SYNOPSIS
Severe combined immunodeficiency (SCID) is the result of genetic defects that impair normal T-cell development. SCID babies typically appear normal at birth, but acquire multiple life-threatening infections within a few months. Early diagnosis and treatment with a bone-marrow transplant markedly improves long-term outcomes.
On January 1, 2008, the newborn screening (NBS) program in Wisconsin became the first in the world to routinely test all newborns for SCID. A real-time quantitative polymerase chain reaction assay measures T-cell receptor excision circles (TRECs), which are formed during the maturation of normal T-cells. A lack or very low number of TRECs is consistent with T-cell lymphopenia. The development and validation of the TREC assay and the results of the first year of screening have been published. This article describes the process used to add SCID to the NBS panel, the establishment of follow-up capacity, and the integration of SCID screening into routine NBS workflows. The development of this expanded NBS program is described so that other states might benefit from the processes used in Wisconsin.
The expansion of a state newborn screening (NBS) panel presents many scientific, technical, ethical, and policy issues that must be addressed prior to the addition of a new condition to the test panel. The recent experience with the addition of severe combined immunodeficiency (SCID) to Wisconsin's existing NBS panel of 46 other conditions provides the opportunity to share the process with others who may be considering adding SCID screening to their NBS programs.1 In this article, we describe the process used in Wisconsin to initiate a pilot project to routinely screen all newborns for SCID.
SCID represents a group of conditions characterized by blocks in T-cell development, which lead to functional deficiencies in both T-cells and B-cells.2–4 As a result, newborns with these immunological conditions lack the ability to produce an effective response to infectious agents. Affected newborns are extremely prone to all forms of infection—viral, bacterial, and fungal. Undiagnosed, these infants typically require treatment for a series of extremely critical conditions, suffer repeated and prolonged hospitalizations, and usually die during the first year of life. Buckley et al. demonstrated that very early diagnosis and treatment by haematopoietic stem-cell transplantation can establish normal immune function.2 If treated before 3.5 months of age, SCID newborns have more than a 95% chance at long-term (>20 years) survival. If diagnosed after 3.5 months of age, even with intensive interventional care and transplantation, the survival rate drops below 70%. These factors suggest that a reliable laboratory procedure compatible with existing routine NBS programs potentially could be immensely beneficial to SCID-afflicted newborns, who typically appear completely normal at birth. Routine screening of newborns based on conventional dried filter-paper blood specimens is now possible with a deoxyribonucleic acid (DNA)-based assay that meets all of the selection criteria commonly used for adding a new test to the state's NBS panel.5,6
Because it is impossible to measure an infant's immune function or the number of T-cells and B-cells directly from dried-blood spots on filter papers (i.e., Guthrie cards), a surrogate marker must be used. During normal T-cell maturation in the thymus, a short segment of DNA for the T-cell receptor gene is excised and subsequently links ends to form a T-cell receptor excision circle (TREC). Based on the work of Douek et al.7 and Chan and Puck,8 we optimized the method and, using de-identified NBS specimens, tested 5,766 consecutive routine NBS samples during October 2007.9 The method was compatible with the laboratory operations in the existing Wisconsin NBS Program, and it yielded acceptably low numbers of false-positive results. Following the development and initial evaluation of our NBS procedure, the Wisconsin NBS Advisory Committee recommended adding SCID to the test panel on a pilot basis. The Wisconsin Secretary of Health and Family Services approved this addition. Routine screening of all infants born in Wisconsin began on January 1, 2008.
In this article, we describe the steps taken by the Wisconsin NBS Program that led to the first routine application of a DNA-based molecular assay being used as a primary NBS technology by a statewide NBS program. Our discussion includes the logistical, technical, and operational issues associated with implementing routine SCID testing, as well as the quality-assurance, follow-up, and cost considerations for NBS programs. The analytical method used in Wisconsin has been described by Baker et al.;9 and Routes et al.10 have reported the clinical results of the screening and follow-up during the first year.
