligation is not, however, limited to nicked DNA, it is also an essential chemical reaction in the repair 19, splicing 20 and editing processes of RNA 21. For example, following the post-transcriptional insertion or removal of polyuridylate tracts to, or from, nascent mitochondrial transcripts of the genus Trypanosoma 22, one of two RNA editing ligases joins the nicked ends of the mRNA, completing the RNA editing process 23.
In light of the fundamental importance of polynucleotide ligation, it is not surprising that both DNA and RNA ligases have experienced a surge of recent interest as chemotherapeutic targets for the treatment of a range of disease, including bacterial infection 24, 25, cancer 26, and African sleeping sickness 27. It is the purpose of this review to detail these recent advances. Specifically, we limit our attention to efforts targeting pathogenic microorganisms. In particular, we focus on bacterial NAD+-dependent DNA ligases, which are promising broad-spectrum antibiotic targets, and ATP-dependent RNA editing ligases from Trypanosoma brucei, the species responsible for the devastating neurodegenerative disease, African sleeping sickness. The defining structural features of each of these inhibitor targets will be discussed in turn, followed by discussion of the inhibitors reported to date for each target, paying particular attention to inhibitor development and subsequent in vitro and in vivo characteristics. The review will conclude with possible future directions.
2. NAD+-dependent DNA Ligases
DNA ligases were initially characterized by several labs in a flurry of activity spanning the years 1967 and 1968 28, 29,30–33 as reviewed in 34. DNA ligases are not only sentries of genomic integrity, participating in vital DNA repair pathways 12–15, they also play a fundamental role during genetic recombination, a process that occurs during both meosis in eukaryotes, and V(D)J shuffling, which produces the diverse antibody specificity central to an effective immune response in vertebrates 35. Furthermore, DNA ligases are essential in genome replication 36. There are two principal subfamiles of DNA ligases whose members can be recognized according to whether they require an ATP or NAD+ nucleotide during the ligation reaction.
ATP-dependent DNA ligases span the greatest range of biological diversity. They have been characterized in all three kingdoms of life, as well as in bacterial and eukaryotic viruses 2. NAD+-dependent DNA ligases, on the other hand, have nearly the same phylogenic distribution but are noticeably absent in Eukarya 2. Also known as LigAs after the name of the E. coli gene that encodes them, NAD+-dependent DNA ligases are essential in Escherichia coli 37 as well as several gram-positive and gram-negative organisms, including: Salmonella typhimurium 38, Haemophilus influenza 39, Bacillus subtilis 40, Streptococcus pneumonia 41, Staphylococcus auerus 42 and Mycobacterium tuberculosis 43, 44. The absence in eukaryotic organisms and essential function in bacteria make LigA an attractive target for the development of novel, broad-spectrum antibiotics. Indeed, several classes of LigA specific inhibitors have been reported. Before reviewing each inhibitor class, we will briefly detail the structural features of the ATP-dependent and NAD+-dependent DNA ligases. We will pay particular attention to their shared and unique structural elements, which in many cases have allowed LigA specific inhibitor development.
2.1 Defining structural features
Both the ATP-dependent and NAD+-dependent DNA ligases share a modular domain architecture forming a clamp-like structure (Fig. 2) whose opening and closing facilitates progression through the catalytic cycle 1, 3, 45. A minimal catalytic core, which consists of the NTBD, and the oligonucleotide-binding domain (OBD) form the fundamental structural unit of the enzyme clamp. As the common structural feature of all NTR superfamily members, the NTBD houses features essential for nucleotide recognition and chemistry. For example, five of the six motifs, which are conserved across all members of the NTR superfamily1 form a deep beta sheet cleft, which sandwiches the AMP between conserved aromatic and hydrophobic residues 1, 6, 46–50. In both bacterial and ATP-dependent DNA ligases, AMP recognition occurs in the deep end of the binding pocket where it interacts with the protein backbone or side-chains, and the exocyclic-adenine substituents 6, 47, 48. Following nucleotide recognition and binding, the catalytic lysine, contained in motif I (KXDG), attacks the nucleotide and forms the enzyme-adenylate intermediate. In ATP-dependent DNA ligases, the formation of the enzyme-adenylate intermediate is critically dependent on the OBD51. The OB fold, which defines the OBD, consists of a five-stranded anti-parallel beta-barrel and contains a sixth conserved motif, which is thought to position the triphosphate leaving group inline with the attacking lysine nucleophile6. In contrast, in NAD+-dependent DNA ligases, the OBD lacks conserved motif VI and is dispensable for enzyme-adenylate formation 52, 53. Instead, a unique bacterial DNA ligase domain, domain Ia (Fig. 2), comprised of two helices that form a NMN binding pocket, is required. Crystal structures imply that domain Ia orients the NMN leaving group in a manner functionally homologous to the role played by the OBD in ATP-dependent DNA ligases5. Its essential function and uniqueness to bacterial LigAs make domain Ia an appealing structural feature to target during rational inhibitor design.
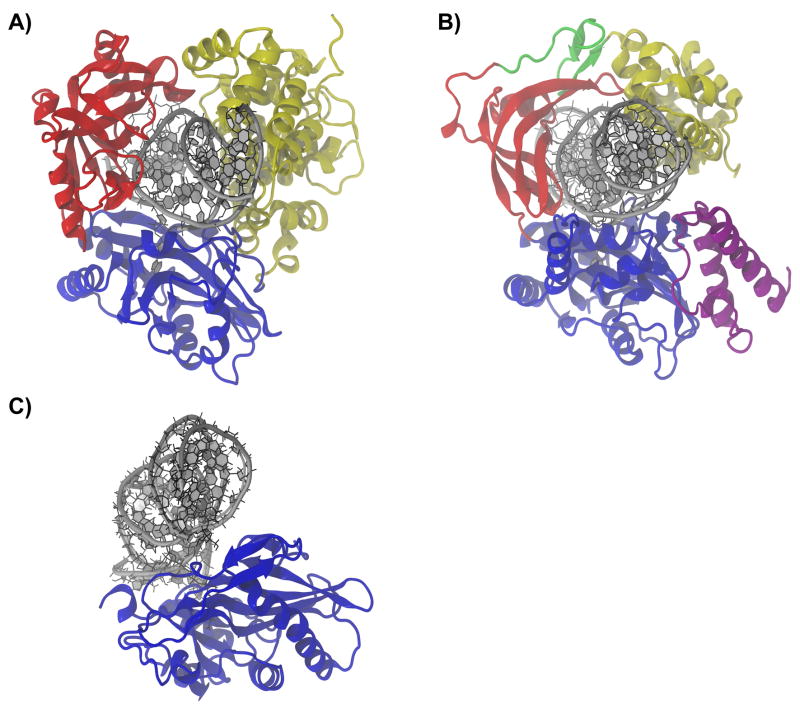
Figure 2. Modular domain architecture in DNA and RNA ligases.
A), B) and C) are ribbon diagrams of Human DNA ligase I (PDB ID 1X9N), E. coli DNA ligase (PDB ID 2OWO), and T. brucei RNA-editing ligase 1 (REL1) (PDB ID 1XDN). Each is shown in complex with their nicked polynucleotide substrates and homologous domains are indicated by color. The clamp-like domain arrangement that facilitates strand joining is clearly seen in (A) and (B). As discussed in the text, a second domain associates non-covalently with the NTBD to form the clamp in REL1. The enzyme-RNA complex in (C) is a homology model as described in Ref. 93. The nucleotide binding domain (NTBD) is shown in blue in (A), (B), and (C). In (B) and (C), the oligonucleotide binding domain (OBD) is shown in red. In (A) the DNA-binding domain (DBD) is shown in yellow, while the homologous helix-hairpin-helix domain (HhH) is shown in yellow in (B). Additional domains, unique to bacterial NAD+-dependent DNA ligases are shown in (B): domain Ia is colored magenta, while the structural-zinc-finger is colored green.
