Abstract
Primate cytomegalovirus (CMV) genomes contain tandemly repeated gene clusters putatively encoding divergent CXC chemokine ligand-like proteins (vCXCLs) and G protein-coupled receptor-like proteins (vGPCRs). In human, chimpanzee and rhesus CMVs, respectively, the vCXCL cluster contains two, three and six genes, and the vGPCR cluster contains two, two and five genes. We report that (i) green monkey CMV strains fall into two groups, containing either eight and five genes or seven and six genes in the respective clusters, and (ii) owl monkey CMV has two and zero genes. Phylogenetic analysis suggested that the vCXCL cluster evolved from a CXCL chemokine gene (probably GRO-α) that was captured in an incompletely spliced form by an ancestor of Old and New World primate CMVs, and that the vGPCR cluster evolved from a GPCR gene captured by an Old World primate CMV. Both clusters appear to have evolved via complex duplication and deletion events.
Keywords: Green monkey cytomegalovirus, Baboon cytomegalovirus, Rhesus cytomegalovirus, Owl monkey cytomegalovirus, Chemokine, Chemokine receptor, GRO-α, Diversity, Gene duplication, Evolution
INTRODUCTION
Several cytomegaloviruses (CMVs) infect Old World primate hosts, including green monkey CMV (GMCMV; alternatively named simian or vervet CMV; species Cercopithecine herpesvirus 5), rhesus macaque CMV (RhCMV; species Macacine herpesvirus 3), baboon CMV (BaCMV; not yet assigned species status), chimpanzee CMV (CCMV; species Panine herpesvirus 2) and human CMV (HCMV; species Human herpesvirus 5). Additional CMVs infect New World primate hosts, including owl monkey CMV (OMCMV; alternatively named aotine herpesvirus 1; not yet assigned species status). These viruses belong, actually or potentially, to the genus Cytomegalovirus, subfamily Betaherpesvirinae, family Herpesviridae, order Herpesvirales. Various strains have been isolated, and, in the description below, their names are given in parentheses immediately after the virus name abbreviations.
One of the features of Old World primate CMV genomes is the presence of two sets of tandemly repeated genes (in the sense of their identification as potential coding regions), one putatively encoding viral CXC chemokine ligand-like proteins (vCXCLs) and the other putatively encoding viral G protein-coupled receptor-like proteins (vGPCRs). Characterization of these gene clusters has not been straightforward.
HCMV. Analysis of the genome sequence of the high passage strain HCMV(AD169) (Chee et al., 1990) showed the presence of two genes (US27 and US28) in the vGPCR cluster (Gao and Murphy, 1994; Michelson et al., 1997; Neote et al., 1993; Vieira et al., 1998), but no vCXCL genes were detected. Later, it became apparent that the HCMV(AD169) genome has suffered a large deletion, and that most or all other HCMV strains encode two genes (UL146 and UL147) in the vCXCL cluster (Cha et al., 1996; Dolan et al., 2004; Murphy et al., 2003; Penfold et al., 1999). The UL146 vCXCL (originally named vCXC-1; Penfold et al., 1999) is extraordinarily variable among strains throughout its length, with 14 widely divergent alleles having been described (Arav-Boger et al., 2006; Dolan et al., 2004). The HCMV UL147 vCXCL (originally named vCXC-2; Penfold et al., 1999) exhibits a similar number of alleles, but variability is less and confined to the N terminus of the protein.
CCMV. The CCMV genome contains orthologs of all of the HCMV vCXCL and vGPCR genes, plus an additional vCXCL gene (UL146A) located between UL146 and UL147 (Davison et al., 2003).
RhCMV. RhCMV(68-1) was initially described as having a single vCXCL gene and RhCMV(190.82) as having none, with both strains having five genes in the vGPCR cluster (Hansen et al., 2003; Penfold et al., 2003; Rivailler et al., 2006). Later, an extensive study (Oxford et al., 2008) of the vCXCL cluster in a low passage strain, RhCMV(22659), and in an unpassaged strain, RhCMV(CNPRC), supported by partial data from two other unpassaged strains, RhCMV(WT2) and RhCMV(WT3), demonstrated that wild-type RhCMV has six genes in the vCXCL cluster, one of which was shown experimentally to be spliced. This study also demonstrated that, although not recognized initially because of rearrangements, RhCMV(68-1) does have three vCXCL genes rather than one as originally reported, but RhCMV(180.92) was confirmed as having all three deleted.
GMCMV. Five genes in the vCXCL cluster (Martin, 1999) and five genes in the vGPCR cluster (Martin, 2000) were described as being present in partial sequence data from a CMV derived by cell culture from a human with neuropathy (Martin et al., 1994; Martin et al., 1995). This agent, dubbed Stealth virus, was interpreted as representing a GMCMV whose genome had supposedly been modified and rearranged via recombination with, and amplification of, immunologically relevant human genes (Martin, 1999; Martin, 2000). However, because additional sequences derived from the virus preparation were highly similar to the major immediate early gene region and other segments of the GMCMV(Colburn) genome (Chang et al., 1995), it appeared likely that Stealth virus was a contaminating GMCMV strain derived from primary green monkey cell cultures. Similarly, GMCMV(Colburn) itself, which was originally thought to be of human origin (Huang et al., 1978), grows very efficiently in human primary fibroblast cell cultures (Jeang and Hayward, 1983; Jeang et al., 1984; LaFemina and Hayward, 1980). An implication of these findings is that CMV strains isolated from green monkeys, as well as GMCMV(Colburn), might encode vCXCL and vGPCR clusters similar to those of GMCMV(Stealth). Indeed, one strain, GMCMV(9610), was found to contain counterparts of the five genes in the GMCMV(Stealth) vGPCR cluster (Sahagun-Ruiz et al., 2004).
Unannotated sequence data totaling 170275 bp (in 22 segments) of the genome of a strain of BaCMV have been deposited in GenBank by Blewett’s group (Oklahoma State University). These data include incomplete versions of both the vCXCL and vGPCR clusters.
In summary, currently available data demonstrate that wild-type, unrearranged primate CMV genomes have the following numbers of genes in the vCXCL and vGPCR clusters, respectively: HCMV, two and two; CCMV, three and two; RhCMV, six and five; and GMCMV, at least five in each. Both clusters are absent from tree shrew herpesvirus (TSCMV, a member of species Tupaiid herpesvirus 1 in subfamily Betaherpesvirinae whose host is believed by some to be a primitive prosimian) and non-primate CMVs.
Although the functions of most primate CMV vCXCLs and vGPCRs are unknown, most of them are dispensible in fibroblast cell cultures. Nonetheless, in our opinion, each paralog is likely to have its own role during infection in vivo, whether as a chemokine or as some other function. The limited information available points to a role in virus spread and dissemination in vivo for HCMV UL146 vCXCL, which binds in a species-specific fashion to human CXCLR2, has potent calcium mobilization and chemotactic properties that are similar to those of cellular GRO-α (also known as CXCL1), and acts in vitro to attract neutrophils (Penfold et al., 1999; Sparer et al., 2004). The CCMV vCXCL1 ortholog has similar properties (Miller-Kittrell et al., 2007). The HCMV US28 vGPCR is similar to cellular CXCR2 and CX3CR1, binding fractalkine and a broad range of other ligands (Kledal et al., 1998; Kuhn et al., 1995). It has been shown to be capable of stimulating chemotactic migration of smooth muscle cells, as well as of sequestering the soluble cellular ligand CCL5 (RANTES) from infected cell culture medium (Bodaghi et al., 1998; Streblow, 1999). All five RhCMV vGPCRs are expressed as surface antigens in DNA-transfected cells, but only the protein encoded by the distal member of the cluster has chemokine ligand-binding properties equivalent in range to those of the HCMV US28 vGPCR, including the ability to bind to cellular CX3CL1 (Penfold et al., 2003). However, cells expressing this RhCMV vGPCR, unlike those expressing the HCMV US28 vGPCR, do not mobilize intracellular calcium ions.
In this paper, we have derived the sequences of the vCXCL and vGPCR clusters in five GMCMV strains and an OMCMV strain. We have compared the organization of the two clusters with those in other known primate CMVs, and attempted to deduce some of the duplication and deletion events that might have occurred during their capture and subsequent divergent evolution into the different viral lineages.
RESULTS
Collation of sequence data
The vCXCL and vGPCR clusters are positioned similarly in each of the Old World primate CMV genomes examined so far. In HCMV, they are located at approximately 0.75 and 0.95 map units, with the vCXCL cluster oriented right to left and the vGPCR cluster oriented left to right. Depicted with transcription proceeding from left to right, the vCXCL cluster is flanked by other genes in the arrangement UL144/UL145/vCXCL cluster/UL147A/UL148, and the vGPCR cluster in the arrangement US24/US26/vGPCR cluster/US29/US30 (Fig 1c). The complete sequences of the clusters in five GMCMV strains, namely GMCMV(Colburn), GMCMV(GR2757), GMCMV(2715), GMCMV(4915) and GMCMV(SA6), were derived by anchoring the data in flanking genes.