METHODS
In May 2007, clinical immunologists from Children's Hospital of Wisconsin presented the concept of screening newborns for SCID to the Wisconsin NBS Advisory Committee. The committee reviewed existing criteria for adding a test to the NBS panel and was convinced that SCID potentially met the criteria used to expand NBS in the past. A new polymerase chain reaction (PCR)-based laboratory method for screening would need to be validated, and protocols for confirmation of positive screening results would need to be available. The University of Wisconsin-Madison Institutional Review Board approved a protocol for the development and evaluation of the screening method using de-identified NBS specimens. The method was subsequently evaluated and optimized for use on dried-blood spots.9 Blood samples that had zero or low TRECs (i.e., Guthrie cards spotted with blood from infants with SCID or depleted of näive T-cells) were used as internal blinded controls to further evaluate the proposed method.
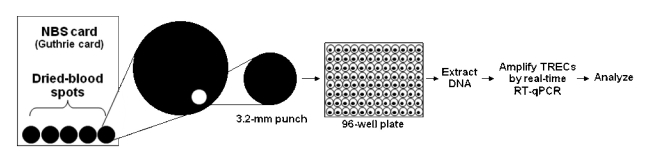
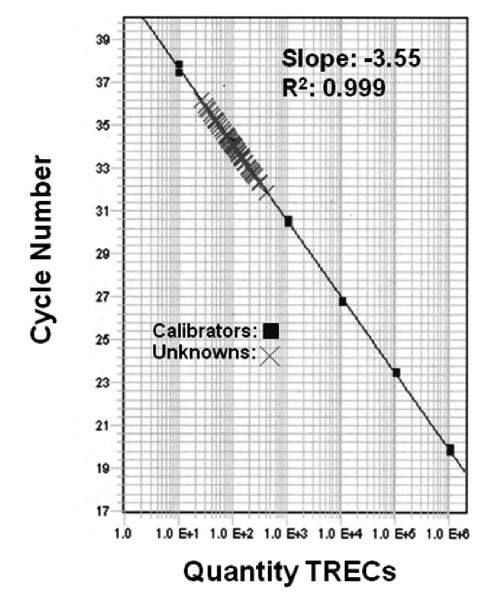
The overall analysis of a 3.2-millimeter (mm) circular punch from a dried-blood spot is shown schematically in Figure 1. Beginning with the newborn blood-collection card that contains five dried-blood spots, a 3.2-mm circular punch containing 3 microliters (μL) of whole blood is processed in a 96-well microtiter plate. The specimen is then analyzed by real-time quantitative PCR (RT-qPCR) to quantify the number of TRECs/μL of blood. The resulting calibration curve (plotted automatically) is shown in Figure 2, with calibrators (run in duplicate) indicated by the solid squares, and patient specimens shown by the crosshatches. Test results on patient specimens were reported as the number of TRECs/μL of blood.
Figure 1.
Graphic representation of SCID-screening method used by the Wisconsin NBS Programa
aIn the method, a 3.2-mm dried-blood spot containing 3 microliters of blood is punched from a filter-paper card and processed in a 96-well microtiter plate, and DNA is extracted, amplified, and analyzed by RT-qPCR.
SCID = severe combined immunodeficiency
NBS = newborn screening
mm = millimeter
DNA = deoxyribonucleic acid
TREC = T-cell receptor excision circle
RT-qPCR = real-time quantitative polymerase chain reaction
Figure 2.
Calibrator curve of serially diluted plasmids containing a known number of TRECs (solid squares) and 82 unknown samplesa
aBaker MW, Grossman WJ, Laessig RH, Hoffman GL, Brokopp CD, Kurtycz DF, et al. Development of a routine newborn screening protocol for severe combined immunodeficiency. J Allergy Clin Immunol 2009;124:522-7. Reproduced with permission from J Allergy Clin Immunol.