Following enzyme-adenylate formation in both bacterial and ATP-dependent DNA ligases, it is thought that large-scale isomerization events occur that move domain Ia and the OBD away from their respective NTBDs, a view consistent with available structural data 4, 5. The motion opens the clamp and allows nicked, double-stranded DNA to bind. After DNA binding has occurred, the clamp shuts again and ligation may proceed. Prior to strand joining, clamp closure in both bacterial and ATP-dependent DNA ligases deforms their DNA substrates, imposing a RNA-like A conformation on nucleotides local to the nick 4, 45, 54, 55. The distortion positions the opposing nicked ends such that nucleophilic attack may occur and seems to be a critical prerequisite to strand joining 45, 54–56. DNA distortion is brought about by a set of homologous protein-substrate interactions as illustrated by a comparison of the crystal structures of both HuLig I and LigA in complex with the adenylated DNA substrate 54, 56. In each structure, the NTBD and OBD engage the minor groove local to the nick, forming two sides to a substrate clamp that enforces the RNA A-like conformation local to the nick (Fig. 2). The key difference between the eukaryotic and bacterial crystal structures is the identity of the domain that forms the clamp base. In ATP-dependent DNA ligases, an alpha helical domain, called the DNA-binding domain (DBD), is covalently linked to the N-terminal of the NTBD and forms the clamp base 54. The clamp base in NAD+-dependent DNA ligases, on the other hand, is formed by a set of four helix-hairpin-helix (HhH) motifs, which are linked to the N-terminal of the NTBD via a structural zinc-binding domain 50, 56. Despite having different folds, the HhH domain and the DBD play analogous roles in strand joining, conveying the major DNA binding activity in representative members of each ligase class 45, 54, 57.
While the domains that form the clamp work cooperatively to efficiently join nicked DNA strands in vitro 53, 54 additional clamp-C-terminal-domain extensions regulate activity in vivo. For example, in mammalian DNA ligase I, a nuclear localization signal and a PCNA binding motif extend from the N-terminal of the DBD and coordinate interactions with the DNA replication fork 36, 58. The other two mammalian isoforms have modulating domains of similar function 58. In bacterial LigAs, a highly mobile BRCT domain can be found extending from the C-terminal domain of the HhH domain. The structure of this domain has not been resolved by crystallography but has been solved independently in solution using NMR spectroscopy (PDB ID 1I7b). However, its importance in nicked strand joining remains a point of contention among scientists 52, 53, 57, 59, 60 and until a consensus has been reached, it is unlikely to command interest as a target receptor in rational inhibitor design.
2.2 Inhibitors
Although the BRCT domain is unlikely to serve as an inhibitor-binding site, Nandakumar, Nair and Shuman pointed out an exceptional opportunity for rational inhibitor design within the NTBD domain 56. In the crystal structure of the step-two adenylate-DNA intermediate (2OWO), they note the presence of a hydrophobic tunnel extending from the exterior of the protein into the adenine-binding pocket of the NTBD domain where the N1, C2 and N3 edge of adenine is exposed 2, 56 (Fig. 3A). Although this pocket is present in all available LigA structures, it is absent in human DNA ligase I (HsLigI), where the space is blocked by a loop connecting motifs IV and V 54, 56. As they note, inhibitors derived from C-2 substituted adenosine, AMP, or NAD+ could prove advantageous by simultaneously utilizing native substrate contacts, while tuning LigA specificity by adding alkyl or aryl substituents strategically positioned to occupy the hydrophobic tunnel 56. Recently, this design strategy has been proven viable. The results of a recently reported high throughput assay describe 2-methylthio derivatives of ADP and ATP that inhibit E. coli LigA with IC50 values of 0.5 and 2.1 μM respectively 61. While structural data illuminating the binding modes of these 2-methylthio derivatives was not reported, it seems likely that the bound state position of the ADP and ATP aromatic adenines overlap with that of the AMP-DNA intermediate adenine such that the 2-methylthio moieties extend into the hydrophobic tunnel. The plausibility that the positions of the bound state aromatic moieties overlap is bolstered by crystal structures of LigA bound to a pyridochromanone derivative, and three pyridopyrimidine derivatives, which are discussed in the next section.
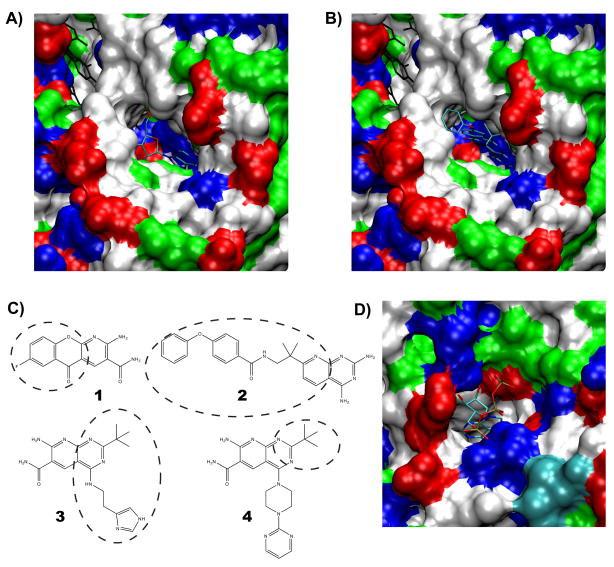
Figure 3. Druggable cavities in the bacterial NAD+-dependent DNA ligase and the T. brucei ATP-dependent RNA editing ligase 1.
(A) The hydrophobic tunnel adjacent to the NAD+-binding pocket. AMP is shown occupying the pocket. Ribose and the N1, C2, N3 adenine atoms (from right to left) are visible. The surface representation of the protein is colored by residue type: blue-basic, red-acidic, green-polar, and white-hydrophobic. The image was constructed using PDB ID 2OWO. (B) Compound 2 shown protruding from the hydrophobic tunnel, PDB ID 3BA9. Protein coloring is as in (A). (C) Structures of pyridochromonone (1) (PDB ID 3BA8), and the three pryidopyrmidine compounds (2, 3, 4) (PDB IDs: 3BA9, 3BAA, 3BAB) which make use of the hydrophobic tunnel. The dashed ovals encompass the portion of the molecule that extends into the tunnel; the remainder of the molecule overlaps with the NAD+ binding pocket. Bold numbers are referenced in the text and tables 1 and 2. (D) The deep ATP binding pocket in T. brucei RNA editing ligase 1 is shown, coloring the same as in (A).
2.2.1 Pyridochromanones, Pyridopyrmidines
Preceding a forthcoming publication, Pinko and coworkers recently deposited four crystal structures into the PDB: 3BA8, which shows the LigA bound state of a fluorinated pyridochromanone derivative, (compound 1, Fig. 4) and a set of 3 pyridopyrimidine derivatives 2, 3, 4 (3BA9, 3BAA, 3BAB). In each crystal structure, the aromatic scaffold of the inhibitor occupies the binding site in a manner similar to the position taken by the DNA-adenylate adenine, illustrated by the crystal structure 2OWO. Furthermore, both the pyridochromanone and the pyridopyrimidine derivatives make use of the hydrophobic tunnel 2 (Fig. 3B & 3C).
Figure 4.
Inhibitors of NAD+ dependent DNA ligases.