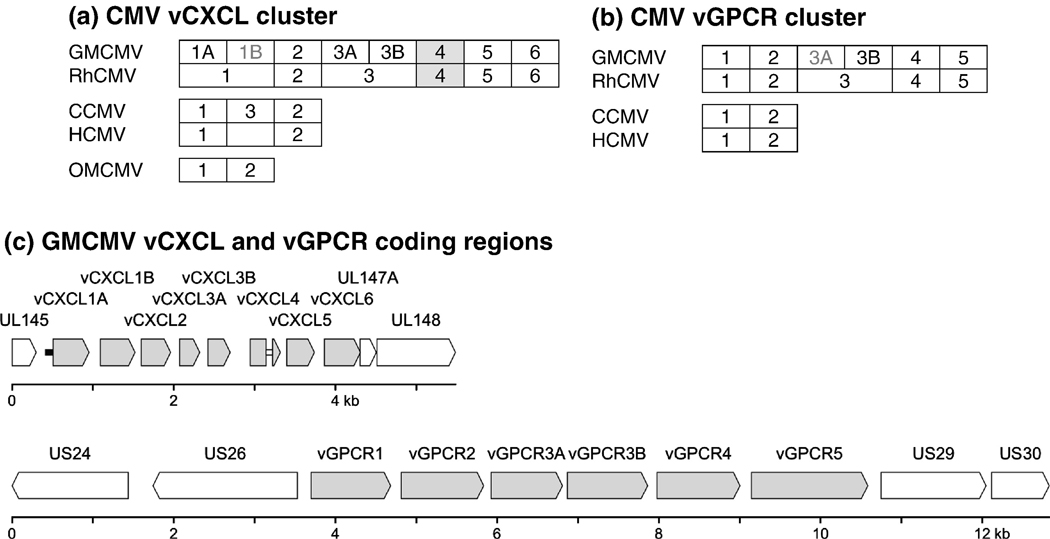
Figure 1. Organization of tandem-repeat vCXCL and vGPCR gene clusters in primate CMV genomes.
The genes are represented as transcribed from left to right. Panels (a) and (b) show schematic representations of the vCXCL and vGPCR clusters, respectively. Genes in grey font are absent from certain GMCMV genomes; vCXCL1B is absent from group II and vGPCR3A is absent from group I. Proposed orthologous relationships are shown for the GMCMV/RhCMV and CCMV/HCMV pairs, but otherwise the numbers used do not imply orthology. The grey shading indicates that the GMCMV and RhCMV vCXCL4 genes are spliced, and the blank box indicates the absence of one of the paralogous genes even within unrearranged, wild type genomes. OMCMV lacks the entire vGPCR cluster. The available data indicate that the gene arrangements in BaCMV are the same as in RhCMV. Panel (c) shows a scale diagram of the vCXCL and vGPCR clusters in GMCMV(2715) (group I) and GMCMV(4915) (group II), respectively, which have the full complement of genes in these regions. vCXCL and vGPCR coding regions are shaded grey and flanking coding regions are shaded white. In the vCXCL cluster, the white bar denotes an intron and the black bar a vestigial vCXCL coding region.
In initial experiments (data not shown), PCR and sequencing of GMCMV(Colburn) and GMCMV(GR2757) using primers based on the GMCMV(Stealth) data (Table 1) showed that both strains had an extra gene internally in the vCXCL cluster, and that GMCMV(2715) had one fewer gene internally in the vGPCR cluster. GMCMV(Colburn) failed to yield PCR products for the vGPCR cluster. Since the GMCMV(Stealth) sequence was anchored only in upstream flanking genes for each cluster, it was recognized that the data obtained were likely to be incomplete. The subsequently derived complete genome sequences of GMCMV(Colburn) and GMCMV(2715) confirmed this, revealing a total of eight genes in the vCXCL cluster for each strain, and five genes in the vGPCR cluster of GMCMV(2715). GMCMV(Colburn) had suffered a 6.8 kbp deletion extending from a position upstream from US26, retaining a single intact vGPCR gene at the distal end of the cluster. Like GMCMV(GR2757), GMCMV(2715) had one more gene in the vCXCL cluster than GMCMV(Stealth), and one less gene in the vGPCR cluster.
Table 1.
PCR and sequencing primers.
| Primer | Locationa | Sequence (5’–3’)b |
|---|---|---|
| Oriented from left to right in the vCXCL cluster | ||
| LGH2579F | 2210 | GAGAACCATGAGCCAACCC* |
| LGH2590F | 2710 | GCAAGATGCTGACGGCTTAC* |
| LGH2870F | 3180 | CGTCAATTCATCTTGGAC |
| LGH2869F | 5785 | GCTTTGGAAGTTGCTCAGAG* |
| LGH2584F | 4020 | GGTAGGGTAAAATGTAAAGC* |
| LGH2593F | 4780 | CCASTTAGAGATGCGATGGG |
| LGH2995F | 5510 | CGTACCATGGTTTTAGAGAGG |
| LGH2992F | 5650 | CTAAAATGGCACCCGTAAGG |
| LGH6415F | 5650 | GTTGGTATTGCGTTATCATG* |
| LGH6416F | 5690 | CTGCTGCTGCTTATATCTATTCTC |
| LGH2581F | 5740 | GAAAAACGGAAATGAAGTTTGCC* |
| Oriented from right to left in the vCXCL cluster | ||
| LGH6408R | 6390 | CCGAGAACGAAAATGAGCACC |
| LGH6414R | 6390 | GGTGCTCATTTTCGTTCTCGG |
| LGH6412R | 6000 | GTGACTAGCAGGGTCACAGAC |
| LGH2580R | 5760 | CCCTCTGTGTTAAGGCAAAC* |
| LGH6407R | 5750 | GTCGTGTCCATGTTGAGACG |
| LGH6413R | 5750 | CGTCTCAACATCCACACGAC |
| LGH2591R | 5440 | GCATTACGGTTTGCTTTTG* |
| LGH2993R | 5520 | CATGGTACGACTACGCATAC |
| LGH2589R | 4925 | GAGCTTTCATGGCGGTTC* |
| LGH2592R | 4270 | GCAATGACAGTGCTGGCGTTG |
| LGH2585R | 4090 | CACATAACGGTTCAATTACCG* |
| LGH2854R | 3620 | GGATAAGTAGCAGCAACAC* |
| LGH6428R | 3390 | CCCATAGTATGCATGTGCATTG* |
| LGH2583R | 2750 | CAACAGTTCACAACATCTCA* |
| Oriented from left to right in the vGPCR cluster | ||
| LGH2856F | 3740 | CAGAGCCGACATGAATTCCAG* |
| LGH2864F | 4270 | GCACATATGCACTTTCTGG |
| LGH6431F | 4900 | CAACTCAACAACACTCTCCAG* |
| LGH6433F | CTGTTTCAACGCATTCCGTAC | |
| LGH2863F | 5870 | CGAATGTTTCCTGTAAACCC* |
| LGH6410F | 6870 | CAAGATCTAGACGCGTGTACG* |
| LGH2902F | 8025 | CAAACATGAACAACTCCTCC* |
| LGH2901F | 8055 | GCAAGTGTTTAAGATCGACG* |
| LGH6409F | GACTGTACCATGCAATCCATG | |
| LGH6400F | CGGTGCGACGCAAGAAAAGG | |
| LGH6401F | CGTGTCTTGTGTGGTCATG | |
| Oriented from right to left in the vGPCR cluster | ||
| LGH2996R | GAAGCAGGTGACAGAATTCG | |
| LGH2997R | CTGGTTTATTGCACGTTTCAC | |
| LGH6432R | CAGAGGATAGCGTAACATCGATC | |
| LGH2999R | GAAGCACGGTTGCTGGCGAACAG | |
| LGH6402R | CTCGTCGGACCAAGTCACA | |
| LGH2865R | 8060 | CGTCGATCTTAAACACTTGC* |
| LGH2861R | 8030 | GGAGGAGTTGTTCATGTTTG* |
| LGH6411R | 8040 | CTTGCAGAAGGTTACAGGAGG* |
| LGH2867R | 6910 | CATGTTGCGGTTTGTGAG* |
| LGH2860R | 5870 | GGGTTTACAGGAAACATTCG* |
| LGH2866R | 5530 | GGCAAAAATTTGCATGAG |
| LGH2857R | 4980 | GATAAAGTTTCCAATCAGTCC* |
Complete sequences for both clusters from GMCMV(GR2757), GMCMV(4915) and GMCMV(SA6) were obtained using additional PCR primers based on the GMCMV(2715) data (Table 1), in some instances in combination with GMCMV(Stealth) primers. GMCMV(SA6) had suffered a 3.3 kbp deletion that removed most of the vCXCL cluster as well as UL147A and part of UL148, leaving the remnant of the distal vCXCL gene as an in-frame fusion with UL148.
