Conjugated polymers have proven of utility in a range of biosensor applications.1-7 Bazan and colleagues have shown, for example, that a nucleic acid-binding cationic conjugated polymer can be used in a micro-array format for the sensitive detection of DNA without the need for target labeling.1,2 Whitten and colleagues have explored the effects of sterics, chemical structure, and electronic properties on the conjugated polymers’ sensory responses through by fluorescence quenching experiments in both solution and solid thin films.3 Typically these sensors rely on signal amplification that occurs when excitation of the conjugated polymer is transferred to a reporting dye,4 or via binding-induced changes in the emission of the conjugated polymer itself.5-7
Motivated by the applications of conjugated polymers in biosensing, several groups have explored the interactions of cationic polymers or oligofluorenes with biomolecules.8 This includes the work of Xu et al., who have argued that both electrostatic and hydrophobic interactions contribute to the formation of complexes comprising cationic conjugated polymer and PNA / DNA heteroduplexes.9-11 Similarly Wang et al.12 have reported that, when normalized by the number of DNA charge units, single-stranded DNA is a more effective quencher than double-stranded DNA of cationic water-soluble oligofluorenes due to the greater flexibility and increased hydrophobicity of the former. Here we further explore this issue by characterizing the interactions of poly [(9,9-bis (6’-N,N,N-trimethylammonium) hexyl) fluorene-alt-1,4-phenylene] bromide (PFP-Br),13,14 a water-soluble cationic conjugated polymer, and several other cationic polymers with both single-stranded DNA and various “folded” DNA structures including both double-stranded DNA and a more complex aptamer fold.
We find that PFP-Br interacts more strongly with single-stranded DNA than double-stranded DNA. This discrepancy is great enough that the polymer can open molecular beacons, a class of optically reporting oligonucleotide hybridization probes that, in the absence of a binding partner, form a stem-loop structure (Figure 1). With a fluorophore covalently linked to one terminus and a quencher covalently linked to the other, molecular beacons do not fluoresce when in their stem-loop structure.15-20 In the presence of PFP-Br, however, they fluoresce brightly (Figure 2 top), suggesting that the polymer preferentially binds to and stabilizes the beacon’s “open,” single-stranded conformation. Studies employing a series of molecular beacons differing only in the stabilities of their stems (Figure 1c and Figure S1 in Supporting Information) provide further evidence in favor of the proposed interaction.21 That is, at a given concentration of PFP-Br the intensity of the molecular beacon fluorescence is dependent on the stability of the stem, with more stable stems leading to reduced emission (Figure 2). This presumably occurs because binding of the PFP-Br to the “open,” single-stranded molecular beacon must compete with an increasingly unfavorable opening equilibrium constant, reducing the affinity with which the polymer binds to the molecular beacon.
Figure 1.
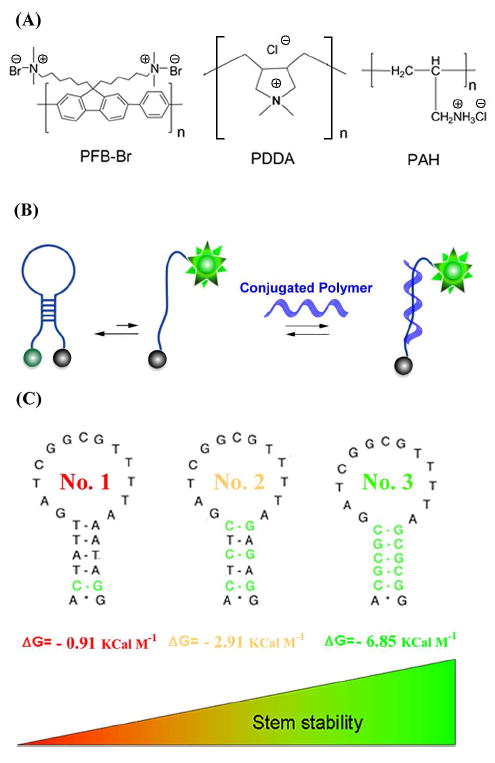
(A) We have explored the interactions between DNA and various cationic polymer including poly [(9,9-bis (6’-N,N,N-trimethylammonium) hexyl) fluorene-alt-1,4-phenylene] bromide (PFP-Br, a water soluble cationic conjugated polymer), and Poly (diallyldimethylammonium chloride) (PDDA) and Poly (allylamine hydrochloride) (PAH), both of which are water soluble cationic polymers. (B) We have done so in part using molecular beacons a fluorophore-and-quencher modified DNA sequence containing complementary termini that forms a stem loop structure. The equilibrium between the non-fluorescent, stem-loop conformation and an extended conformation is perturbed when a target binds the latter, leading to an increase in fluorescence emission. Traditionally the target is a DNA sequence complementary to the loop sequence. Here, instead, we have monitored the ability of various cationic polymers to bind and open a set of (C) related molecular beacons differing only in the sequence, and thus stability, of their stems. Molecular beacon No. 1 through 3 differ in structure and stability.