The occupancy of the fluorinated pyridochromanone derivative in the NAD+-adenine-binding pocket is consistent with the experimental characterization of a set of pyridochromanone derivatives carried out by Brötz-Oesterhelt et al 62. These compounds were identified in a high throughput screen and inhibited LigA via a NAD+ competitive binding mechanism. In two separate assays, carried out using both E. coli and S. pneumoniae LigA, IC50 values ranged from 40 to 100 nM. Interestingly, compound 1, crystallized by Pinko et al., showed the best in vitro inhibition characteristics with an IC50 value of 40 nM (Table 1). When compared to HsLigI inhibition effects, all of the pyridochromanone derivatives showed a roughly 1000-fold specificity toward LigA. Additionally, the pydridochromanone derivatives were active in vivo, but primarily against gram-positive bacteria. While MIC values reported for the gram-positive bacteria S. aureus and B. subtilis were in the low μg mL−1 range, gram-negative E. coli, were unaffected until treated by the permeablizing agent polymycin B nonapeptide; this implies that gram-negative outer membrane permeability is low. Consequently, increasing membrane permeability while maintaining binding affinity and selectivity must be achieved before members of the current pyridochromanone derivative set can find use as broad-spectrum antibiotics. Nevertheless, in their current state, they boast nanomolar IC50 values and bind LigA with 1000-fold selectivity compared to HsLigI, promising characteristics in a novel gram-positive antibiotic lead compound.
Table 1.
| Compound no. | LigA IC50, (μM) | † HsLig1 IC50, (μM) | NAD+ competitive? | MIC (μg mL−1) |
|---|---|---|---|---|
| 1 | 0.04 | >75 | Yes | *6 |
| 2–4 | N/A | N/A | N/A | N/A |
| 5 | 0.5 | >100 (μg/mL) | Yes | 64 |
| 6 | 53 | >720 (T4) | No | 2.5×104 |
| 7 | 63 | >2000 (T4) | No | N/A |
| 8 | 2.6 (n=0) | 16 (n=0) | No | 2.3×104 |
| 9 | 10.2 (n=0) | 49 (n = 0) | No | N/A |
| 10 | 46.2 | 320 | Yes | *0.2 |
| 11 | 260 | 72 | N/A | N/A |
| 12 | 11.4 | 27 | N/A | N/A |
| 13 | 85 | 380 | Yes | *0.1 |
| 14 | 225 | 94 | N/A | N/A |
| 15 | 13.5 | 212 | Yes | *7 |
IC50 values determined using the T4-phage ligase in the the place of HsLig1 are noted: (T4).
The membrane permeablyzing agent polymxin nonapeptide was added to the gram-negative bacterial growth media.
Pyridopyrmidine derivatives, similar to those crystallized in complex with LigA, have been reported as possible chemotherapies spanning a diverse range of afflictions from possible anti-diarrheal agents to bacterial drug efflux pumps 63–67. Despite the diversity of their potential applications, a thorough characterization of their antibacterial potential has not been published, yet the LigA-bound crystal structures show promising features. In addition to aromatic moieties that overlap with the NAD+ adenine, each makes use of the hydrophobic tunnel adjacent to the NAD+ binding pocket (Fig. 3A, 3B & 3C), two features that lend themselves to competitive binding with NAD+ and selectivity toward LigA.
2.2.2 Pyrimidopyrmidines
Similar in structure to the pyridopyrimidine derivatives, a pyrimidopyrimidine derivative, 2,4-diamino-7-dimethylamino-pyrimido[4,5-d]pyrimidine, or DDPP, 5, was recently reported as a LigA inhibitor by researchers at Eli Lilly (Fig. 4). DDPP inhibits the Streptococcus pneumonia LigA with an IC50 of 0.5 μM by competitively binding with NAD+ 68 (Table 1). Selectivity was determined by comparing LigA DDPP IC50 values to those measured for T4-phage ligase and HsLigI. DDPP demonstrated 600 and 1000 fold selectivity toward LigA when compared to the phage and human ligase, respectively. Additionally, DDPP, like the pyridochromanone derivatives, demonstrated in vivo activity. MIC values reported for S. pneumoniae, S. aureus, H. influenza were 64, 128 and 128 μg mL−1, respectively, indicating that the compound is effective against both gram-positive and gram-negative human pathogens 68. As further validation of the compounds in vivo activity, S. pneumoniae was grown in the presence of tritium labeled thymidine both with and without DDPP. Incorporation of labeled thymidine into DNA was significantly reduced in the presence of DDPP. Similar incorporation inhibition was observed with novobiocin, a known DNA replication inhibitor 68, consistent with the role of DDPP as a DNA replication inhibitor. While crystal structures of the LigA bound DDPP could not be obtained 68, the similarity of the DDPP and pyridopyrimidine molecular scaffolds (compare 2, 3 and 4, to 5) makes it likely that DDPP shares a binding mode similar to that observed for the pyridopyrimidines. Furthermore, while the IC50 of DDPP is roughly an order of magnitude greater than that of the pyridochromanone, DDDP demonstrates similar selectivity and better membrane permeability, as inferred by lower DDPP MIC values in gram-negative bacteria. Without the need to overcome membrane permeability issues, pyrimidopyrimidine derivatives, like DDPP, may serve as better leads to broad-spectrum antibiotics.
2.2.3 Arylamino compounds
While pyridochromanones, pyridopyrimidines, and pyrimidopyrmidines all bind competitively with NAD+, a third class of compounds exerts their inhibitory effects via a non-competitive mechanism. Arylamino compounds, including derivatives of chloroquine, a well-known anti-malarial agent suffering from emerging resistance issues 69, and bisquinoline, a potential scaffold for new anti-malarial agents 70–72, were the first compound class reported that specifically target LigAs 73. Chloroquine, 6, and hydroxychloroquine, 7, both quinoline derivatives, showed IC50 values of 53 and 63 μM respectively, with order of magnitude selectivity toward LigA when inhibitor activity was compared to that for HsLigI or the homologous ATP-dependent DNA ligase from T4 phage (Fig. 4 and Table 1). The C-6 (8) and C-8 (9) bisamide-linked bisquinolines, whose quinoline groups were substituted by C-4 diamino moieties, exhibit IC50 values ranging from 2.6 to 10.2 μM, an improvement over the chloroquines. While various lengths of the bisamide linker were tested (n = 0, 6, 8 for 8; n = 0, 4 for 9), the inhibitory effects showed little variability, changing by no more than 0.8 μM, an amount only slightly larger than the reported experimental error. However, IC50 values improved by roughly five-fold when the bisamide linker spanned the quinoline C-6 positions, rather than the C-8 positions. For example, compound 8, with n = 0, has a reported IC50 of 2.6μM, while compound 9, with n = 0, has a reported IC50 of 10.2μM. Inspection of the structures of chloroquine, hydroxychloroquine and the assayed bisquinoline derivatives shows a conserved diamino group extending from C-4 of the quinoline moieties. This diamino group was deemed an important mediator of inhibitor binding. When a chloro group was substituted for the diamino group at the C-4 position, a 1000-fold increase in measured IC50 values were reported in strand joining assays. Without a crystal structure to confirm, one might speculate that the quinoline derivatives make use of the NAD+ binding site and compete with NAD+ binding. Despite this, and though a detailed kinetic analysis is absent, evidence suggests that quinoline derivatives exert the majority of their effects through DNA interactions 73.
