Abstract
Metabolite-sensing regulatory RNAs, oft referred to as riboswitches, are widely used among eubacteria for control of diverse biochemical pathways and transport mechanisms. Great strides have been made in understanding the general structure and biochemistry of individual riboswitch classes. However, along with these advancements, it has become clear that metabolite-sensing riboswitches respond to an increasingly structurally diverse range of metabolite and metal ligands. Moreover, the recent accruement of new riboswitches has uncovered individual examples and classes that utilize unique regulatory strategies or employ a regulatory logic other than simple feedback inhibition.
Background
Since the initial description of the lac operon over fifty years ago, genetic control in eubacteria has been presumed to result mostly from protein factors as the agents of regulatory function. However, discoveries over the past ten years have illuminated an ever-expanding role for noncoding RNAs in the control of bacterial gene expression. These RNAs employ a variety of mechanisms to exert their regulatory functions but can be broadly segregated into several distinct classes. Specifically, small noncoding regulatory RNAs (sRNAs) are generally transcribed as independent transcripts that interact with target mRNAs through base-pairing interactions [1, 2]. Other sRNAs affect gene expression by associating with specific RNA-binding proteins or with RNA polymerase [3,4]. In contrast, regulatory RNAs can also be enslaved within the target transcript that they regulate (cis-acting regulatory RNAs). In bacteria, examples of the latter category are largely located within the 5′ untranslated region (5′ UTR) of the transcripts that they regulate. These cis-acting regulatory elements individually respond to many different types of effector signals, including fluctuations in temperature, RNA-binding proteins and trans-encoded RNAs (tRNAs or sRNAs) (Figure 1) [5,6,7]. Additionally, a growing collection of metabolite-sensing regulatory RNAs is important for regulation of fundamental biochemical pathways and essential cellular functions. Multiple classes of these metabolite-sensing RNAs, commonly called riboswitches, have been analyzed through extensive biochemical, biophysical, and structural methodology [8,9,10]. However, equally significant to these biochemical advancements are recent discoveries that highlight the diversity of their metabolic ligands, genetic mechanisms, and regulatory logic. The intent of this review is to briefly summarize these latter advancements while speculating on what these insights may portend for future riboswitch discoveries.
Figure 1.
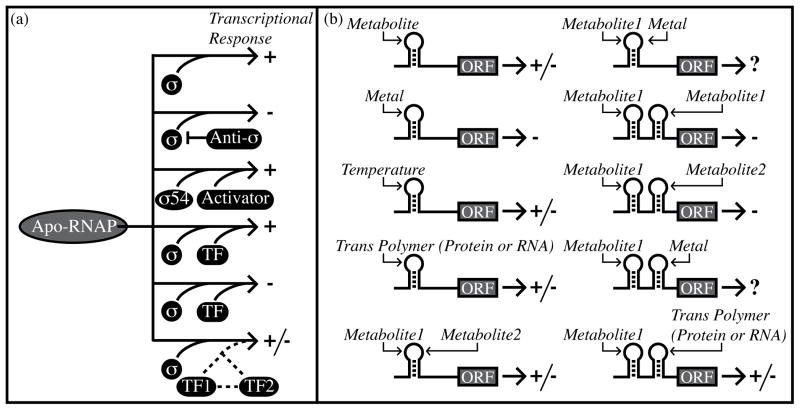
Regulation of bacterial gene expression by transcriptional and posttranscriptional mechanisms. (a) Simplified view of transcriptional regulation using alternative sigma factors and DNA-binding transcription factors. Arrows point to the genetic outcome. Plus and minus signs indicate transcriptional activation and repression, respectively. Sigma factors promote transcription initiation through sequence-specific DNA contacts. Sigma 54 is an exception in that it requires a protein factor for transcriptional activation. Anti-sigma factors prevent sigma function. Transcription factors (“TF”) can individually activate or repress expression. Also, transcription factors can coordinate with one another to produce complex regulatory outcomes. (b) Simplified view of posttranscriptional regulation. Plus and minus symbols indicate increased or decreased gene expression. Hairpin structures denote individual cis-regulatory RNAs, which can respond to temperature, metabolites, metals, or trans-encoded polymers (RNAs or proteins). Alternatively, two different metabolites can associate with the same cis-regulatory RNA [25]. It is also possible that a particular, hypothetical cis-regulatory RNA may respond to a combination of metals and metabolite concentrations. Metabolite-sensing riboswitches may be arranged in tandem to respond to the same metabolite or to two different metabolite ligands [25,28]. It is also possible for multiple, distinct posttranscriptional regulatory mechanisms to cooperate in controlling downstream expression [30].