Figure 2.
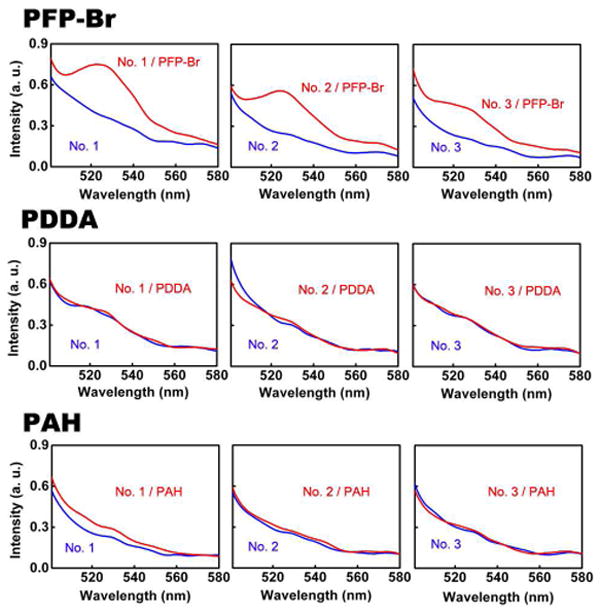
PFP-Br (Top Panel) significantly enhances the fluorescence of molecular beacons, suggesting that this conjugated polymer preferentially binds to (and thus stabilizes) the open, single-stranded conformation of this probe, PDDA (Middle Panel) and PAH (bottom panel), in contrast, do not produce significantly enhanced fluorescence, suggesting that the electrostatic attraction is not the only important interaction that regulates the binding between the water-soluble conjugated polymer and DNA. All experiments were conducted with the molecular beacons at 96 nM and the polymer at 154 nM. The fluorescence spectra are excited at 490 nm with (red) and without (blue) cationic polymers.
Because the electrostatic attraction between the cationic PFP-Br and the anionic backbone of the molecular beacon likely also plays a role in the formation of the proposed complex, we have investigated the behavior of two other cationic polymers: poly (diallyldimethylammonium chloride) PDDA and poly (allylamine hydrochloride) PAH. We find, however, that neither of these polymers enhances the fluorescence of molecular beacons despite the fact that their charge densities are, respectively, 8 and 10-times that of PFP-Br (Figure 2). Thus, electrostatic attraction alone appears insufficient to generate the observed binding between the water-soluble conjugated polymer and single-stranded DNA.
Studies of the interaction of linear DNA with PFP-Br provide further evidence that the conjugated polymer binds preferentially to single-stranded DNA. To test this, we tagged both single- and double-stranded DNA with the luminescent chromophore, Cy3, allowing us to monitor interactions with the conjugated polymer via FRET from it to the dye. Energy transfer from PFP-Br to Cy3-modified, double-stranded DNA is less efficient than the energy transfer process from PFP-Br to the equivalently modified single-stranded DNA (solid curves in Figure 3). Consistent with this, the ability of PFP-Br to open molecular beacons is not inhibited by the presence of a 10-fold excess of double-stranded DNA (Figure S2 in Supporting Information). These results confirm that PFP-Br binds more tightly to single-stranded DNA than to double-stranded DNA (Figure S3 in Supporting Information).