On the basis of DNA sequence relatedness and similarities in gene organization, the five strains sequenced and the two for which data had been published fell into two groups: group I consisting of GMCMV(Colburn), GMCMV(GR2757), GMCMV(2715) and GMCMV(SA6), and group II consisting of GMCMV(Stealth), GMCMV(4915) and GMCMV(9610). Overlooking GMCMV(Colburn) and GMCMV(SA6), in which deletions had occurred, and GMCMV(Stealth) and GMCMV(9610), for which the published data are incomplete, group I strains are characterized by eight genes in the vCXCL cluster and five genes in the vGPCR cluster, and group II strains by seven genes in the vCXCL cluster and six genes in the vGPCR cluster.
Simplified depictions of the arrangements of genes in the vCXCL and vGPCR clusters of GMCMV and other primate CMVs are compared in Fig. 1a and Fig 1c, respectively. The nomenclature employed effectively follows the approach by which HCMV vCXC-1 and vCXC-2 were first named on the basis of sequence similarity, in the absence of functional information (Penfold et al., 1999). We emphasize that this is an attempt at a unitary scheme for the purposes of the discussion below, and that it carries no implications about functionality. On the basis of positional and sequence relationships (explained below), the numbering scheme for the GMCMV and RhCMV genes indicates proposed orthology, with the vCXCL genes named sequentially from right to left in the direction that they are transcribed. Similarly, perceived orthology is inherent within the CCMV and HCMV nomenclature. However, an orthologous relationship is not implied among or between the GMCMV/RhCMV, CCMV/HCMV or OMCMV groups, where the high level of divergence confounds its establishment in most cases. Fig. 1 emphasizes the fact that GMCMV strains in group I lack the vGPCR3A gene, and those in group II lack the vCXCL1B gene. As described below, BaCMV sequences are incomplete for both clusters, but the available data indicate that the gene number and arrangements are the same as in RhCMV.
In conducting the phylogenetic analyses described below, limitations in the published sequences were registered. Data for GMCMV(Stealth) do not extend beyond a point in vCXCL4, and they also do not extend beyond a point in vGPCR4. Data for GMCMV(9610) do not extend beyond vGPCR4, and are not available for the vCXCL cluster. A frameshift in RhCMV(180.92) vGPCR5 was corrected for the analysis, and two frameshifts in RhCMV(68-1) vGPCR3A (Hansen et al., 2003) were amended by reference to an apparently correct version (Penfold et al., 2003). Complete sequences were not available for vCXCL1, vCXCL5 and vCXCL6 in RhCMV(WT2), and for vCXCL6 in RhCMV(WT3). The sequences available for the BaCMV vCXCL cluster terminate within vCXCL6, and those for the vGPCR cluster extend from a point within vGPCR1 to a point within vGPCR5. The BaCMV vGPCR4 sequence contains an in-frame termination codon, which was corrected (TGA to AGA) for the analysis.
Relationships in the vCXCL cluster
The precursors of cellular vCXCLs consist of 100–110 amino acid residues, with a cleavable signal peptide sequence just prior to the CXC motif. The structure of the mature protein depends upon four conserved C residues. The shortest GMCMV vCXCL, vCXCL3A, contains only 81–82 residues and lacks the third and fourth C residues. Initially, the vCXCL4 coding region appeared to be even smaller (62 residues) and to lack the fourth C residue in all five of the GM-CMV strains, but this appears to be the only example where the function as an active chemokine may have been compromized. However, utilization of conserved potential splice donor and acceptor sites introduced a second exon that is similar to the C-terminus of vCXCL5 and contains the additional C residue, resulting in a precursor of 92 residues. These splice sites are located identically in RhCMV vCXCL4 (Oxford et al., 2008) and BaCMV vCXCL4.
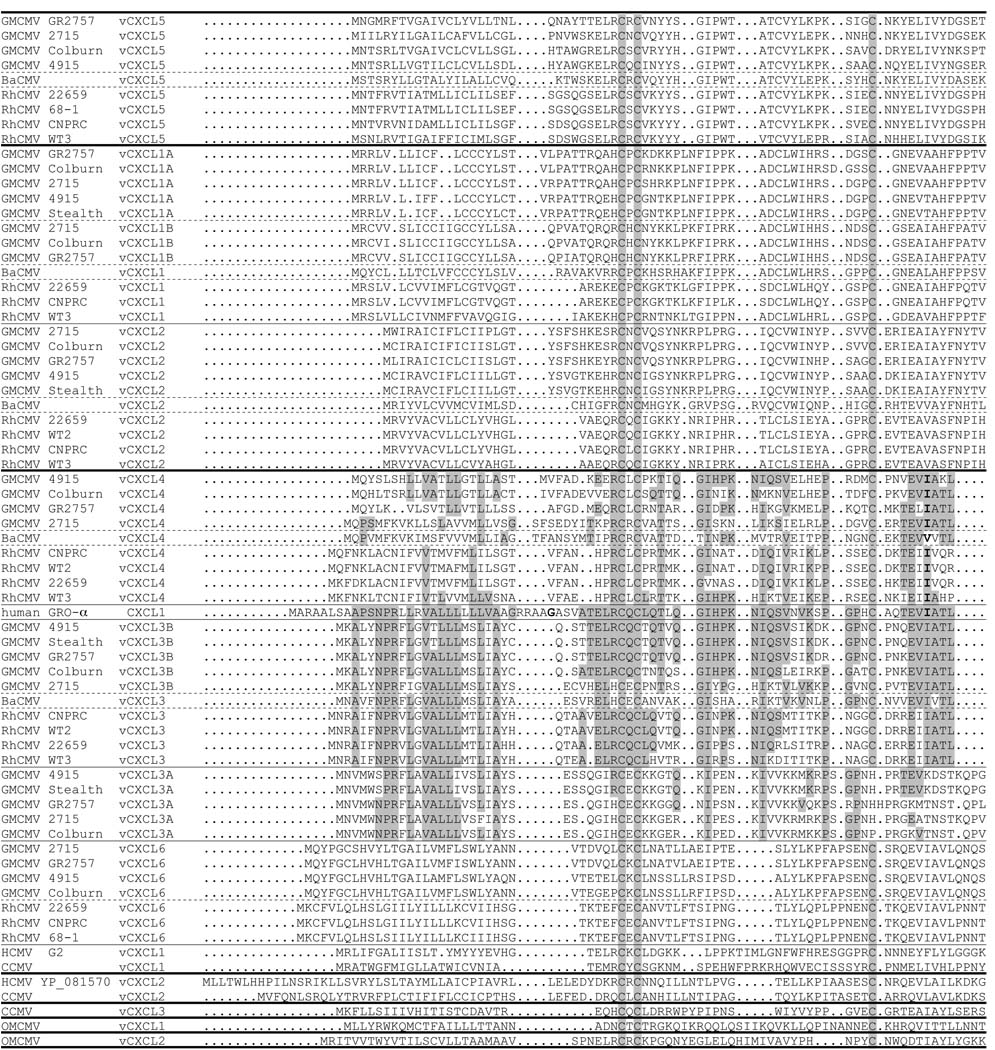
Among the four GMCMV strains for which sequences of the complete vCXCL cluster were derived (i.e. GMCMV(Colburn), GMCMV(2715) and GMCMV(GR2757) in group I, and GMCMV(4915) in group II), pairwise alignments of the orthologs showed a wide spectrum (36–97%) of sequence conservation. That between GMCMV(2715) and GMCMV(GR2757) ranged from 42% (vCXCL4) to 97% (the highest observed in any pairwise comparison; vCXCL1B), and that between GMCMV(2715) and GMCMV(4915) ranged from 36% (the lowest observed in any pairwise comparison; vCXCL4) to 54% (vCXCL1B/vCXCL1). The amino acid sequences of the GMCMV vCXCLs were aligned with those of other primate CMV vCXCLs, including the major variants of the HCMV vCXCLs and incorporating primate versions of GRO-α, which is the cellular CXCL most closely related to the vCXCLs. All versions of Old World primate vCXCL3/vCXCL3B and vCXCL5 proteins retain the ELRCXC motif characteristic of GRO-α (or its minor derivative, ELHCXC), but the others vary considerably. A subset consisting of most of the sequences from the complete alignment is shown in Fig. 2.
Figure 2. Sequence alignment comparisons among primate CMV vCXCL proteins.
The alignment shows predicted primary translation products encoded by the primate CMV vCXCL cluster, plus human GRO-α (CXCL1). Dots indicate gapping characters. The four conserved C residues are shaded, as are residues conserved between human GRO-α and vCXCL3A/vCXCL3B/vCXCL3/vCXCL4 in GMCMV, BaCMV and RhCMV. Bold characters in human GRO-α and GMCMV, BaCMV and RhCMV vCXCL4 correspond to the codons in which exon boundaries are located.