The arylamino compounds may suffer from poor membrane permeability, an issue that would hinder their development as broad-spectrum antibiotics. For example, based on in vitro IC50 values, the in vivo activity was considerably less than expected. 25 mg mL−1 of chloroquine were required to reduce log-phase growth of Salmonella typhimurium by ~50%, a concentration approximately three orders of magnitude larger than required for comparable in vitro inhibition (IC50 = 53 μM or 27 μg mL−1). One explanation for this, as the authors point out, is the weakly basic nature of the quinoline derivatives, which results in partial protonation at pH 7 73, 74. Charged compounds have reduced hydrophobicity and ability to partition into the outer membrane of gram-negative bacteria, which in turn may lead to the higher observed concentration requirements. While it is possible that membrane permeability could be improved with appropriate structural modifications, the majority of the in vitro activity would still likely be due to non-specific DNA interactions, which, at least until a ternary inhibitor-DNA-ligase structure is solved, eliminates structure-based inhibitor design as a viable alternative and increases the difficulty of the inhibitor refinement process.
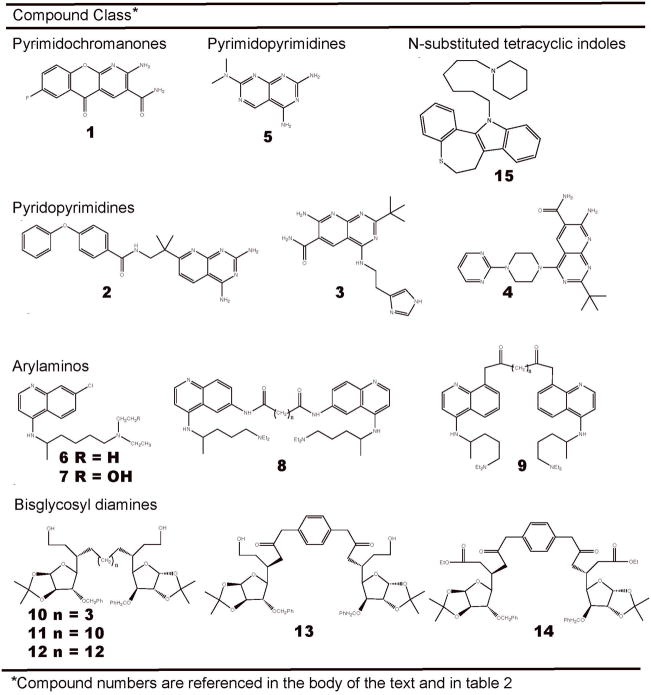
2.2.4 Bisglycosyl diamines
Another class of compounds that shows potential to carry forward in the drug discovery pipeline is the bisglycosyl diamine derivatives, as reported by Srivastava and coworkers 75, 76. Using in silico screening of an in-house compound database, two classes of bis-xylofuranosylated diamines were identified: those with variable length diamino alkyl spacers, e.g. compound 10, and those with phenylene carbomyl spacers, compounds 13 and 14 (Fig. 4). Diamino alkyl spacers of 3, 10 and 12 methylene units (compounds 10, 11, and 12) were synthesized and tested. The two phenylene carbomyl variants tested, 13 and 14, had fixed spacer lengths, but different oxidation states. Compounds 10, 11 and 12 were assayed in vitro for inhibitory effects and LigA selectivity using recombinant LigA from M. tuberculosis (MtuLigA). IC50 values of 46.2, 260 and 11.4 μM, respectively, were reported for compound 10, 11, and 12 (Table 1). MtuLigA selectivity, while present, was modest. When compared to HsLigI, compounds 10, 11 and 12 showed 7.5, 2.4 and 4.5 fold selectivity, respectively, toward MtuLigA. In order to gain insight into the reported IC50 values, AutoDock was used to dock each of the three compounds into the LigA crystal from M. tuberculosis (Mtu) 75. The results demonstrated that the binding modes of compounds 10 and 12 were largely contained in a wire-mesh surface demarcating the binding volume of NAD+. On the other hand, the majority of compound 11 resided outside of the NAD+ binding volume 76. A similar phenomenon was observed with the two tested variants of the phenyelene carbamoyl linked bis-xylofuranosylated diamino compounds, 13 and 14. Compound 13, which made the most effective use of the NAD+ binding volume, had an IC50 value roughly 3 times lower than compound 14, whose binding mode showed little overlap with the NAD+ binding volume, i.e. IC50 values of 85 and 225 μM, respectively. When MtuLigA and HsLigI inhibitory efficacy were compared, compound 13 selectively favored MtuLigA inhibition by roughly 4 fold. Interestingly, while compounds 10, 12 and 13 each made effective use of the NAD+ binding volume, none took advantage of the adjacent hydrophobic tunnel (Fig. 3A). The overlap of the AutoDock predicted inhibitor binding modes and the binding mode of NAD+ imply that the inhibitors bind competitively with NAD+. Kinetic analysis confirmed these predictions, indicating that compounds 10 and 13, the only two compounds tested, both bind LigA in a NAD+ competitive mechanism. Moreover, ethidium bromide displacement assays showed that these compounds did not intercalate DNA. While only a crystal structure can confirm the predicted detailed atomic inhibitor-receptor interactions, when considered collectively, the kinetic analysis and the ethidium bromide displacement assays support the AutoDock predicted overlap of the inhibitor and NAD+ binding volumes. In order to test whether the in vitro efficacy of the compounds held in vivo, MIC values were determined in E. coli and S. typhimurim. While MIC values were found to be in the low μg mL−1 for both compounds 10 and 13, again the only two compounds tested, the membrane permeablyzing agent polymxin nonapeptide was used, indicating poor membrane permeability. In summary, while compound 10, has an IC50 value approximately four times that of compound 12, it is a more effective inhibitor than compound 13. Furthermore, compound 10 shows greater LigA selectivity than both compounds 12 and 13. Those observations may make it the most promising broad-spectrum-antibiotic lead presented.
2.2.5 N-substituted tetracyclic indoles
In a more recent work by Srivastava and coworkers, in silico screens identified roughly half a dozen N-substituted tetracyclic indoles 53. The initial set of in silico compounds were sorted for LigA selectivity by testing at high concentrations against MtuLigA and T4Lig. The high concentration selectivity screen resulted in the identification of compound 15, which was further experimentally characterized (Fig. 4). When joining assays were carried out in the presence of increasing concentrations of compound 15 using both recombinant MtuLigA and HsLigI, IC50 values for MtuLigA were 13.5, and those for HsLigI were 204.0, a roughly 15 fold selectivity for LigA (Table 1). While extensive attempts at crystallizing the compound proved unsuccessful, independent docking studies performed using four different software packages (AutoDock, Gold, FlexX, and Affinity) were used to predict the compounds binding mode. The docking studies resulted in a consensus orientation of the compound, in which the tetracyclic indole occupied the adenine-binding cleft in the NTBD and the N-hexylpiperidine moiety extended toward domain Ia. Consistent with the predicted binding pose, kinetic studies indicated that compound 15 bound competitively with NAD+. The ability of compound 15 to inhibit bacterial growth was also independently determined in the gram-negative E. coli and S. typhimurim, with MIC values reported in the low μg mL−1 range. While these values are the same order of magnitude as the in vitro IC50 values, polymxin nonapeptide was used to permeablize the membrane indicating that membrane translocation may be a problem. The reported μM IC50 value, 15-fold LigA selectivity compared to HsLigI, make compound 15 a promising candidate for further refinement.
3. ATP-dependent RNA ligases
The final promising therapeutic target that we will review here are the ATP-dependent RNA editing ligases from the pathogenic Trypanosoma species. The development of new and effective drugs to treat the diseases caused by these protozoan parasites has been relatively nonexistent. Each year, millions of people in the poorest countries in the world suffer needlessly from the infectious tropical diseases borne by these pathogens, including African sleeping sickness (T. brucei), Chagas disease (T. cruzi), and leishmaniasis (Leishmania major). These “orphan diseases” cause not only death, but also promote a crippling cycle of poverty within the Americas, Asia, and Africa, which collectively harbor a disproportionate amount of the neglected disease burden 77. Existing drugs are too costly or difficult to deliver, induce debilitating or fatal side effects, and are showing increased signs of resistance 78.