An increasingly structurally diverse repertoire of riboswitch ligands
All riboswitches include an aptamer portion, an intricately structured and highly selective receptor domain that is conformationally altered upon ligand association. For most riboswitches these conformational changes result in sequestration of sequences involved in formation of mutually exclusive secondary structures within the downstream region. The latter secondary structure elements oftentimes share a physical relationship to either transcription termination signals or the ribosomal binding region, suggesting a basic mechanism for ligand-induced control of gene expression (Figure 2a–b). However, for certain riboswitches, nucleotides from the ribosomal binding region are intimately or directly involved in ligand recognition. For these riboswitches, metabolite association is directly coupled with gene control.
Figure 2.
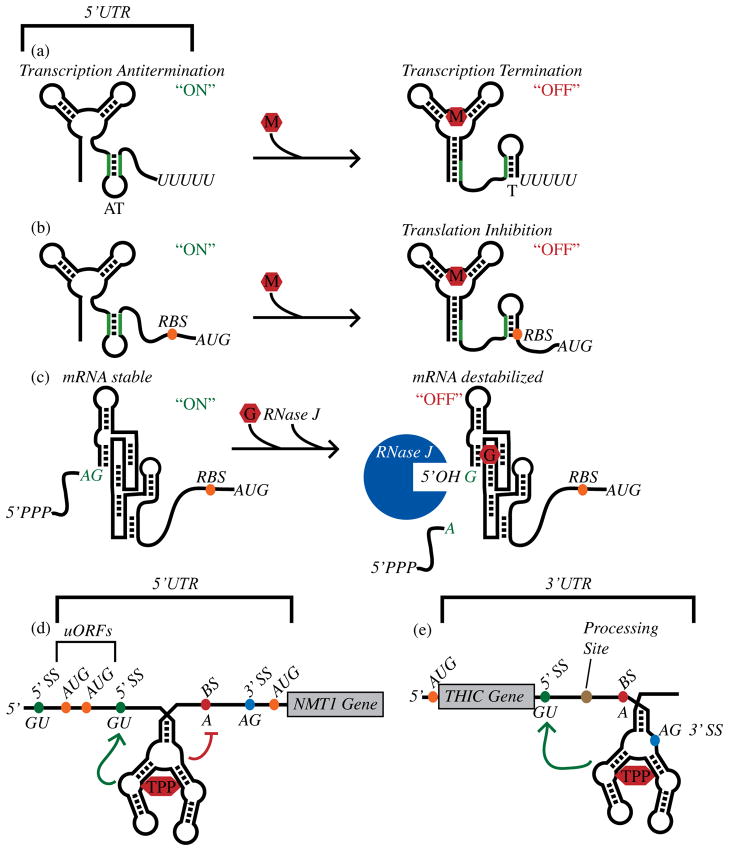
Genetic control by metabolite-sensing riboswitches. (a) Control of formation of an intrinsic transcription terminator by a metabolite-binding riboswitch. “M” denotes the metabolite ligand. (b) Control of translation initiation efficiency by a metabolite-sensing riboswitch. (c) Control of mRNA stability by the GlcN6P-sensing ribozyme as described in the text. “G” denotes the GlcN6P riboswitch ligand. (d) An example of a TPP riboswitch that controls alternative splicing of the NMT1 gene in Neurospora crassa [37], as discussed in the text. “5′ SS” indicates the 5′ splice sites. “BS” indicates a branch site. (e) As discussed in the text, a TPP riboswitch from the 3′ UTR of Arabidopsis thaliana THIC controls mRNA processing and stability [38]. Green arrows signify regions where structural rearrangements occur upon TPP binding and that activate splicing elements. The red line identifies a region where structural rearrangements occur upon TPP binding and that repress splicing elements.