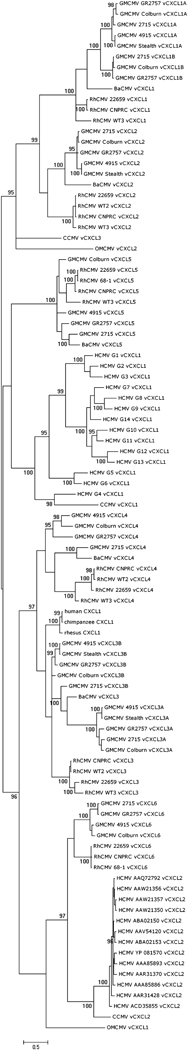
A Bayesian phylogenetic tree derived from the complete alignment is shown in Fig. 3. The versions of GMCMV vCXCL1A group into a clade (100% of trees), as do those of vCXCL1B (100%). These two clades group (100%), and are sister to RhCMV vCXCL1 (100%). The versions of RhCMV vCXCL2 group (100%), and those of GMCMV vCXCL2 group less convincingly (95%). The grouping of these two clades is poorly supported (57%), but they are sister to vCXCL1A/vCXCL1B/vCXCL1 (99%). The grouping of vCXCL3A, vCXCL3B, vCXCL3 and vCXCL4 is supported moderately (97%). The three examples of mammalian cellular GRO-α genes fall loosely as a clade in this section of the tree (see Fig. 2 for the degree of conservation). The versions of GMCMV vCXCL3A group (100%), as do those of RhCMV vCXCL3 (100%). However, grouping of GMCMV vCXCL3B versions is not supported. The RhCMV vCXCL4 versions group (100%), but clustering of the GMCMV vCXCL4 versions is less convincing (94%). The grouping of RhCMV vCXCL4 with GMCMV vCXCL4 is also not strongly supported (82%), although these genes share the same splicing pattern, which is not found in any other vCXCL gene. The vCXCL6 versions form individual groups for GMCMV and RhCMV (100% for each), but the evidence that these two clades are sisters is less convincing (74%). vCXCL6 is sister to vCXCL3A/vCXCL3B/vCXCL3/vCXCL4 in the majority of trees (96%). The GMCMV and RhCMV vCXCL5 versions form a clade (100%). The BaCMV vGPCRs generally group with those of GMCMV rather than RhCMV.
Figure 3. Phylogenetic grouping of primate CMV vCXCL proteins.
Bayesian phylogenetic tree of proteins encoded by the primate CMV vCXCL cluster, rooted at the midpoint. Nodes supported by at least 95% of trees are indicated. The tree includes all recognized versions of HCMV vCXCL1 (G1–G14) and vCXCL3 (as accession numbers), plus primate versions of GRO-α (CXCL1).
The gene complement in the vCXCL cluster is simpler in CCMV and HCMV, consisting of three and two genes, respectively (Fig. 1). The highly divergent versions of HCMV vCXCL1 group with the single known version of CCMV vCXCL1 (100%), with allele G4 showing the closest similarity (98%). Similarly, CCMV vCXCL2 groups with HCMV vCXCL2 (100%), and this clade is sister to GMCMV and RhCMV vCXCL6 (97%). This part of the tree represents the only (moderately) convincing relationship between the GMCMV/RhCMV and CCMV/HCMV groups. The origins of CCMV vCXCL3 are not discernible. The New World primate virus, OMCMV, has two genes in the vCXCL cluster, though their origins in relation to Old World primate viruses are also not discernible.
Relationships in the vGPCR cluster
Cellular GPCRs consist of about 320 amino acid residues encompassing seven hydrophobic transmembrane domains (TMDs), with the cytoplasmic C-terminal tail involved in G-protein signaling and serine/threonine kinase interactions. Key functional features include a cytoplasmic [DE]RY motif. All genes within the Old World primate vGPCR clusters appear functionally intact and the paralogs within each cluster are much more diverged than any of the orthologs.
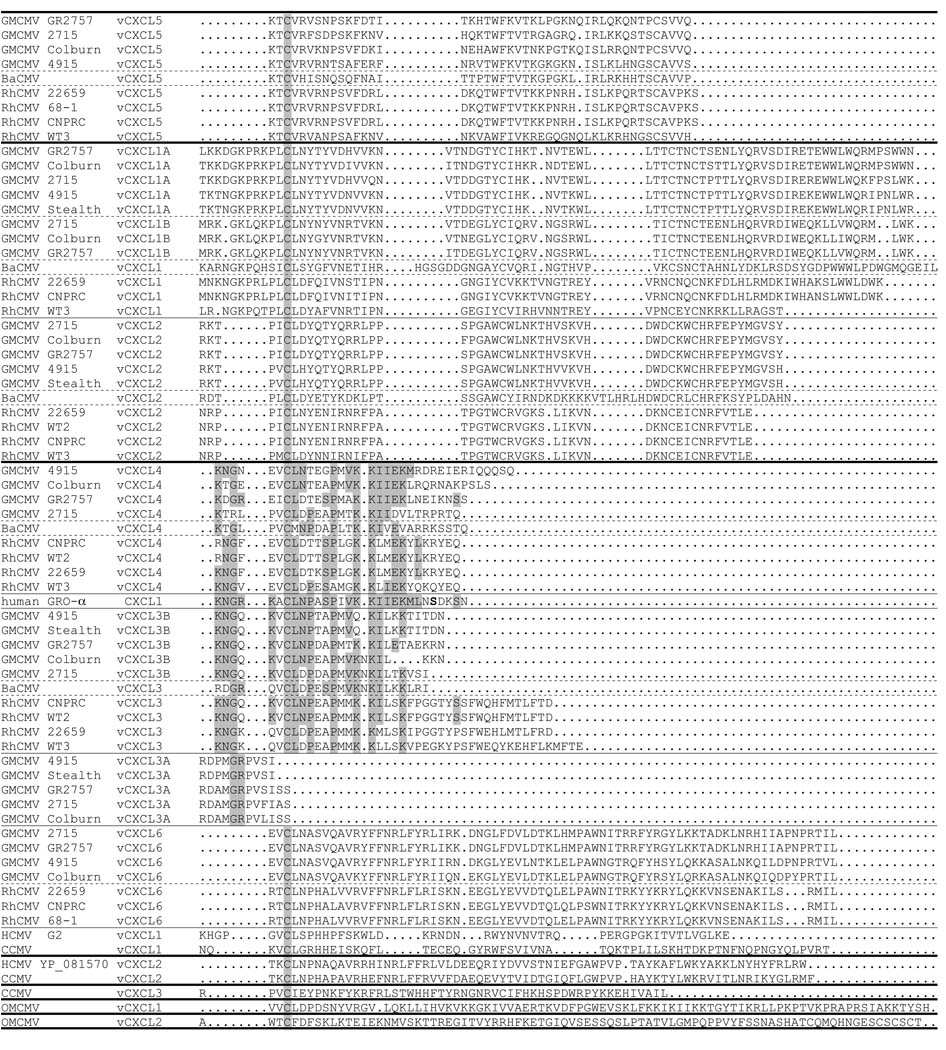
Among the four GMCMV strains for which sequences of the complete vGPCR cluster were derived (i.e. GMCMV(SA6), GMCMV(2715) and GMCMV (GR2757) in group I, and GMCMV(4915) in group II), pairwise alignments showed that vGPCR5 is least conserved (91% of residues). However, the majority of the variation in vGPCR5 is present in an S/T-rich N-terminal extension of about 140 residues (truncated in Fig. 4), which lacks counterparts in other vGPCRs, with 57–85% of residues conserved. Conservation in the GPCR domains ranges from 90% (vGPCR3B) to 99% (vGPCR2). The amino acid sequences of GMCMV vGPCRs were aligned with those of other primate CMV vGPCRs. OMCMV was not included because it lacks the vGPCR cluster. A subset consisting of some of the sequences from the complete alignment is shown in Fig. 4.