The targeting of RNA editing ligase in these species attacks a unique aspect of trypanosomal biology, which is the extensive editing of their mitochondrial mRNAs 21. Through the insertion and deletion of uridylates (U’s), the editing process transforms premature mitochondrial RNA (pre-mRNA) to mature mRNA in a multi-protein complex known as the editosome 22, 79. The exact composition of the editosome complex has yet to be fully characterized, although 20S core complexes have a Mw of 1.6 MDa and appear to be comprised of 16–20 proteins 80. It has recently been demonstrated that at least three different 20S editosomes of heterogeneous composition and distinct specificity are involved in the editing process 81, 82, possibly reflecting compositional changes of this dynamic multicatalyst complex at different stages in the editing process 79.
The mRNA editing process begins in the trypanosomal mitochondrial DNA, which consists of a topologically linked network of thousands of minicircles and dozens of maxicircles. It is the transcripts of these maxicircles, which encode components of respiratory complexes and energy transduction systems that undergo extensive RNA editing. The editing process begins when guide RNAs (gRNAs) are transcribed from the minicircles in the mt genome and subsequently base-pair with pre-mRNA sequences through a conserved “anchor sequence” 83, 84. Endonucleolytic cleavage of the pre-mRNA strand occurs at a point of mismatch between the trans-acting gRNA and its cognate pre-mRNA, and the type of RNA mismatch determines which 20S editosome catalyzes the cleavage reaction. As specified by the gRNA sequence, U’s are then either added, by the terminal uridylyl transferase (TUTase) TbRET2, or deleted, by a U-specific 3′-exoribonuclease. The processed RNA fragments are then religated by one of two RNA ligases, RNA editing ligase 1 (TbREL1) or 2 (TbREL2). Interestingly, TbREL1 is essential for the process 23 whereas TbREL2 is not 85, implying that TbREL1 can complement for the loss of TbREL2, but not vice versa. This religation of the now completely base-paired double-stranded RNA strands occurs in a three-step process, which is mechanistically conserved among the NTR superfamily (Fig. 1).
3.1 Defining Structural Features
Many of the core features of the NTR superfamily structure are also conserved in TbREL1. TbREL1 is comprised of a catalytic N-terminal adenylation domain and a C-terminal domain that facilitates non-covalent interaction with another editosome protein, KREPA2 8. In this system, the OBD, which is usually associated with DNA ligases and capping enzymes in cis, appears to be provided by KREPA2 in trans and has been predicted to act as a conformational switch regulating various steps in the editing process 8. In 2004, Deng et al. published a 1.2 Å resolution crystal structure of the adenylation domain, TbREL1, from T. brucei with the bound ATP ligand. This structure revealed a deep ATP binding pocket with three buried water molecules that could potentially be displaced by novel ligands (Fig. 3D). The closest known relative to TbREL1 is the RNA ligase 2 from T4 phage, which has crystal structures available for the AMP-intermediate, and RNA bound states 55, 86. The established similarity between these two enzymes is important, as much of this information for the T4 phage system can be used to help interpret, understand, and direct strategic studies for the enzymatic activity of TbREL1. Steady-state and pre-steady-state kinetic analysis coupled with strategic mutagenesis of the T4 RNA ligase 2 has established functional roles for the highly conserved binding site residues and mapped many of the important interactions of the RNA ligase active site 86, 87.
3.2 Inhibitors
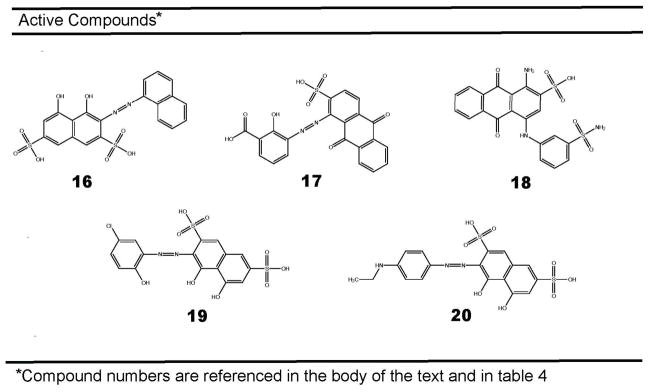
Recently, Amaro et al. used an integrated computational and experimental approach called the “relaxed complex scheme” to identify a set of micromolar inhibitors of TbREL 27. An initial virtual screen was first performed against the TbREL1 crystal structure. In order to account for protein-receptor flexibility, a molecular dynamics (MD) simulation of the target protein followed 88. Each of the top hits from the initial virtual screen was then docked into multiple representative protein configurations extracted from the MD trajectory, yielding a spectrum of binding energies for each compound. For each compound, predicted binding energies were then averaged over the ensemble, giving an ensemble-average docking score that may better predict the ligand binding affinity than a single docking score based on the crystal structure alone. Measuring formation of TbREL1-[32P]AMP by SDS-PAGE and autoradiography, the study was looking for inhibitors of the adenylylation reaction in step 1. The experimental assays were carried out at 10 μM compound concentration in the presence of 0.1% Triton X-100 as a first measure to select against any promiscuous, aggregate-based inhibitors 89. Testing in round 1 identified three compounds representing two different chemical scaffolds, the most potent inhibitor 16 based on a polysulfonated naphtyl dye-like scaffold and compounds 17 and 18, which are sulfonated diketoanthracene based scaffolds (Fig. 5 and Table 2). A second Tanimoto-based similarity search identified three additional compounds 18, 19, 20 which all belong to the same family of dye-like compounds as compound 16. The top three compounds were tested for selectivity against the T4 phage and human DNA Lig IIIβ enzymes and were shown to have 6- to 36-fold selectivity for the trypanosomal target (Table 2). The compounds were not tested against whole cell assays, however, it is almost certain the charged sulfonic acid moieties will cause issues permeating cell membranes, and additionally, the azo linkage and sulfonate ester groups are subjects of metabolic concern. An additional challenge for the TbREL1 target is that crystallization is extremely challenging and so far only the ATP-bound receptor has been resolved.
Figure 5.
Inhibitors of ATP-dependent RNA ligase 1.
Table 2.
| Compound no. | TbREL1 IC50, (μM) | HsLIG3β IC50, (μM) | ATP competitive? |
|---|---|---|---|
| 16 | 1.95 | 27.49 | Yes |
| 17 | ~ 10 * | N/A | Yes |
| 18 | ~ 100 * | N/A | Yes |
| 19 | 1.95 | 36.89 | Yes |
| 20 | 1.01 | 6.59 | Yes |
IC50’s were not determined; % activity at 10 μM inhibitor concentration showed 87.2% activity for compound 17 and 68.5% activity for compound 18
4. Expert Opinion Section
LigA is ideally suited for rational inhibitor design due in part to published high throughput assays 41, 61, the availability of high quality crystal structures, unique structural domains, and the extent of biochemical characterization. For example, the work of Srivastava et al. targeted the NAD+ binding pocket formed by the AMP binding site in the NTBD and the NMN binding groove, housed in domain, Ia. By exploiting domain Ia, which is unique to bacterial LigAs, their target-structure based approach resulted in the discovery of LigA specific bisglycosyl diamines 75 76, and the N-substituted tetracyclic indoles 53. The results of Srivastava et al. proved that LigA specific inhibitors could be constructed using a structure based design strategy, yet the conserved hydrophobic tunnel adjacent to the NAD+ binding pocket (Fig. 3A) went unutilized. The usefulness of this tunnel in inhibitor design has been substantiated by four crystal structures (3BA8, 3BA9, 3BAA and 3BAB), recently deposited in the PDB by Pinko and colleagues, preceding a forthcoming publication. These structures excite the possibility of optimizing inhibitors, which primarily make use of the NAD+-binding pocket, by strategic substitutions that direct alkyl or aryl groups into the hydrophobic tunnel. Even in the absence of a bound inhibitor crystal structure, in silico docking could conceivably provide a qualitatively correct picture of the bound state that could be used during the optimization process.