TREC = T-cell receptor excision circle
R2 = correlations coefficient
E = exponential notation
Because the purpose of this NBS assay is to identify newborns with very low or zero TRECs, we chose two quality-control monitors to assess the performance of the test and to satisfy Clinical Laboratory Improvement Amendments performance requirements. The first was to monitor the slope of the calibration plot. A reproducible slope is consistent with good control of the analytical process. During 75 consecutive runs, the slope was −3.55 ± 0.070 (coefficient of variation = 2%). The second quality-control parameter uses one of two forms of “true zero control” (i.e., an NBS positive, as represented by zero or low numbers of TRECs): whole blood spotted on filter paper from a known SCID infant (severe T-cell lymphopenia confirmed by flow cytometry) or an adult blood sample depleted of näive T-cells (e.g., CD45RA+CD3+T-cells). These zero controls not only monitor the method's performance in the critical area, they serve to alert the analyst of potential cross-contamination.
Our method relied on manual pipetting by chemists. One chemist can process one plate containing 82 specimens in three hours, and two plates in four hours using a single RT-qPCR system. Two analysts and two systems can comfortably process our current workload of 1,500 specimens per week, which includes punching, all extraction/analysis steps, reporting, method, quality control, and maintenance. Current studies to automate the manual pipetting steps will result in greater throughput within the NBS laboratory, which will be necessary for other laboratories with higher birth rates than Wisconsin.
Funding for the startup of SCID screening was initially provided by the Jeffrey Modell Foundation, Children's Hospital of Wisconsin, and the Wisconsin State Laboratory of Hygiene. The startup funding was used to hire additional staff, optimize the test methodology, purchase high-throughput PCR equipment and reagents, and cover the cost of confirmatory testing by flow cytometry. We spent approximately $630,000 from the development phase through the first nine months of the full-scale pilot test of routine SCID screening. Current screening for SCID is supported by a three-year grant from the Centers for Disease Control and Prevention (CDC). The estimated cost to screen 70,000 infants is $420,000.
The Administrator of the Wisconsin Division of Public Health and the Secretary of the Department of Health and Family Services were convinced that SCID screening could and should be brought on line in Wisconsin. An emergency public health rule was approved by the Secretary, which allowed the pilot project for SCID screening to begin on January 1, 2008. The Wisconsin Administrative Code was later amended to add SCID screening to the 46 other conditions that were part of the existing test panel.11
Prior to screening for SCID, the collaborators contacted many of the state and local medical associations, particularly those representing pediatricians, neonatologists, and family practitioners, and provided them with information on SCID and the proposed addition of SCID to the NBS test panel. A letter to physicians from the Wisconsin NBS Program and an announcement in the Wisconsin Medical Journal described the addition of SCID to the NBS panel and the proposed follow-up for infants who are screened positive,12 and project collaborators presented lectures and grand rounds on the topic. Once the program began, clinical consultants designated by the NBS program were available to respond to clinician questions.
RESULTS
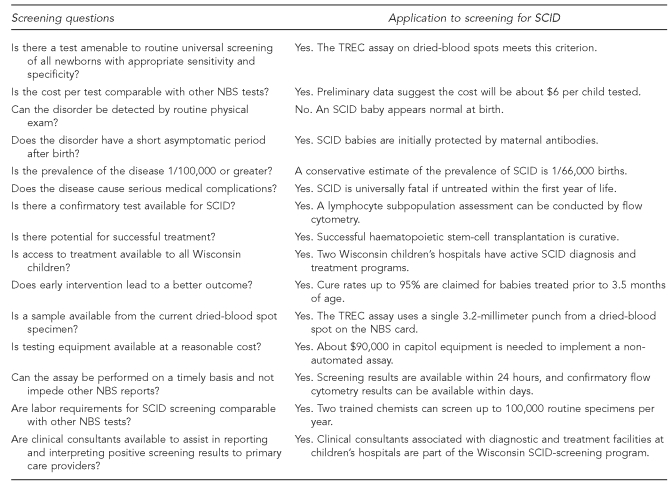
Figure 3 summarizes the programmatic criteria the Wisconsin NBS Program considered prior to adding SCID to the test panel. The responses to these 15 questions made it clear that a screening program for SCID could be successful if effective screening and follow-up protocols were established.