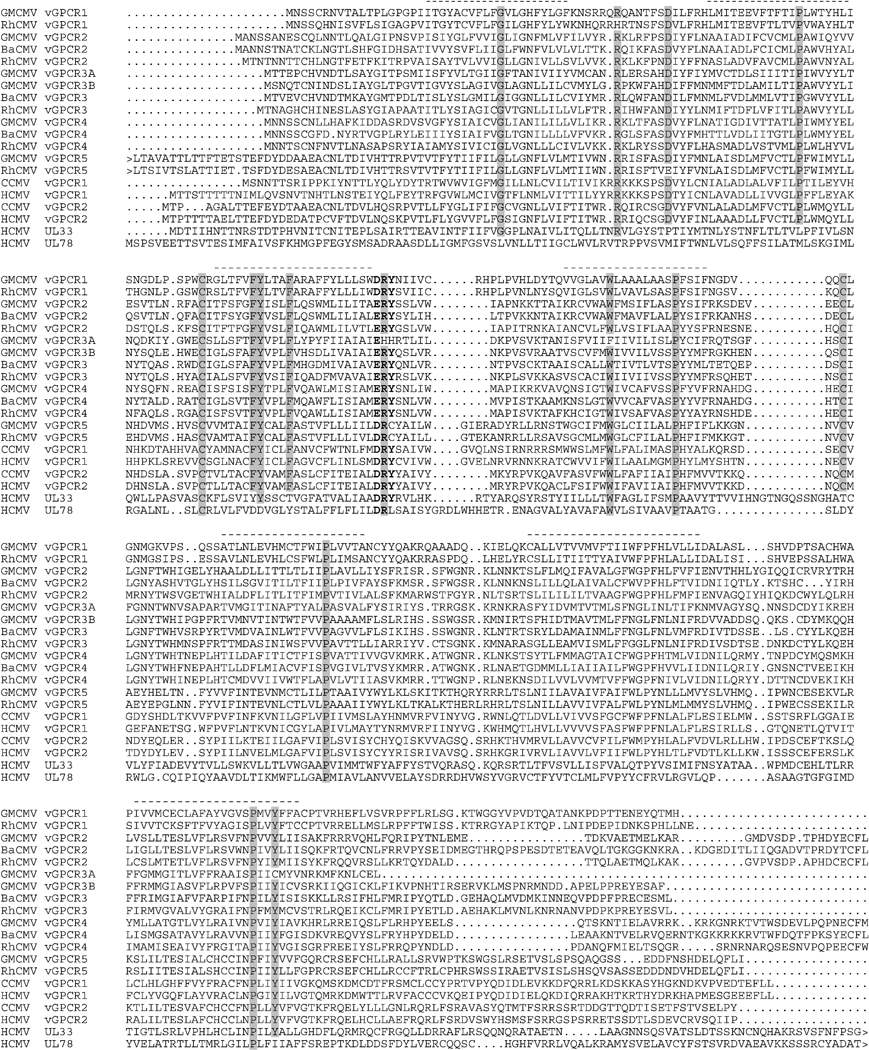
Figure 4. Sequence alignment comparisons among primate CMV vGPCR proteins.
The alignment shows predicted primary translation products encoded by the primate CMV vGPCR cluster, plus those from the primate CMV UL33 and UL78 GPCRs. For GMCMV and RhCMV, only sequences from GMCMV(4915) and RhCMV(22659) are included. The functionally important [DE]RY motif is in bold type. Dots indicate gapping characters, and highly conserved residues are shaded. Arrowheads indicate N- or C-terminal extensions (not shown) in certain proteins. The seven predicted TMDs in GMCMV vGPCR1 (as an example) are indicated by dashes above the alignment.
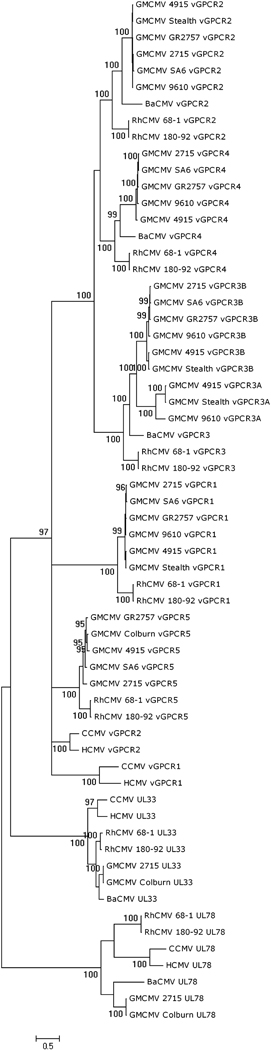
A Bayesian phylogenetic tree derived from the complete alignment is shown in Fig. 5. The tree includes Old World primate CMV data for two vGCPRs encoded by genes UL33 and UL78, which are not part of the cluster and, since orthologs are present in non-primate CMVs, probably represent genes captured earlier in the evolution of subfamily Betaherpesvirinae. All three versions of vGPCR3A are truncated so that, although they retain the seven TMDs, they lack most of the C-terminal intracellular domain. Two of these three also lack the [DE]RY motif, which is marked in Fig. 4 and in all of the vGPCR5 genes this motif is modified to DRCY.
Figure 5. Phylogenetic grouping of primate CMV vGPCR proteins.
Bayesian phylogenetic tree of proteins encoded by the primate CMV vGPCR cluster, rooted at the midpoint. Nodes supported by at least 95% of trees are indicated. The tree includes primate CMV versions of vGPCRs encoded by genes UL33 and UL78.
The greater length and conservation of vGPCR sequences provides a more reliable view of relationships than was derived for the vCXCLs. In Fig. 5, each vGPCR groups into a fully supported (100%) clade containing all the orthologous GMCMV and RhCMV members. vGPCR3A is sister to vGPCR3B (100%), and together they are sister to vGPCR3 (100%). The vGPCR3A/vGPCR3B/vGPCR3, vGPCR4 and vGPCR2 clades group (100%). Again, the BaCMV vGPCRs generally group with those of GMCMV rather than RhCMV. CCMV and HCMV vGPCR1 group together (100%), as do CCMV and HCMV vGPCR2 (100%). All of the vGPCRs in the cluster form a clade separate from UL33 and UL78 (97%).
DISCUSSION
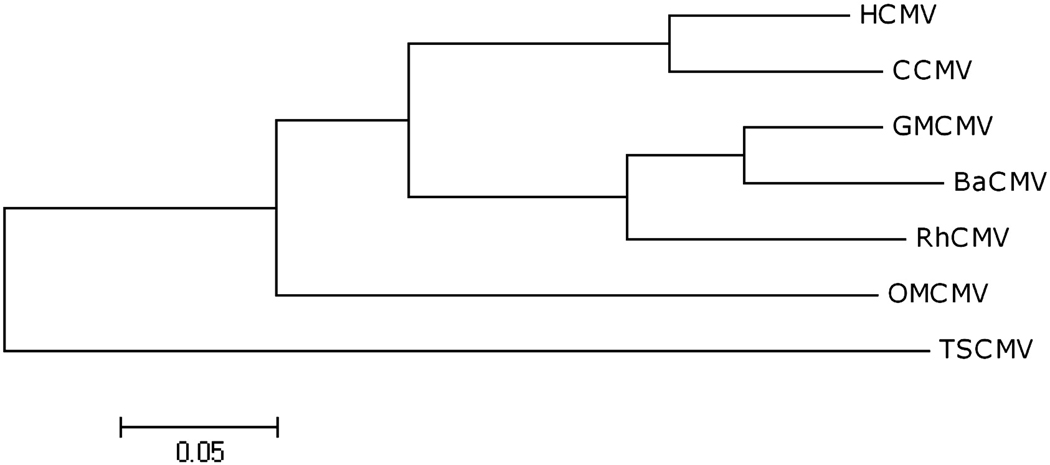
The prevailing view is that in general primate CMVs have evolved along with their hosts throughout primate evolution. Divergence dates inferred from host phylogeny are 42.9 millions of years ago (Mya) for Old World monkeys from New World monkeys, 30.5 Mya for apes from Old World monkeys, 9.9 Mya for macaques and baboons from green monkeys, 6.6 Mya for macaques from baboons and 6.6 Mya for humans from chimpanzees (Steiper and Young, 2006). This speciation order is largely congruent with CMV phylogeny based on a conserved gene (DNA polymerase), except that BaCMV is more closely related to GMCMV than RhCMV (Fig. 7). BaCMV is also generally more closely related to GMCMV than RhCMV in the vCXCL and vGPCR clusters. From a parsimonious standpoint, GMCMV seems to have proliferated extra genes in each cluster since its divergence from BaCMV, which has a gene layout in each cluster like that of RhCMV rather than GMCMV.
Figure 7. Phylogenetic grouping of complete primate CMV DNA polymerase proteins.
The ClustalW-based tree is rooted with TSCMV as the outgroup. Bootstrap values were 100% at each node.
The evolutionary outcome of the interactions between virus and host is clearly discernible in the vCXCL and vGPRC clusters, both in terms of sequence diversity, even among orthologs (especially in the vCXCL cluster), and in terms of variation in gene number between virus species. Several factors are important in discerning the evolutionary steps that may have occurred. First, an accurate understanding of the gene arrangement must be derived for each virus. This includes accounting for errors or mutations in published sequences, recognizing the effects of large deletions or rearrangements that may occur during passage of virus in vitro, anchoring derived sequences in flanking genes to ensure that the whole cluster is defined, and identifying protein-coding regions optimally. Second, as reliable a sequence alignment as achievable must be used to derive a statistically supported phylogeny. Relationships can then be assessed with discrimination, and strengthened by other features shared between genes, such as relative position or splicing. Third, gene loss, as well as gene duplication, must be considered as an evolutionary step. These alternatives may sometimes be distinguished on the basis of residual similarities between DNA sequences. Respectively, GMCMV, BaCMV, RhCMV, CCMV, HCMV and OMCMV have seven or eight, six, six, three, two and two genes in the vCXCL cluster and five or six, five, five, two, two and zero genes in the vGPCR cluster. Since only a subset of the events that have occurred are likely to be represented in extant viruses, any view on the evolution of the clusters will be necessarily incomplete.