Although utilizing the hydrophobic tunnel may result in inhibitors with greater LigA specificity, it may also stimulate resistance mutations more rapidly than inhibitors that specifically target essential residues within the NAD+ binding pocket. The recently proposed “substrate envelope hypothesis”, which offers an explanation of HIV-protease inhibitor resistance, is consistent with this view 90, 91. In this hypothesis, the protease substrate occupies a “consensus volume”, which encompasses enzyme residues essential for catalysis. Inhibitor resistance mutations are more frequent in regions where the inhibitor falls outside of the consensus volume, contacting non-essential residues. Rapid generation time, and selective pressure are shared causes of antibiotic and antiviral resistance, making it likely that the principles entailed in the substrate envelope hypothesis are also applicable to antibiotic resistance. It follows that residues within the LigA hydrophobic tunnel may be more prone to resistance mutations when targeted by an inhibitor than residues essential for catalysis. Despite this, its size, and proximity to the NAD+-binding pocket make the hydrophobic tunnel the most promising structural element to target for optimized LigA inhibitor binding.
Targeting the RNA editing ligases in the pathogenic Trpanosoma species is another relevant and promising avenue for antiparasitic therapeutics. Economic factors are likely to play a role in the continued neglect of these targets by industry; yet, strides made in the academic arena are promising in that more attention is being paid to the orphan diseases thanks to shifts in the current funding climate as well as the increased participation of academic groups in drug discovery and design. A high throughput REL1 assay is under development 92 and will assist in diversifying the chemistry of known hits, which is an important next step before identification of any promising lead compounds. Although the REL1 enzymes do not have a human homologue, the high similarity to human ATP-dependent DNA ligases will require the simultaneous cross-optimization against these enzymes. In this regard, targeting REL1-RNA bound complexes may facilitate the development of inhibitors that take advantage of the RNA to achieve greater specificity over the related human enzymes. Targeting the hybrid complex would also allow the strategic development of high affinity compounds that will bind to and inhibit different complexes along the three-step ligation reaction pathway. Although high-resolution structures for REL1 in complex with RNA are not yet available, structural dynamics of homology models based on similarity to the available RNA-bound T4 phage RNA ligase crystal structure 55 have been pursued 93.
Abbreviations
- NTR
Nucleotidyltransferase
- PPi
pyrophosphate
- NMN
nicotinamide mononucleotide
- AMP
adenosine monophosphate
- ATP
adenosine triphosphate
- NTBD
nucleotide-binding domain
- OBD
oligonucelotide-binding domain
- gRNA
guide RNA
- TbREL1
RNA editing ligase 1 from Trypanosma brucei
- TUTase
terminal uridyl transferase
- HsLigI
human DNA ligase I
- DBD
DNA-binding domain
- HhH
helix-hairpin-helix
- LigA
NAD+-dependent DNA ligase
Footnotes
Declaration of interest
This work was funded in part by NIH GM077729 and MRAC CHE060073N to R.E.A.
References
- 1.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol. 2004 Dec;14(6):757–64. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Shuman S. DNA ligases: progress and prospects. J Biol Chem. 2009 Jun 26;284(26):17365–9. doi: 10.1074/jbc.R900017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem Rev. 2006 Feb;106(2):687–99. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 4.Nair PA, Nandakumar J, Smith P, Odell M, Lima CD, Shuman S. Structural basis for nick recognition by a minimal pluripotent DNA ligase. Nat Struct Mol Biol. 2007 Aug;14(8):770–8. doi: 10.1038/nsmb1266. [DOI] [PubMed] [Google Scholar]
- 5.Gajiwala KS, Pinko C. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure. 2004 Aug;12(8):1449–59. doi: 10.1016/j.str.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 6.HÂkansson K, Doherty AJ, Shuman S, Wigley DB. X-Ray Crystallography Reveals a Large Conformational Change during Guanyl Transfer by mRNA Capping Enzymes. Cell. 1997;89(4):545–53. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 7.Swift RV, McCammon JA. Substrate induced population shifts and stochastic gating in the PBCV-1 mRNA capping enzyme. J Am Chem Soc. 2009 Apr 15;131(14):5126–33. doi: 10.1021/ja808064g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnaufer A, Ernst NL, Palazzo SS, O’Rear J, Salavati R, Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol Cell. 2003 Aug;12(2):307–19. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993 Apr 22;362(6422):709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 10.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007 Apr 1;6(4):429–42. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA--general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007 Apr 1;6(4):505–16. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Fortini P, Pascucci B, Parlanti E, Sobol RW, Wilson SH, Dogliotti E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998 Mar 17;37(11):3575–80. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- 13.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009 Mar;66(6):981–93. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci. 2009 Mar;66(6):1039–56. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–51. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 16.Teo IA, Arlett CF, Harcourt SA, Priestley A, Broughton BC. Multiple hypersensitivity to mutagens in a cell strain (46BR) derived from a patient with immuno-deficiencies. Mutat Res. 1983 Feb;107(2):371–86. doi: 10.1016/0027-5107(83)90177-x. [DOI] [PubMed] [Google Scholar]
- 17.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992 May 1;69(3):495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 18.Mackenney VJ, Barnes DE, Lindahl T. Specific function of DNA ligase I in simian virus 40 DNA replication by human cell-free extracts is mediated by the amino-terminal non-catalytic domain. J Biol Chem. 1997 Apr 25;272(17):11550–6. doi: 10.1074/jbc.272.17.11550. [DOI] [PubMed] [Google Scholar]
- 19.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. Embo J. 1987 Aug;6(8):2499–503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998 May 22;273(21):12685–8. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 21.Stuart K, Allen TE, Heidmann S, Seiwert SD. RNA editing in kinetoplastid protozoa. Microbiol Mol Biol Rev. 1997 Mar;61(1):105–20. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson L, Sbicego S, Aphasizhev R. Urididine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–76. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Jr, Wirtz E, Salavati R, et al. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001 Mar 16;291(5511):2159–62. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 24.Dwivedi N, Dube D, Pandey J, Singh B, Kukshal V, Ramachandran R, et al. NAD(+)-dependent DNA ligase: a novel target waiting for the right inhibitor. Med Res Rev. 2008 Jul;28(4):545–68. doi: 10.1002/med.20114. [DOI] [PubMed] [Google Scholar]