Figure 3.
Programmatic criteria considered by the Wisconsin NBS Advisory Committee in evaluating the addition of SCID screening to its NBS test panel
NBS = newborn screening
SCID = severe combined immunodeficiency
TREC = T-cell receptor excision circle
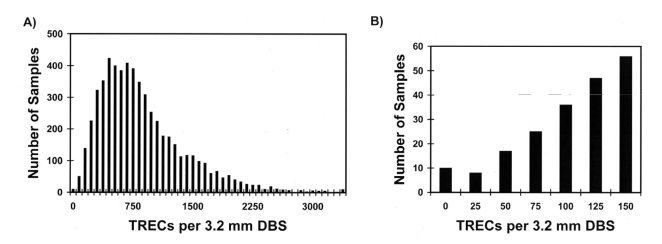
Figure 4A shows the distribution of the number of TRECs extracted from 5,766 de-identified dried-blood spot samples submitted to the NBS program and analyzed during the proof-of-concept pilot study in 2007. The number of TRECs is shown as TRECs/3.2-mm circular dried-blood spot. A 3.2-mm circular punch from a dried-blood spot contains 3 μL of whole blood. The number of TRECs/3 μL ranged from 0 to 3,900. In this sample, the mean number of TRECs/3 μL was 827, and the median number was 708. The shape of the distribution function, skewed (i.e., exhibiting a “tail” of values) to the right, is typical of many clinical analytes; however, the region of interest when screening for SCID and immunodeficiencies lies with the zero and low values at the left end of the distribution. A newborn exhibiting a significant number of TRECs is, by definition, producing T-cells and, therefore, is negative for SCID and many other T-cell immunodeficiencies. Figure 4B shows the distribution of TRECs from newborns who had ≤150 TRECs on initial testing. Only about 1.06% of newborns have <75 TRECs/3 μL or the equivalent of 25 TRECs/μL. Using data from this pilot study, we set the internal (intra-laboratory) cutoff at <25 TRECs/μL. Specimens with ≥25 TRECs/μL are considered to be normal (i.e., negative for SCID and other immunodeficiencies). The cutoff of <25 TRECs/μL clears 98.94% of all initial specimens for SCID on the initial tests.
Figure 4.
Results of initial RT-qPCR TREC assay conducted by the Wisconsin NBS Program on 5,766 NBS cards during the 2007 SCID-screening pilot studya,b
aBaker MW, Grossman WJ, Laessig RH, Hoffman GL, Brokopp CD, Kurtycz DF, et al. Development of a routine newborn screening protocol for severe combined immunodeficiency. J Allergy Clin Immunol 2009;124:522-7. Reproduced with permission.
b(A) TREC distribution, where the mean is 827 TRECs/3.2-mm DBS and the median is 708 TRECs/3.2-mm DBS; and (B) number of samples with ≤150 TRECs/3.2-mm DBS
RT-qPCR = real-time quantitative polymerase chain reaction
TREC = T-cell receptor excision circle
NBS = newborn screening
SCID = severe combined immunodeficiency
mm = millimeter
DBS = dried-blood spot
From these pilot-study results, a screening algorithm was developed to move forward for prospective analyses. Specimens not reported as normal (i.e., ≥25 TRECs/μL) fall into one of two categories: inconclusive or abnormal. If the blood spots are of poor quality for NBS, have low TRECs but are from newborns of very low gestational age, or, in the judgment of the laboratory personnel, cannot be declared negative, results are deemed to be inconclusive. The primary care provider is requested to provide a second filter-paper specimen for retesting of all 47 NBS parameters, including SCID. This category represents approximately 0.17% of all births. The abnormal category represents specimens that on the initial test exhibited <25 TRECs/μL. For these, the original filter paper is re-punched and retested in duplicate. If confirmed to have zero or low numbers of TRECs, the clinical consultant to the NBS program is notified of the results. The consultant (a physician who diagnoses and treats immunodeficiencies in newborns, including those with SCID) contacts the newborn's primary care physician, and they jointly decide to test either a repeat filter-paper specimen or an immediate, micro whole-blood -specimen for immune cell subset levels by flow cytometry. If T-cell markers are absent, an immediate referral to an immunodeficiency clinic and full workup for SCID is recommended. If the repeat filter-paper specimen is again abnormal, a liquid whole-blood specimen is requested. Specimens that are normal on repeat testing (either filter paper or flow) are considered screening false positives.