From a parsimonious standpoint, certain evolutionary steps can be proposed for the vCXCL cluster in GMCMV and RhCMV, on a scale of increasing phylogenetic support. In an ancestor of Old and New World primate CMVs (more than 42.9 Mya), there was a single vCXCL gene that in the lineage leading to GMCMV, BaCMV and RhCMV gave rise to four daughters, one becoming vCXCL1/vCXCL2, one vCXCL3/vCXCL4, one vCXCL5, and one vCXCL6. vCXCL3 was duplicated in the GMCMV lineage (possibly after divergence from the BaCMV lineage) to generate vCXCL3A and vCXCL3B, and then, apparently as the most recent duplication event based on levels of paralogous divergence, vCXCL1A and vCXCL1B were generated from vCXCL1 in GMCMV group I. However, events might not have been parsimonious. For example, the most recent event in the GMCMV vCXCL cluster might not have involved a duplication event to give rise to vCXCL1A and vCXCL1B in group I. Instead, this duplication might have occurred in a parent of both lineages, followed by deletion of vCXCL1B in group II. Examination of DNA sequence alignments (Fig. 6) showed that the insertion/deletion event involved the complete coding region of vCXCL1B plus 150 bp extending upstream to the termination codon of vCXCL1A. It did not affect the 78 bp sequence between vCXCL1B and vCXCL2, which is conserved in groups I and II and is unrelated to the region upstream from vCXCL1B. In group I, the initiation codons of vCXCL1A and vCXCL1B are located in a conserved sequence of 29 bp, and the termination codons in a different conserved sequence of 21 bp. These sequences are also conserved at the initiation and termination codons of vCXCL1A in group II. Thus, loss of vCXCL1B might have involved straightforward homologous recombination between the duplicated motifs at the 3’-ends of vCXCL1A and vCXCL1B.
Figure 6. Vestigial genes provide evidence for recent deletion events in the GMCMV vCXCL and vGPCR gene clusters.
Both parts show an alignment of the relevant DNA sequences of GMCMV(2715) and GMCMV(4915), which belong to groups I and II, respectively. The alignment for the vCXCL cluster proceeds from the end of the UL145 coding region, through a vestigial vCXCL, the inititation and then the termination codon of vCXCL1A, and the initiation and termination codon of vCXCL1B. Repeated sequences that are largely conserved at the 5’-end of vCXCL1A and in the vestigial vCXCL are underlined, and those conserved at the initiation and termination codons of vCXCL1A and vCXCL1B are underlined in bold. The alignment for the vGPCR cluster proceeds from the end of vGPCR2, through the initiation and then the termination codon for vGPCR3A, to the initiation codon for vGPCR3B. The vestigial vGPCR sequence is underlined. In both alignments, tildes indicate coding sequences (lengths in bp) that are not shown, and dots indicate gapping characters.
In further support of the occurrence of gene deletion during evolution of the vCXCL cluster, the six GMCMV strains for which relevant sequence data are available contain an 83 bp sequence at the 5’-end of the vCXCL1 coding region that is essentially repeated in the region close upstream from vCXCL1A, in the non-coding region between this gene and UL145; that is, a vestigial vCXCL gene (Fig. 6). This repeated sequence contains a third copy of the conserved sequence at the initiation codons of vCXCL1A and vCXCL1B, and is not present in the RhCMV or BaCMV vCXCL clusters. Therefore, it is plausible that the lineage leading to GMCMV groups I and II, but not the other two lineages, once possessed, and later lost, an additional vCXCL gene upstream from vCXCL1A.
A final feature of the vCXCL data is the unexpectedly large divergence of GMCMV(2715) vCXCL4 from all other GMCMV vCXCL4 versions. Indeed, Fig. 2 and Fig 3 show that both GMCMV(2715) vCXCL3B and vCXCL4 are more similar to BaCMV vCXCL3 and vCXCL4, respectively, than to the other GMCMV versions of these genes. Furthermore, a linked 66 bp non-coding segment in an unusually large gap immediately upstream of the BaCMV vCXCL4 gene showed 85% nucleotide identity with GMCMV(2715), but minimal identity with the equivalent region from any other GMCMV (data not shown). This pattern could be the result of a homologous recombination exchange event between a progenitor of the GMCMV(2715) virus and a BaCMV-like virus. However, it is also possible that this additional complexity instead reflects a further level of expansion and differential deletion events involving vCXCL3 and vCXCL4 that occurred in an ancestor of one or both lineages.
A parsimonious approach also permits certain evolutionary steps to be proposed for the vGPCR cluster in GMCMV and RhCMV, with full phylogenetic support. In an ancestor of Old World primate CMVs (42.9-32.5 Mya), there was a single vGPCR gene that in the lineage leading to GMCMV and RhCMV gave rise to three daughters, one becoming vGPCR1, one vGPCR2/vGPCR3/vGPCR4, and one vGPCR5. vGPCR3 then duplicated in GMCMV (possibly after divergence from the BaCMV lineage) to give rise to vGPCR3A and vGPCR3B in group II. However, similar to the situation in the vCXCL cluster, evidence is available from DNA sequence alignments for a less parsimonious route, involving earlier generation of vGPCR3A and vGPCR3B in a progenitor of both GMCMV lineages and then loss of vGPCR3A to give rise to group I. In comparison with group II, group I sequences lack a region of 851 bp that extends from 35 bp upstream from the vGPCR3A initiation codon to 63 bp upstream from the termination codon (Fig. 6). In group I, the region between vGPCR2 and vGPCR3B is highly similar to coding and non-coding sections at the 3’-end of vGPCR3A in group II, effectively resulting in a vestigial vGPCR3A gene located between vGPCR2 and vGPCR3B. The RhCMV and BaCMV vGPCR clusters lack this feature, thus confirming that the events involving vGPCR3A are specific to the GMCMV lineage.
GMCMV strains in group I have eight genes in the vCXCL cluster and five in the vGPCR cluster, whereas those in group II have seven and six genes, respectively. Multiple strains in each group were detected, indicating that the differences in gene number originated from genuine evolutionary events, rather than from deletions taking place in cell culture, although the latter may account for the unique deletion mutations found in the GMCMV(SA6) vCXCL and GMCMV(Colburn) vGPCR clusters. The existence of two distinct GMCMV groups was also supported by data from gene UL144, which is located in the flanking sequence upstream from the vCXCL cluster: members of group I differ from members of group II at the same seven residues (data not shown). The precise origins of the groups are not understood. In a broad sense, green monkeys consist of several subspecies of Cercopithecus aethiops (or, in the alternative taxonomy, species of the genus Chlorocebus). The two virus groups may have evolved in the same host subspecies or in different subspecies, perhaps spreading to other subspecies in primate housing facilities.
It is more problematic to extend the findings from Old World monkey CMVs to include the evolution of CCMV, HCMV and OMCMV. Little can be said beyond that vCXCL2 in CCMV and HCMV may be orthologs of vCXCL6 in GMCMV and RhCMV. In the absence of convincing phylogenetic data, and given the potential involvement of both duplications and deletions, it is unrewarding to speculate in detail about additional evolutionary events in more ancient ancestral viruses. Nonetheless, the fact that the closest cellular relative of the vCXCL cluster is GRO-α suggests that this gene was acquired by capture (lateral transfer), and that its descendents in the GMCMV and RhCMV vCXCL3A/vCXCL3B/vCXCL3/vCXCL4 lineage have been selected to retain greatest similarity to the parental gene. Alternative explanations, that GRO-α was inserted into the vCXCL cluster more recently, and has founded only the vCXCL3A/vCXCL3B/vCXC3/vCXC4 lineage, or that the vCXCL cluster was derived from a captured gene other than GRO-α (for example, the interleukin-8 (IL-8) gene), with the vCXCL3A/vCXCL3B/vCXC3/vCXC4 lineage converging to resemble GRO-α, seem less plausible. Functional aspects of CMV vCXCLs remain largely unexplored, and it is not known how many are CXC chemokines. There is a considerable degree of variation among human CXC chemokines, and many lack the ELR element of the ELRCXC motif. Both types exhibit chemokine functions, those possessing the ELR element tending to promote the attraction of neutrophils, and those lacking it tending to promote the attraction of lymphocytes and monocytes (Laing and Secombes, 2004). Therefore, the observation that most vCXCLs lack the ELR element (Fig. 2) does not rule out chemokine or chemokine-related functions.