Figure 3.
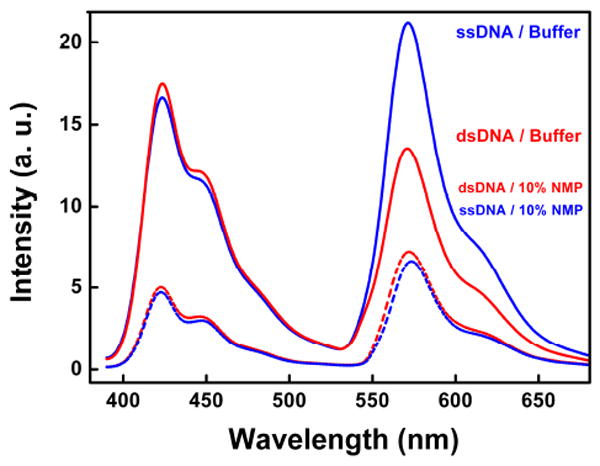
FRET from PFP-Br is more efficient to single-stranded DNA than double-stranded DNA, indicating the higher affinity binding with the former (solid curves). The addition of an organic solvent, however, reduces FRET efficiency to both double-stranded and single-stranded DNA (dashed curves), presumably because hydrophobic interactions between the conjugated polymer and the DNA are reduced. Emission spectra of PFP-Br / DNA complex by excitation of the polymer PFP-Br (excited at 380 nm) in non-organic solvent (solid curves) or in 10 % N-methyl-pyrrolidone (dashed curves).
PFP-Br is significantly more hydrophobic than either PDDA or PAH (Figure 1a), suggesting that hydrophobic interactions likely play an important role in the formation of the proposed complexes with DNA. That is, the observation that single-stranded DNA is more flexible and exposes significantly more hydrophobic surface than double-stranded DNA12 suggests a mechanism by which the conjugated polymer might preferentially bind to the single-stranded form. If this hypothesis is true, reduction of the hydrophobic interactions between PFP-Br and DNA should therefore reduce the affinity between the conjugated polymer and single-stranded DNA. Consistent with this prediction, the presence of 10 % N-methyl-pyrrolidone (NMP) reduces the efficiency of energy transfer from PFP-Br to Cy3-modified, single-stranded DNA (Figure 3). Indeed, under these conditions transfer to Cy3-modified, double-stranded DNA is a bit more efficient than to the equivalent single-stranded DNA (dashed curves in Figure 3), presumably because, when the hydrophobic interactions are weakened, the greater charge density of double-stranded DNA causes a corresponding increase in the electrostatic attraction to the cationic PFP-Br. Likewise, we do not observe any change in molecular beacon fluorescence when PFP-Br is added in the presence of 10 % NMP (Figure S4 in Supporting Information). These lines of evidence suggest that the preferential interaction between PFP-Br and single-stranded DNA is driven by hydrophobic interactions that are largely abolished with double-stranded DNA.
PFP-Br also binds single-stranded DNA in preference to more complex DNA folds. To demonstrate this we have employed the cocaine-binding aptamer of Stojanovich,22 which we have cut into two fragments, one of which is tagged with Cy3 (Top in Figure 4). In the absence of cocaine, the two parts largely dissociate into single-stranded DNA. These re-associate upon cocaine binding to form a well-folded three-way junction.23 We find that energy transfer from PFP-Br to Cy3 is more efficient in the absence of cocaine (solid curves in Figure 4 Bottom and Figure S3 in Supporting Information), again consistent with the conclusion that PFP-Br binds the two single-stranded aptamer fragments with greater affinity than it binds the folded three-way junction. Once again, the addition of 10 % NMP greatly reduces the observed FRET efficiency and again inverts the pattern: under these conditions the observed FRET is higher to the folded DNA than the single-stranded aptamer fragments (dashed curves in Figure 4 Bottom).
Figure 4.