The intra-laboratory reflexive testing of the initial filter-paper specimen in duplicate enables us to clear 99.98% of full-term newborns from whom we have obtained satisfactory specimens. Similarly, for premature births, the clearance rate is 99.97%. These rates are as good, or better, than the other tests in the current NBS panel; however, the number of inconclusive specimens is much higher in premature newborns, reflecting, in part, on low-birthweight babies in intensive care units, where a final discharge specimen will be forthcoming.
Results from the 2008 prospective study of full-term newborns and premature infants are shown in the Table. Twelve full-term newborns had abnormal (positive screening) results. Six whole-blood samples and four repeat filter-paper specimens were collected from these full-term newborns and retested. Two of the full-term newborns who initially had abnormal results were not retested (one died, one refused). Of the 10 full-term newborns who were retested, two had normal flow results (false positives), four exhibited non-SCID-related T-cell lymphopenia, and four were found to be normal. None were diagnosed with true SCID. Only 23 of 6,603 premature newborns had abnormal screening results reported during 2008. Five of these infants died before a repeat specimen could be obtained. Of the remaining 18, 12 tested normal on repeat screening, three had normal flow cytometry results (false positives), and three had an abnormal result at the equivalent age of 37 weeks gestation. The follow-up flow cytometry on these three newborns exhibited non-SCID T-cell lymphopenia. Although none of the premature newborns was diagnosed with SCID, eight infants were identified as having clinically significant T-cell lymphopenia, one of whom underwent successful bone-marrow transplantation. These results illustrate that the TREC assay used as part of routine NBS is also a test for T-cell lymphopenia, not just SCID.
Table.
Wisconsin Newborn Screening Program severe combined immunodeficiency screening results, 2008 (n=71,000)
Importantly, screening for SCID did not delay reporting of the other 46 NBS tests. Results are available in less than two days from receipt of the initial specimen in the laboratory and are reported as the number of TRECs/μL, along with the interpretation (normal, abnormal, or inconclusive). The addition of SCID screening has been accomplished without significant impact on the existing NBS program.
DISCUSSION
The Wisconsin NBS Program initiated the first full-scale, routine, population-based screening for SCID in the world and demonstrated one of the first instances of using an RT-qPCR (i.e., DNA or molecular) testing methodology as the primary test in an NBS program. The expansion of the NBS program was the result of collaboration among the NBS laboratory, the NBS follow-up program, the clinical consultants, two children's hospitals, and a private foundation. As new technologies evolve, NBS programs will continue to be challenged by the need to expand the number of tests in the standard test panel. The decision to add one or more new tests is typically based upon a set of criteria that relate to the characteristics of the disease or condition, the availability of reliable screening and confirmatory test procedures, the estimated cost to detect an affected infant, the availability of an effective treatment or clinical intervention, and the availability of clinical consultants.
On January 21, 2010, the Health Resources and Services Administration Advisory Committee on Heritable Disorders in Newborns and Children voted unanimously to recommend adding SCID to the core panel for universal screening of all newborns in the United States.13 The Advisory Committee's policy recommendation will now be presented to the Secretary of Health and Human Services, who has 180 days to consider and respond to the committee's proposal. Our data during 2008 indicate that the TREC assay identifies newborns with clinically important causes of T-cell lymphopenia apart from SCID, a factor that should also be considered when determining whether the TREC assay should be added to the NBS panel.