Gene capture has evidently been a prominent feature of herpesvirus evolution, and there are multiple examples in all herpesviruses of genes that have clear cellular counterparts. Most captured genes lack introns, but some are expressed by splicing patterns that, at least partially, resemble those of the cellular genes. In principle, gene capture may be achieved by recombination with cDNA copies (presumably retrovirus-mediated) of cellular transcripts in unspliced, partially spliced or fully spliced forms, or by recombination with cellular genomic DNA to acquire genes in unspliced form. In addition, special mechanisms have been proposed in certain instances, such as that of a cluster of nine adjacent, unspliced genes in Kaposi’s sarcoma-associated herpesvirus (a member of the Gammaherpesvirinae), which may have been captured by sequential, accidental initiation by cellular mRNAs of DNA strand growth at a bidirectional origin of DNA replication (Nicholas et al., 1997; Nicholas et al., 1998).
The observation that GMCMV and RhCMV vCXCL4 contain an equivalent of the second intron located within the I codon (V codon in BaCMV) of a largely conserved motif (EVIATLKNG) encoded by GRO-α (Fig. 2) and IL-8 (data not shown) is strong evidence that the cellular gene was captured in an unspliced or partially spliced form (with subsequent loss of the first intron), and that the second intron was present in the vCXCL that underwent the initial duplication event. This intron has been retained in lineages leading to GMCMV, BaCMV and RhCMV vCXCL4 (which, incidentally, along with vCXCL3A, vCXCL3B and vCXCL3 retain strong sequence similarity to GRO-α; Fig. 2), but has been lost from other lineages, perhaps on more than one occasion. The vCXCL family thus joins other examples of captured CMV genes that have retained cellular splicing patterns. The cyclooxygenase-2 (vCOX-2) gene is present as a partially spliced gene in RhCMV (Hansen et al., 2003; Rue et al., 2004), as well as in GMCMV with one fewer intron (A. Dolan, unpublished data), but is absent from CCMV and HCMV. The interleukin-10 (vIL-10) gene is present as a partially spliced gene in RhCMV and GMCMV and with one fewer intron in HCMV, but has evidently been lost from CCMV (Davison et al., 2003; Lockridge et al., 2000). These examples lend further credence to the idea that more captured genes than are apparent in extant CMVs may have been acquired in unspliced or partially spliced forms, via RNAs or from the cellular genome, with subsequent loss of introns, rather than via fully spliced RNAs.
Finally, it is of interest to note that nine human CXCL genes (including GRO-α and IL-8) form a tandemly repeated cluster of largely similarly spliced genes on chromosome 4. The processes of duplication and deletion that appear to have been involved in evolution of the primate CMV vCXCL cluster may also have occurred in the generation of the host CXCL cluster. Furthermore, similar processes may have operated throughout CMV genome evolution, as evidenced by the many sets of multicopy gene paralogs, clustered or otherwise, that occur in primate and non-primate CMV genomes.
MATERIALS AND METHODS
Viruses
Low passage strains GMCMV(2715) and GMCMV(4915), and the original vervet strain GMCMV(SA6) isolated in 1957, were obtained in 1982 from Richard Heberling at the Southwest Foundation for Research and Education in San Antonio, Texas. High passage strain GMCMV(Colburn), which was isolated ostensibly from a human, was obtained in 1976 from Milan Fiala at UCLA (Huang et al., 1978; LaFemina and Hayward, 1980). GMCMV(GR2757), for which the passage history is unknown, was obtained in 1974 from Eng-Sheng Huang, University of North Carolina at Chapel Hill (Huang and Pagano, 1974; Jeang and Hayward, 1983). OMCMV(S34E) was isolated from an owl monkey (Daniel et al., 1971), and was obtained in 2004 from the American Type Culture Collection (ATCC) as VR-606.
Virus growth and DNA purification
The GMCMV viruses were passaged at low MOI in diploid human fibroblast cells under the standard conditions used to avoid accumulation of defective virus genomes (LaFemina et al., 1989). OMCMV was passaged similarly on the owl monkey kidney cell line OMK(637-69), which was obtained from the ATCC as CRL-1556. DNA was isolated from virus particles obtained by standard procedures from infected cell cultures exhibiting maximal CPE. Briefly, GMCMV virions were purified from culture medium by sucrose density gradient ultracentrifugation, pelleted, incubated with DNase I, and lysed in SDS/Tris-HCl/EDTA. OMCMV virions were isolated in the cytoplasmic fraction of infected cells, incubated with DNase I, pelleted and lysed in SDS/Tris-HCl/EDTA. DNA was purified by phenol extraction, ethanol precipitation and dialysis.
PCR DNA sequence data
The primers listed in Table 1 were used in appropriate pairwise combinations to amplify and sequence overlapping fragments encompassing the vCXCL and vGPCR clusters. Strategically, the design of PCR primers for anchoring the upstream ends of the vCXCL and vGPCR clusters of the five GMCMV strains was based on published GMCMV(Stealth) sequence data. These primers were then supplemented or replaced by additional primers designed for each strain on the basis of primer-walking. The design of PCR primers for anchoring the downstream ends of both clusters in GMCMV(GR2757), GMCMV(4915) and GMCMV(SA6) was based on the complete genome sequences of GMCMV(2715) and GMCMV(Colburn). PCR products were sequenced on both strands using an ABI310 instrument.
Whole genome sequence data
The sequences of the vCXCL and vGPCR clusters in GMCMV(2715), GMCMV(Colburn) and OMCMV were extracted from complete genome sequences, which were derived in our laboratories by standard plasmid shotgun methods. There were no differences between the GMCMV(2715) or GMCMV(Colburn) genome sequences and comparable, incomplete sequences obtained for these strains by PCR.
Genbank accession numbers
The accession numbers for new DNA sequences containing the vCXCL cluster were: GMCMV(GR2757), FJ883003; GMCMV(4915), FJ883005; and GMCMV(SA6), FJ883007. Those for the vGPCR cluster were GMCMV(GR2757), FJ883004; GMCMV(4915), FJ883006; and GMCMV(SA6), FJ883008.The accession numbers of previously published DNA sequences containing all or part of the vCXCL gene cluster were: GMCMV(Stealth), AF145588; RhCMV(22659), EU130540; RhCMV(CNPRC), EF990255; RhCMV(WT2), EF990256; and RhCMV(WT3), EU003822. Those for the vGPCR gene cluster were: GMCMV(Stealth), U27802; and GMCMV(9610), AY340790, AY340791, AY340792, AY340793 and AY340794. The accession numbers of genome sequences from which the vCXCL and vGPCR clusters and DNA polymerase genes were extracted were: GMCMV(Colburn), FJ483969; GMCMV(2715), FJ483968; BaCMV (OCOM4-37), AC090446 (working draft version 27); RhCMV(68-1), AY186194; RhCMV(180.92), DQ120516; CCMV(Heberling), AF480884; HCMV(Merlin), AY446894; and OMCMV(S34E), FJ483970. Additional sequences for vCXCL genes in other HCMV strains were: additional alleles of HCMV UL146 (accession numbers in Dolan et al., 2004); and additional alleles of HCMV UL147 identified by Blast searches of GenBank. The accession numbers of the sequences of rhesus macaque, chimpanzee and human CXCL1 were NP_001028050, XP_001156038 and NP_001502, respectively. The locations of introns in the human GRO-α and IL-8 genes were derived from the annotation for human chromosome 4 (NT_006216).