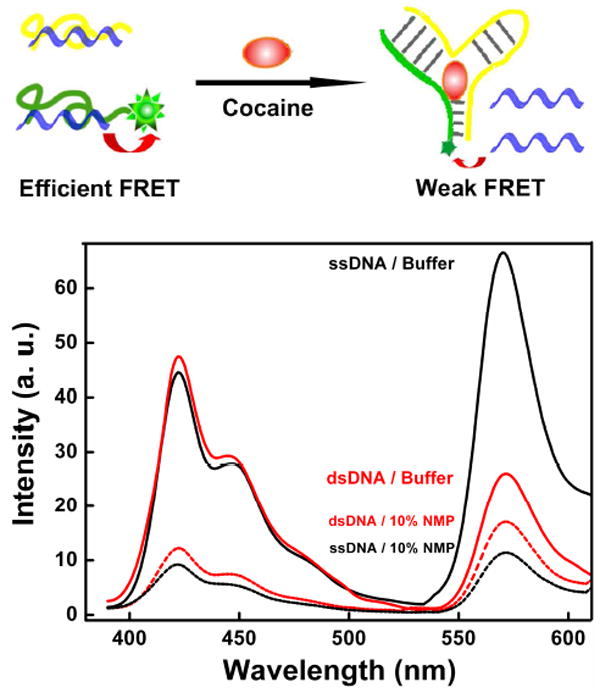
PFP-Br preferentially binds single-stranded DNA relative to more complex DNA folds. To demonstrate this we have employed an aptamer sequence split into two fragments. In the absence of its binding partner, cocaine, the two parts dissociate into single-stranded DNA, leading to enhanced interactions with PFP-Br and efficient FRET from conjugated polymer to an attached dye. Upon cocaine binding, the two single-stranded elements associated to form a folded aptamer, leading to reduced fluorescence (solid curves). In 10 % NMP, however, (dashed curves), this effect is largely eliminated.
In conclusion, water soluble, cationic conjugated polymer PFP-Br binds single-stranded DNA with greater affinity than it binds double-stranded or otherwise “folded” DNAs due to the greater hydrophobicity of the former. This appears to reflect the greater hydrophobicity of single-stranded DNA. Consistent with this, two other, less hydrophobic cationic polymers, PDDA and PAH, do not exhibit this property. Given that binding-induced changes in conjugated polymer emission have been used to support a number of biosensor platforms,1-7, 24 these observations suggest there is potential for this differential affinity to be incorporated into routine characterization protocols in molecular biology applications.
Supplementary Material
Acknowledgments
This research was supported by the Heeger presidential Chair Funds, the National Science Foundation under NSF-FMR 0602280, Institute for Collaborative Biotechnologies through Grant DAAA19-03-D-0004 from the U. S. Army Research Office, and by the National Institutes of Health (EB007689-02).
Footnotes
Supporting Information Available: Materials and methods, including fabrication of sensors and all DNA sequences used; the thermo-stability of the molecular beacons, the interaction between PFP-Br and molecular beacons is not competed by double-stranded DNA and the reduction of the hydrophobic effect prevents PFP-Br from opening molecular beacons. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sun CJ, Gaylord BS, Hong JW, Liu B, Bazan GC. Nat Protoc. 2007;2:2148–2151. doi: 10.1038/nprot.2007.307. [DOI] [PubMed] [Google Scholar]
- 2.Liu B, Bazan GC. P Natl Acad Sci USA. 2005;102:589–593. doi: 10.1073/pnas.0408561102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim Y, Whitten JE, Swager TM. J Am Chem Soc. 2005;127:12122–12130. doi: 10.1021/ja052828x. [DOI] [PubMed] [Google Scholar]
- 4.Pu KY, Liu B. Biosens Bioelectron. 2009;24:1067–1073. doi: 10.1016/j.bios.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 5.(a) Feng F, Wang H, Han L, Wang S. J Am Chem Soc. 2008;130:11338–11343. doi: 10.1021/ja8011963. [DOI] [PubMed] [Google Scholar]; (b) Fang H, Tang Y, Wang S, Li Y, Zhu D. J Am Chem Soc. 2005;127:12343–12346. doi: 10.1021/ja051507i. [DOI] [PubMed] [Google Scholar]; (c) Ho H-A, Boissinot M, Beigeron MG, Corbeil G, Doré K, Boudreau D, Leclerc M. Angew Chem Int Ed. 2002;41:1548–1551. doi: 10.1002/1521-3773(20020503)41:9<1548::aid-anie1548>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]; (d) Béra-Abérem M, Ho H-A, Leclerc M. Tetrahedrom. 2004;60:11169–11173. [Google Scholar]; (e) Duan X, Li Z, He F, Wang S. J Am Chem Soc. 2007;129:4154–4155. doi: 10.1021/ja070452c. [DOI] [PubMed] [Google Scholar]; (f) Peter K, Nilsson R, Inganäs O. Nature. 2003;2:419–424. doi: 10.1038/nmat899. [DOI] [PubMed] [Google Scholar]
- 6.Heeger AJ. 2000 http://www.nobel.se.