The addition of an NBS test for SCID was potentially an expensive endeavor. The Wisconsin NBS Program was fortunate to obtain $250,000 from the Jeffrey Modell Foundation to hire key staff and cover some of the costs associated with equipment, supplies, and training. Children's Hospital of Wisconsin contributed another $250,000 to cover portions of the startup costs to bringing the TREC screening assay on line and provide confirmatory flow cytometry testing. The Wisconsin State Laboratory of Hygiene (University of Wisconsin—Madison) also contributed nearly $300,000 to the startup costs for staff, equipment, and supplies. Beginning in October 2008, CDC awarded the Wisconsin State Laboratory of Hygiene $1.5 million during a three-year period to continue the initial pilot screening for SCID and to assist other state NBS programs with the addition of SCID screening by sharing methods and expertise.
Our SCID testing protocol, beginning with a single 3.2-mm punch from the filter-paper card and processing in 96 well plates, is completely compatible with our existing NBS program and 46-test panel. For programs with up to 100,000 specimens per year, two full-time chemists and two RT-qPCR systems, in part for redundancy and in part to smooth workflow, are optimal. We have reduced the current manual pipetting steps, which has resulted in increased staff efficiency. Incorporation of a molecular test (i.e., RT-qPCR-based test) as the primary test procedure for routine use has not proven to be a problem with either staff training or integration into NBS production. Cross-training with other NBS program chemists was accomplished without difficulty. Based on one year's experience, our per-sample cost is slightly less than $6 and is similar to other single tests in the panel. We estimate that with three to four weeks of hands-on training in our laboratory, another state NBS program could become fully operational within six months.
We selected a cutoff of <25 TRECs/μL to isolate the lowest 1.06% of all specimens for retesting. In the case of the Wisconsin NBS Program, about 700 specimens per year are being retested. Because a true SCID newborn would have a TREC value near zero, the high cutoff is designed to ensure that an SCID-affected newborn, as well as some newborns with other immunodeficiencies, will be in the subpopulation selected for further study. Early in the Wisconsin pilot study, the clinical consultants determined that a finding of low numbers of TRECs is a nonspecific result that should be brought to the attention of the newborn's primary care provider.
CONCLUSIONS
The true incidence of SCID is unknown; estimates range from 1/40,000 to 1/100,000.14 The data from a prospective SCID-prevalence study in Australia suggested an incidence of 1/69,000 live births.15 In part, the Wisconsin SCID-testing pilot program is attempting to answer this question. Even if the true rate for SCID is 1/70,000, there is a significant probability that a true SCID would not appear in the first year of screening. Because the true incidence of SCID in Wisconsin newborns is still unknown, it is not surprising that an infant with classical SCID has not yet been identified.
From the first year's experience with 71,000 primary screens, we are confident that the test is able to pick up newborns with no or low numbers of TRECs that may be associated with SCID and other immunodeficiencies, as it effectively picked up several infants with associated T-cell lymphopenia conditions. In 2008, eight infants with non-SCID immunodeficiencies were identified.10 No infant born in Wisconsin since the initiation of statewide screening for SCID has been diagnosed with SCID. About 20 infants who had low or no TRECs on initial screening died before follow-up testing could be conducted. Further evaluation of these infant deaths will determine if any of these infants had lymphopenia or lacked a thymus, which is a possible indicator of immunodeficiency. Years two and three of our pilot study, as well as data from other states that begin screening newborns for SCID, will help to determine the true incidence of SCID and other immunodeficiencies in newborns.
Footnotes
This work was supported in part by the Jeffrey Modell Foundation, the Children's Hospital of Wisconsin Foundation, the Children's Research Foundation, the Wisconsin State Laboratory of Hygiene, and grant #1U01EH000365-1 from the National Center for Environmental Health, Centers for Disease Control and Prevention (CDC). The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of CDC.
The death of one of the coauthors, Ronald H. Laessig, PhD, occurred during the early implementation of severe combined immunodeficiency testing in Wisconsin. Dr. Laessig, former Director of the Wisconsin State Laboratory of Hygiene, played an important part in all phases of the work described in this article.