Phylogenetic analysis
ClustalW (Thompson et al., 1994) was used to produce alignments of nucleic acid or predicted amino acid sequences, and the latter were adjusted manually. Bayesian inference of phylogeny was carried out on aligned sequences using MrBayes 3 (Ronquist and Huelsenbeck, 2003). For each analysis, the program was run for 2 million generations, during which 20001 trees were sampled, with the initial 15001 discarded as burn-in. The Jones-Taylor-Thornton amino acid substitution matrix was used with a gamma-plus-invariant site categories model and variable rates molecular clock model. Phylogenetic analysis of primate CMV DNA polymerases was performed using Mega4 (ClustalW alignment followed by neighbour-joining; Tamura et al., 2007). Phylogenetic trees were drawn using Mega4. TMDs were predicted from predicted primary amino acid sequences using the Phobius web server (Käll et al., 2007).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Derrick Dargan for providing OMCMV DNA, Duncan McGeoch for commenting on the manuscript, and Sarah Heaggans for assistance with PCR sequence editing and assembly. The studies at Johns Hopkins University were supported by National Institutes of Health Research Grant R01 A124576 to GSH from the National Institute of Allergy and Infectious Disease. The studies at the MRC Virology Unit were supported by the UK Medical Research Council. We thank Susan Neubauer and Earl Blewett (Oklahoma State University) for making unpublished DNA sequence data for BaCMV available.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arav-Boger R, Foster CB, Zong JC, Pass RF. Human cytomegalovirus-encoded α-chemokines exhibit high sequence variability in congenitally infected newborns. J. Infect. Dis. 2006;193:788–791. doi: 10.1086/500508. [DOI] [PubMed] [Google Scholar]
- Bodaghi B, Jones TR, Zipeto D, Vita C, Sun L, Laurent L, Arenzana-Seisdedos F, Virelizier J-L, Michelson S. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J. Exp. Med. 1998;188:855–866. doi: 10.1084/jem.188.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-N, Jeang K-T, Lietman T, Hayward GS. Structural organization of the spliced immediate-early gene complex that encodes the major acidic nuclear (IE1) and transactivator (IE2) proteins of African green monkey cytomegalovirus. J. Biomed. Sci. 1995;2:105–130. doi: 10.1007/BF02253062. [DOI] [PubMed] [Google Scholar]
- Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Hornsnell T, Hutchinson CA, III, Kouzarides T, Martignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston KM, Barrell BG. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 1990;154:126–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Daniel MD, Meléndez LV, King NW, Fraser CE, Barahona HH, Hunt RD, Garcia FG, Trum BF. Herpes virus aotus: a latent herpesvirus from owl monkeys (Aotus trivirgatus) isolation and characterization. Proc. Soc. Exp. Biol. Med. 1971;138:835–845. doi: 10.3181/00379727-138-36002. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Dolan A, Akter P, Addison C, Dargon DJ, Alcendor DJ, McGeoch DJ, Hayward GS. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 2003;84:17–28. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GWG, Davison AJ. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 2004;85:1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- Gao J-L, Murphy PM. Human cytomegalovirus open reading frame US28 encodes a functional β chemokine receptor. J. Biol. Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cytomegalovirus. J. Virol. 2003;77:6620–6636. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ES, Kilpatrick B, Lakeman A, Alford CA. Genetic analysis of a cytomegalovirus-like agent isolated from human brain. J. Virol. 1978;26:718–723. doi: 10.1128/jvi.26.3.718-723.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ES, Pagano JS. Human cytomegalovirus. II. Lack of relatedness to DNA of herpes simplex I and II, Epstein-Barr virus, and nonhuman strains of cytomegalovirus. J. Virol. 1974;13:642–645. doi: 10.1128/jvi.13.3.642-645.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K-T, Cho M-S, Hayward GS. Abundant constitutive expression of the immediate-early 94K protein from cytomegalovirus (Colburn) in a DNA-transfected mouse cell line. Mol. Cell. Biol. 1984;4:2214–2223. doi: 10.1128/mcb.4.10.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang KT, Hayward GS. A cytomegalovirus DNA sequence containing tracts of tandemly repeated CA dinucleotides hybridizes to highly repetitive dispersed elements in mammalian cell genomes. Mol. Cell. Biol. 1983;3:1389–1402. doi: 10.1128/mcb.3.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction – the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kledal TN, Rosenkilde MM, Schwartz TW. Selective recognition of the membrane-bound CX3C chemokine, fractalkine, by the human cytomegalovirus-encoded broad-spectrum receptor US28. FEBS Lett. 1998;441:209–214. doi: 10.1016/s0014-5793(98)01551-8. [DOI] [PubMed] [Google Scholar]
- Kuhn DE, Beall CJ, Kolattukudy PE. The cytomegalovirus US28 protein binds multiple CC chemokines with high affinity. Biochem. Biophys. Res. Comm. 1995;211:325–330. doi: 10.1006/bbrc.1995.1814. [DOI] [PubMed] [Google Scholar]
- LaFemina RL, Hayward GS. Structural organization of the DNA molecules from human cytomegalovirus. ICN-UCLA Symp. Mol. Biol. 1980;18:39–55. [Google Scholar]
- LaFemina RL, Pizzorno MC, Mosca JD, Hayward GS. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology. 1989;172:584–600. doi: 10.1016/0042-6822(89)90201-8. [DOI] [PubMed] [Google Scholar]
- Laing KJ, Secombes CJ. Chemokines. Dev. Comp. Immunol. 2004;28:443–460. doi: 10.1016/j.dci.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Lockridge KM, Zhou S-S, Kravitz RH, Johnson JL, Sawai ET, Blewett EL, Barry PA. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology. 2000;268:272–280. doi: 10.1006/viro.2000.0195. [DOI] [PubMed] [Google Scholar]
- Martin WJ. Melanoma growth stimulatory activity (MGSA/GRO-α) chemokine genes incorporated into an African green monkey simian cytomegalovirus-derived stealth virus. Exp. Mol. Pathol. 1999;66:15–18. doi: 10.1006/exmp.1999.2240. [DOI] [PubMed] [Google Scholar]
- Martin WJ. Chemokine receptor-related genetic sequences in an African green monkey simian cytomegalovirus-derived stealth virus. Exp. Mol. Pathol. 2000;69:10–16. doi: 10.1006/exmp.2000.2305. [DOI] [PubMed] [Google Scholar]
- Martin WJ, Ahmed KN, Zeng LC, Olsen J-C, Seward JG, Seehrai JS. African green monkey origin of the atypical cytopa8thic 'stealth virus' isolated from a patient with chronic fatigue syndrome. Clin. Diagn. Virol. 1995;4:93–103. doi: 10.1016/0928-0197(95)00020-9. [DOI] [PubMed] [Google Scholar]
- Martin WJ, Zeng LC, Ahmed K, Roy M. Cytomegalovirus-related sequence in an atypical cytopathic virus repeatedly isolated from a patient with chronic fatigue syndrome. Amer. J. Pathol. 1994;145:440–451. [PMC free article] [PubMed] [Google Scholar]
- Michelson S, Dal Monte P, Zipeto D, Bodaghi B, Laurent L, Oberlin E, Arenzana-Seisdedos F, Virelizier JL, Landini MP. Modulation of RANTES production by human cytomegalovirus infection of fibroblasts. J. Virol. 1997;71:6495–6500. doi: 10.1128/jvi.71.9.6495-6500.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Kittrell M, Sai J, Penfold M, Richmond A, Sparer TE. Functional characterization of chimpanzee cytomegalovirus chemokine, vCXCL-1CCMV. Virology. 2007;364:454–465. doi: 10.1016/j.virol.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neote K, DiGregorio D, Mak JY, Horuk R, Schall TJ. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- Nicholas J, Ruvolo V, Zong J-C, Ciufo D, Guo H-G, Reitz MS, Hayward GS. A single 13-kilobase divergent locus in Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains at least nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas J, Zong J-C, Alcendor DJ, Ciufo DM, Poole LJ, Sarisky RT, Chiou C-J, Zhang X, Wan X, Guo H-G, Reitz MS, Hayward GS. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J. Natl. Canc Inst. Monogr. 1998;23:79–88. doi: 10.1093/oxfordjournals.jncimonographs.a024179. [DOI] [PubMed] [Google Scholar]
- Oxford KL, Eberhardt MK, Yang K-W, Strelow L, Kelly S, Zhou S-S, Barry PA. Protein coding content of the ULb' region of wild-type rhesus cytomegalovirus. Virology. 2008;373:181–188. doi: 10.1016/j.virol.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold MET, Dairaghi DJ, Duke GM, Saederup N, Mocarski ES, Kemble GW, Schall TJ. Cytomegalovirus encodes a potent α chemokine. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold MET, Schmidt TL, Dairaghi DJ, Barry PA, Schall TJ. Characterization of the rhesus cytomegalovirus US28 locus. J. Virol. 2003;77:10404–10413. doi: 10.1128/JVI.77.19.10404-10413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivailler P, Kaur A, Johnson RP, Wang F. Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J. Virol. 2006;80:4179–4182. doi: 10.1128/JVI.80.8.4179-4182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rue CA, Jarvis MA, Knoche AJ, Meyers HL, DeFilippis VR, Hansen SG, Wagner M, Fruh K, Anders DG, Wong SW, Barry PA, Nelson JA. A cyclooxygenase-2 homologue encoded by rhesus cytomegalovirus is a determinant for endothelial cell tropism. J. Virol. 2004;78:12529–12536. doi: 10.1128/JVI.78.22.12529-12536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahagun-Ruiz A, Sierra-Honigmann AM, Krause P, Murphy PM. Simian cytomegalovirus encodes five rapidly evolving chemokine receptor homologues. Virus Genes. 2004;28:71–83. doi: 10.1023/B:VIRU.0000012265.33168.b5. [DOI] [PubMed] [Google Scholar]
- Sparer TE, Gosling J, Schall TJ, Mocarski ES. Expression of human CXCR2 in murine neutrophils as a model for assessing cytomegalovirus chemokine vCXCL-1 function in vivo. J. Interferon Cytokine Res. 2004;24:611–620. doi: 10.1089/jir.2004.24.611. [DOI] [PubMed] [Google Scholar]
- Steiper ME, Young NM. Primate molecular divergence dates. Mol. Phylogenet. Evol. 2006;41:384–394. doi: 10.1016/j.ympev.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Streblow DN, Soderberg-Naucler C, Vieira J, Smith P, Wakabayashi E, Ruchti F, Mattison K, Altschuler Y, Nelson JA. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell. 1999;99:511–520. doi: 10.1016/s0092-8674(00)81539-1. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, Schall TJ, Corey L, Geballe AP. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J. Virol. 1998;72:8158–8165. doi: 10.1128/jvi.72.10.8158-8165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.