- 7.Tang YL, Teng F, Yu MH, An LL, He F, Wang S, Li YL, Zhu DB, Bazan GC. Adv Mater. 2008;20:703–705. [Google Scholar]
- 8.Fan CH, Plaxco KW, Heeger AJ. J Am Chem Soc. 2002;124:5642–5643. doi: 10.1021/ja025899u. [DOI] [PubMed] [Google Scholar]
- 9.Xu QH, Gaylord BS, Wang S, Bazan GC, Moses D, Heeger AJ. P Natl Acad Sci USA. 2004;101:11634–11639. doi: 10.1073/pnas.0404574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu QH, Wang S, Korystov D, Mikhailovsky A, Bazan GC, Moses D, Heeger AJ. P Natl Acad Sci USA. 2005;102:530–535. doi: 10.1073/pnas.0408568102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaylord BS, Heeger AJ, Bazan GC. J Am Chem Soc. 2003;125:896–900. doi: 10.1021/ja027152+. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Liu B, Gaylord BS, Bazan GC. Adv Funct Mater. 2003;13:463–467. [Google Scholar]
- 13.He F, Tang Y, Yu M, Wang S, Li Y, Zhu D. Adv Funct Mater. 2006;16:91–94. [Google Scholar]
- 14.Hoven C, Yang R, Garcia A, Heeger AJ, Nguyen TQ, Bazan GC. J Am Chem Soc. 2007;129:10976–10977. doi: 10.1021/ja072612q. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi S, Kramer FR. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 16.Bonnet G, Tyagi S, Libchaber A, Kramer FR. P Natl Acad Sci USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 18.Han MY, Gao XH, Su JZ, Nie S. Nat Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 19.Tyagi S, Marras SAE, Kramer FR. Nat Biotechnol. 2000;18:1191–1196. doi: 10.1038/81192. [DOI] [PubMed] [Google Scholar]
- 20.El-Hajj HH, Marras SAE, Tyagi S, Shashkina E, Kamboj M, Kiehn TE, Glickman MS, Kramer FR, Alland D. J Clin Microbiol. 2009;47:1190–1198. doi: 10.1128/JCM.02043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallee-Belisle A, Ricci F, Plaxco KW. P Natl Acad Sci USA. 2009;106:13802–13807. doi: 10.1073/pnas.0904005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stojanovic MN, de Prada P, Landry DW. J Am Chem Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 23.(a) Zuo XL, Xiao Y, Plaxco KW. J Am Chem Soc. 2009;131:6944–6945. doi: 10.1021/ja901315w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang J, Wang LH, Pan D, Song SP, Boey FYC, Zhang H, Fan CH. Small. 2008;4:1196–1200. doi: 10.1002/smll.200800057. [DOI] [PubMed] [Google Scholar]
- 24.Chen LH, McBranch DW, Wang HL, Helgeson R, Wudl F, Whitten DG. P Natl Acad Sci USA. 1999;96:12287–12292. doi: 10.1073/pnas.96.22.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