- 25.Korycka-Machala M, Rychta E, Brzostek A, Sayer HR, Rumijowska-Galewicz A, Bowater RP, et al. Evaluation of NAD(+) -dependent DNA ligase of mycobacteria as a potential target for antibiotics. Antimicrob Agents Chemother. 2007 Aug;51(8):2888–97. doi: 10.1128/AAC.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong S, Chen X, Zhu X, Dziegielewska B, Bachman KE, Ellenberger T, et al. Identification and validation of human DNA ligase inhibitors using computer-aided drug design. J Med Chem. 2008 Aug 14;51(15):4553–62. doi: 10.1021/jm8001668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2008 Nov 11;105(45):17278–83. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gefter ML, Becker A, Hurwitz J. The enzymatic repair of DNA. I. Formation of circular lambda-DNA. Proc Natl Acad Sci U S A. 1967 Jul;58(1):240–7. doi: 10.1073/pnas.58.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gellert M. Formation of covalent circles of lambda DNA by E. coli extracts. Proc Natl Acad Sci U S A. 1967 Jan;57(1):148–55. doi: 10.1073/pnas.57.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivera BM, Lehman IR. Linkage of polynucleotides through phosphodiester bonds by an enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1967 May;57(5):1426–33. doi: 10.1073/pnas.57.5.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss B, Richardson CC. Enzymatic breakage and joining of deoxyribonucleic acid, I. Repair of single-strand breaks in DNA by an enzyme system from Escherichia coli infected with T4 bacteriophage. Proc Natl Acad Sci U S A. 1967 Apr;57(4):1021–8. doi: 10.1073/pnas.57.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cozzarelli NR, Melechen NE, Jovin TM, Kornberg A. Polynucleotide cellulose as a substrate for a polynucleotide ligase induced by phage T4. Biochem Biophys Res Commun. 1967 Aug 23;28(4):578–86. doi: 10.1016/0006-291x(67)90353-1. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl T, Edelman GM. Polynucleotide ligase from myeloid and lymphoid tissues. Proc Natl Acad Sci U S A. 1968 Oct;61(2):680–7. doi: 10.1073/pnas.61.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson CC. Enzymes in DNA metabolism. Annu Rev Biochem. 1969;38:795–840. doi: 10.1146/annurev.bi.38.070169.004051. [DOI] [PubMed] [Google Scholar]
- 35.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–70. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 36.Martin IV, MacNeill SA. ATP-dependent DNA ligases. Genome Biol. 2002;3(4):REVIEWS3005. doi: 10.1186/gb-2002-3-4-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horiuchi T, Sato T, Nagata T. DNA degradation in an amber mutant of Escherichia coli K12 affecting DNA ligase and viability. J Mol Biol. 1975 Jun 25;95(2):271–87. doi: 10.1016/0022-2836(75)90395-2. [DOI] [PubMed] [Google Scholar]
- 38.Park UE, Olivera BM, Hughes KT, Roth JR, Hillyard DR. DNA ligase and the pyridine nucleotide cycle in Salmonella typhimurium. J Bacteriol. 1989 Apr;171(4):2173–80. doi: 10.1128/jb.171.4.2173-2180.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petit MA, Ehrlich SD. The NAD-dependent ligase encoded by yerG is an essential gene of Bacillus subtilis. Nucleic Acids Res. 2000 Dec 1;28(23):4642–8. doi: 10.1093/nar/28.23.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaczmarek FS, Zaniewski RP, Gootz TD, Danley DE, Mansour MN, Griffor M, et al. Cloning and functional characterization of an NAD(+)-dependent DNA ligase from Staphylococcus aureus. J Bacteriol. 2001 May;183(10):3016–24. doi: 10.1128/JB.183.10.3016-3024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen XC, Hentz NG, Hubbard F, Meier TI, Sittampalam S, Zhao G. Development of a fluorescence resonance energy transfer assay for measuring the activity of Streptococcus pneumoniae DNA ligase, an enzyme essential for DNA replication, repair, and recombination. Anal Biochem. 2002 Oct 15;309(2):232–40. doi: 10.1016/s0003-2697(02)00302-0. [DOI] [PubMed] [Google Scholar]
- 42.Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):966–71. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003 Apr;48(1):77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 44.Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J Biol Chem. 2004 May 14;279(20):20594–606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- 45.Pascal JM. DNA and RNA ligases: structural variations and shared mechanisms. Curr Opin Struct Biol. 2008 Feb;18(1):96–105. doi: 10.1016/j.sbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J Mol Biol. 2004 Oct 22;343(3):601–13. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 47.Subramanya HS, Doherty AJ, Ashford SR, Wigley DB. Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell. 1996 May 17;85(4):607–15. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- 48.Odell M, Sriskanda V, Shuman S, Nikolov DB. Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining. Mol Cell. 2000 Nov;6(5):1183–93. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 49.El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, LeGrice SF, et al. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J Biol Chem. 2006 Jan 20;281(3):1573–9. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 50.Lee JY, Chang C, Song HK, Moon J, Yang JK, Kim HK, et al. Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications. Embo J. 2000 Mar 1;19(5):1119–29. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sriskanda V, Shuman S. Mutational analysis of Chlorella virus DNA ligase: catalytic roles of domain I and motif VI. Nucleic Acids Res. 1998 Oct 15;26(20):4618–25. doi: 10.1093/nar/26.20.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang LK, Nair PA, Shuman S. Structure-guided mutational analysis of the OB, HhH, and BRCT domains of Escherichia coli DNA ligase. J Biol Chem. 2008 Aug 22;283(34):23343–52. doi: 10.1074/jbc.M802945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava SK, Dube D, Kukshal V, Jha AK, Hajela K, Ramachandran R. NAD+-dependent DNA ligase (Rv3014c) from Mycobacterium tuberculosis: novel structure-function relationship and identification of a specific inhibitor. Proteins. 2007 Oct 1;69(1):97–111. doi: 10.1002/prot.21457. [DOI] [PubMed] [Google Scholar]
- 54.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004 Nov 25;432(7016):473–8. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 55.Nandakumar J, Shuman S, Lima CD. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006 Oct 6;127(1):71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 56.Nandakumar J, Nair PA, Shuman S. Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate. Mol Cell. 2007 Apr 27;26(2):257–71. doi: 10.1016/j.molcel.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 57.Jeon HJ, Shin HJ, Choi JJ, Hoe HS, Kim HK, Suh SW, et al. Mutational analyses of the thermostable NAD+-dependent DNA ligase from Thermus filiformis. FEMS Microbiol Lett. 2004 Aug 1;237(1):111–8. doi: 10.1016/j.femsle.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 58.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem. 2008;77:313–38. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng H, Parker JM, Lu J, Cao W. Effects of deletion and site-directed mutations on ligation steps of NAD+-dependent DNA ligase: a biochemical analysis of BRCA1 C-terminal domain. Biochemistry. 2004 Oct 5;43(39):12648–59. doi: 10.1021/bi049451c. [DOI] [PubMed] [Google Scholar]
- 60.Lim JH, Choi J, Kim W, Ahn BY, Han YS. Mutational analyses of Aquifex pyrophilus DNA ligase define essential domains for self-adenylation and DNA binding activity. Arch Biochem Biophys. 2001 Apr 15;388(2):253–60. doi: 10.1006/abbi.2001.2291. [DOI] [PubMed] [Google Scholar]