REFERENCES
- 1.Baker M, Brokopp C, Hoffman G, Laessig R, Kurtycz D, Grossman W, et al. Newborn screening for severe combined immunodeficiencies (SCID)—a 2008 Wisconsin perspective. Proceedings of the 2008 Newborn Screening and Genetic Testing Symposium; 2008 Nov 3–6; San Antonio. Silver Spring (MD): Association of Public Health Laboratories; 2008. [Google Scholar]
- 2.Buckley RH. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu Rev Immunol. 2004;22:625–55. doi: 10.1146/annurev.immunol.22.012703.104614. [DOI] [PubMed] [Google Scholar]
- 3.Puck JM. Neonatal screening for severe combined immunodeficiency. Curr Opin Allergy Clin Immunol. 2007;7:522–7. doi: 10.1097/ACI.0b013e3282f14a2a. [DOI] [PubMed] [Google Scholar]
- 4.Geha RS, Notarangelo LD, Casanova JL, Chapel H, Conley ME, Fischer A, et al. Primary immunodeficiency diseases: an update from the International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee. J Allergy Clin Immunol. 2007;120:776–94. doi: 10.1016/j.jaci.2007.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson JMG, Junger G. Principles and practice of screening for disease. Geneva: World Health Organization; 1968. [Google Scholar]
- 6.Newborn screening: toward a uniform screening panel and system. Genet Med. 2006;8(Suppl 1):S1–252. doi: 10.1097/01.gim.0000223891.82390.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douek DC, Vescio RA, Betts MR, Brenchley JM, Hill BJ, Zhang L, et al. Assessment of thymic output in adults after haematopoietic stem-cell transplantation and prediction of T-cell reconstitution. Lancet. 2000;355:1875–81. doi: 10.1016/S0140-6736(00)02293-5. [DOI] [PubMed] [Google Scholar]
- 8.Chan K, Puck JM. Development of population-based newborn screening for severe combined immunodeficiency. J Allergy Clin Immunol. 2005;115:391–8. doi: 10.1016/j.jaci.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Baker MW, Grossman WJ, Laessig RH, Hoffman GL, Brokopp CD, Kurtycz DF, et al. Development of a routine newborn screening protocol for severe combined immunodeficiency. J Allergy Clin Immunol. 2009;124:522–7. doi: 10.1016/j.jaci.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Routes JM, Grossman WJ, Verbsky J, Laessig RH, Hoffman GL, Brokopp CD, et al. Statewide newborn screening for severe T-cell lymphopenia. JAMA. 2009;302:2465–70. doi: 10.1001/jama.2009.1806. [DOI] [PubMed] [Google Scholar]
- 11.Wisconsin Administrative Code, Department of Health Services. Criteria for adding or deleting conditions (DHS 115.06) (revised) 2009.
- 12.Katcher ML, Brokopp CD. Wisconsin first state to screen all newborns for “bubble boy disease”. WMJ. 2008;107:113. [PubMed] [Google Scholar]
- 13.Health Resources and Services Administration (US). Advisory Committee on Heritable Disorders and Genetic Diseases in Newborns and Children. [cited 2009 Nov 20]. Available from: URL: http://www.hrsa.gov/heritabledisorderscommittee. [DOI] [PubMed]
- 14.Lipstein E, Knapp AA, Perrin JM. Evidence review: severe combined immunodeficiency (SCID) [cited 2009 Nov 20]. Available from: URL: ftp://ftp.hrsa.gov/mchb/genetics/reports/SCIDevidencereviewfinal.pdf.
- 15.Yee A, De Ravin SS, Elliott E, Ziegler JB, Contributors to the Australian Paediatric Surveillance Unit Severe combined immunodeficiency: a national surveillance study. Pediatr Allergy Immunol. 2008;19:298–302. doi: 10.1111/j.1399-3038.2007.00646.x. [DOI] [PubMed] [Google Scholar]