- 61.Miesel L, Kravec C, Xin AT, McMonagle P, Ma S, Pichardo J, et al. A high-throughput assay for the adenylation reaction of bacterial DNA ligase. Anal Biochem. 2007 Jul 1;366(1):9–17. doi: 10.1016/j.ab.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 62.Brotz-Oesterhelt H, Knezevic I, Bartel S, Lampe T, Warnecke-Eberz U, Ziegelbauer K, et al. Specific and potent inhibition of NAD+-dependent DNA ligase by pyridochromanones. J Biol Chem. 2003 Oct 10;278(41):39435–42. doi: 10.1074/jbc.M306479200. [DOI] [PubMed] [Google Scholar]
- 63.Tanifum EA, Kots AY, Choi BK, Murad F, Gilbertson SR. Novel pyridopyrimidine derivatives as inhibitors of stable toxin a (STa) induced cGMP synthesis. Bioorg Med Chem Lett. 2009 Jun 1;19(11):3067–71. doi: 10.1016/j.bmcl.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 64.Kots AY, Choi BK, Estrella-Jimenez ME, Warren CA, Gilbertson SR, Guerrant RL, et al. Pyridopyrimidine derivatives as inhibitors of cyclic nucleotide synthesis: Application for treatment of diarrhea. Proc Natl Acad Sci U S A. 2008 Jun 17;105(24):8440–5. doi: 10.1073/pnas.0803096105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Debenham JS, Madsen-Duggan CB, Wang J, Tong X, Lao J, Fong TM, et al. Pyridopyrimidine based cannabinoid-1 receptor inverse agonists: Synthesis and biological evaluation. Bioorg Med Chem Lett. 2009 May 1;19(9):2591–4. doi: 10.1016/j.bmcl.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 66.Martin-Martinez M, Marty A, Jourdan M, Escrieut C, Archer E, Gonzalez-Muniz R, et al. Combination of molecular modeling, site-directed mutagenesis, and SAR studies to delineate the binding site of pyridopyrimidine antagonists on the human CCK1 receptor. J Med Chem. 2005 Jul 28;48(15):4842–50. doi: 10.1021/jm0501127. [DOI] [PubMed] [Google Scholar]
- 67.Nakayama K, Kawato H, Watanabe J, Ohtsuka M, Yoshida K, Yokomizo Y, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 3: Optimization of potency in the pyridopyrimidine series through the application of a pharmacophore model. Bioorg Med Chem Lett. 2004 Jan 19;14(2):475–9. doi: 10.1016/j.bmcl.2003.10.060. [DOI] [PubMed] [Google Scholar]
- 68.Meier TI, Yan D, Peery RB, McAllister KA, Zook C, Peng SB, et al. Identification and characterization of an inhibitor specific to bacterial NAD+-dependent DNA ligases. FEBS J. 2008 Nov;275(21):5258–71. doi: 10.1111/j.1742-4658.2008.06652.x. [DOI] [PubMed] [Google Scholar]
- 69.Foley M, Tilley L. Quinoline antimalarials: mechanisms of action and resistance and prospects for new agents. Pharmacol Ther. 1998 Jul;79(1):55–87. doi: 10.1016/s0163-7258(98)00012-6. [DOI] [PubMed] [Google Scholar]
- 70.Ridley RG, Matile H, Jaquet C, Dorn A, Hofheinz W, Leupin W, et al. Antimalarial activity of the bisquinoline trans-N1,N2-bis (7-chloroquinolin-4-yl)cyclohexane-1,2-diamine: comparison of two stereoisomers and detailed evaluation of the S,S enantiomer, Ro 47–7737. Antimicrob Agents Chemother. 1997 Mar;41(3):677–86. doi: 10.1128/aac.41.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dascombe MJ, Drew MG, Morris H, Wilairat P, Auparakkitanon S, Moule WA, et al. Mapping antimalarial pharmacophores as a useful tool for the rapid discovery of drugs effective in vivo: design, construction, characterization, and pharmacology of metaquine. J Med Chem. 2005 Aug 25;48(17):5423–36. doi: 10.1021/jm0408013. [DOI] [PubMed] [Google Scholar]
- 72.Raynes K. Bisquinoline antimalarials: their role in malaria chemotherapy. Int J Parasitol. 1999 Mar;29(3):367–79. doi: 10.1016/s0020-7519(98)00217-3. [DOI] [PubMed] [Google Scholar]
- 73.Ciarrocchi G, MacPhee DG, Deady LW, Tilley L. Specific inhibition of the eubacterial DNA ligase by arylamino compounds. Antimicrob Agents Chemother. 1999 Nov;43(11):2766–72. doi: 10.1128/aac.43.11.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith KT, Dawes IW. The preferential inhibition of Bacillus subtilis spore outgrowth by chloroquine. Arch Microbiol. 1989;152(3):251–7. doi: 10.1007/BF00409659. [DOI] [PubMed] [Google Scholar]
- 75.Srivastava SK, Tripathi RP, Ramachandran R. NAD+-dependent DNA Ligase (Rv3014c) from Mycobacterium tuberculosis. Crystal structure of the adenylation domain and identification of novel inhibitors. J Biol Chem. 2005 Aug 26;280(34):30273–81. doi: 10.1074/jbc.M503780200. [DOI] [PubMed] [Google Scholar]
- 76.Srivastava SK, Dube D, Tewari N, Dwivedi N, Tripathi RP, Ramachandran R. Mycobacterium tuberculosis NAD+-dependent DNA ligase is selectively inhibited by glycosylamines compared with human DNA ligase I. Nucleic Acids Res. 2005;33(22):7090–101. doi: 10.1093/nar/gki1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.WHO Report on Global Surveillance of Epidemic-prone Infectious Diseases. World Health Organization; 2003. [Google Scholar]
- 78.Croft SL, Barrett MP, Urbina JA. Chemotherapy of trypanosomiases and leishmaniasis. Trends Parasitol. 2005 Nov;21(11):508–12. doi: 10.1016/j.pt.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 79.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005 Feb;30(2):97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 80.Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003 Nov 15;31(22):6392–408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. Rna. 2006 Jun;12(6):1038–49. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008 Jan;28(1):122–30. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blum B, Simpson L. Formation of guide RNA/messenger RNA chimeric molecules in vitro, the initial step of RNA editing, is dependent on an anchor sequence. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11944–8. doi: 10.1073/pnas.89.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996 Mar 22;84(6):831–41. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 85.Drozdz M, Palazzo SS, Salavati R, O’Rear J, Clayton C, Stuart K. TbMP81 is required for RNA editing in Trypanosoma brucei. Embo J. 2002 Apr 2;21(7):1791–9. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho CK, Wang LK, Lima CD, Shuman S. Structure and mechanism of RNA ligase. Structure. 2004 Feb;12(2):327–39. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 87.Yin S, Ho CK, Shuman S. Structure-function analysis of T4 RNA ligase 2. J Biol Chem. 2003 May 16;278(20):17601–8. doi: 10.1074/jbc.M300817200. [DOI] [PubMed] [Google Scholar]
- 88.Amaro RE, Swift RV, McCammon JA. Functional and structural insights revealed by molecular dynamics simulations of an essential RNA editing ligase in Trypanosoma brucei. PLoS Neglected Tropical Diseases. 2007 doi: 10.1371/journal.pntd.0000068. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ryan AJ, Gray NM, Lowe PN, Chung CW. Effect of detergent on “promiscuous” inhibitors. J Med Chem. 2003 Jul 31;46(16):3448–51. doi: 10.1021/jm0340896. [DOI] [PubMed] [Google Scholar]
- 90.Chellappan S, Kiran Kumar Reddy GS, Ali A, Nalam MN, Anjum SG, Cao H, et al. Design of mutation-resistant HIV protease inhibitors with the substrate envelope hypothesis. Chem Biol Drug Des. 2007 May;69(5):298–313. doi: 10.1111/j.1747-0285.2007.00514.x. [DOI] [PubMed] [Google Scholar]
- 91.Chellappan S, Kairys V, Fernandes MX, Schiffer C, Gilson MK. Evaluation of the substrate envelope hypothesis for inhibitors of HIV-1 protease. Proteins. 2007 Aug 1;68(2):561–7. doi: 10.1002/prot.21431. [DOI] [PubMed] [Google Scholar]
- 92.Schnaufer A. Personal Communication. 2009.
- 93.Swift RV, Durrant J, Amaro RE, McCammon JA. Toward understanding the conformational dynamics of RNA ligation. Biochemistry. 2009 Feb 3;48(4):709–19. doi: 10.1021/bi8018114. [DOI] [PMC free article] [PubMed] [Google Scholar]