Prior to 2006, ten classes of metabolite-sensing riboswitches were discovered, individually responding to a variety of metabolites [8,9,11]. Three of these classes were found to respond to enzymatic cofactors (adenosylcobalamin (AdoCbl), thiamine pyrophosphate (TPP), and flavin mononucleotide (FMN)). Three separate riboswitch classes were found to respond to S-adenosylmethionine (SAM). Riboswitches had also been identified that sense amino acids (glycine and lysine) as well as an amino sugar, glucosamine-6-phosphate (GlcN6P). Finally, a riboswitch class had been demonstrated to respond to either a guanine or adenine nucleobase. These discoveries spurred further development of bioinformatics-based approaches for riboswitch identification, resulting in discovery of a plethora of ‘orphan’ riboswitches that closely resemble classical riboswitches but whose metabolite ligands remain unidentified. During the past several years, analyses of these orphan RNAs have led to identification of seven new riboswitch classes that respond to small organic molecules [12–19]. Four of these new riboswitch classes sense nucleobase-containing ligands (deoxyguanosine (dG), cyclic diguanosine monophosphate (cyclic di-GMP), preQ1 – an intermediate of queuosine biosynthesis). Two classes sense enzymatic cofactors (S-adenosylhomocysteine (SAH), molybdenum cofactor (Moco)) and the remaining class is proposed to function as an additional amino acid sensor, responsive to arginine.
A minimal natural aptamer domain
Riboswitch aptamer domains typically vary between ~70–200 nt in length [6]. Indeed, based upon the average size (~150 nt) of the first three biochemically validated riboswitch classes (AdoCbl, TPP, FMN) one might have assumed all aptamer domains would require similar lengths and secondary structure complexity. However, recently, a few riboswitch aptamers have been identified that appear to exhibit more simplified secondary structure arrangements and shorter sequence requirements. For example, an aptamer domain for one of the SAM riboswitch classes is only ~55 nt in length and has been shown to adopt a common, compact H-type pseudoknot fold [20]. More recently, a riboswitch class that responds to an intermediate (7-aminomethyl-7-deazaguanine (preQ1)) in the biosynthesis of queuosine, a hypermodified nucleoside in certain tRNAs, sets a new standard for ‘small’ riboswitches as it requires only ~34 nt [13]. Yet, despite its relatively simple secondary structure and short length this riboswitch can still attain high selectivity and affinity (~50 nM) for the appropriate ligand (preQ1). Small, simple motifs such as these RNA elements are notoriously difficult to identify through bioinformatics alone. Therefore, these discoveries signal that it may be possible that many other small riboswitch aptamers may still await discovery, perhaps even including signal-responsive versions of similarly sized, ubiquitous elements such as ribosomal frameshifting H-type pseudoknot folds.
RNA-mediated metalloregulation
Most chemicals sensed by riboswitches are metabolic byproducts and enzymatic cofactors. However, several riboswitch classes sense metal ions as an important recognition determinant of the appropriate metabolite ligands. For example, molybdenum cofactor (Moco) is a tricyclic pyranopterin that coordinates one atom of the transition metal molybdenum and that participates in redox reactions in the carbon, nitrogen, and sulphur metabolic cycles [21]. Recently, a Moco riboswitch candidate was discovered for regulation of (Moco) biosynthesis and availability [12,15]. Interestingly, certain variants of the Moco-sensing riboswitch are thought to specifically recognize tungsten cofactor (Tuco), which resembles Moco except for substitution of tungsten for molybdenum. Similarly, chelation of the appropriate metal is important for ligand recognition by AdoCbl-sensing riboswitches. However, recent data also indicate that riboswitches can function as direct sensors of intracellular metals, suggesting that RNA polymers may functionally substitute for metalloregulatory proteins. Specifically, two separate Mg2+-responsive RNA elements have been discovered within the 5′ UTR of transcripts encoding for Mg2+ transport genes [22,23]. For Salmonella enterica, a Mg2+-responsive regulatory RNA has been identified within the mgtA 5′ UTR that may control feedback repression of downstream gene expression via Mg2+-induced transcription termination [22]. A structurally distinct Mg2+-responsive regulatory RNA coined the M-box was identified in the Bacillus subtilis mgtE 5′ UTR [23••]. Association of Mg2+ with M-box RNAs induces a compacted tertiary structural configuration that directly sequesters nucleotides otherwise required for antiterminator helix formation. Functional biochemical analyses reveal that at least six Mg2+ are required for this conformation (Wakeman CA and Winkler WC, unpublished data), establishing the M-box as a multi-ligand sensor. In total, structural and biochemical analyses of the M-box RNA reveal that it shares many common structural and mechanistic features with metabolite-sensing riboswitches, suggesting that metal-sensing aptamer domains may be indistinguishable from those that respond to small organic molecules. Intriguingly, multiple additional classes of orphan riboswitches are predicted to regulate metal transport genes suggesting that there may be other classes of metalloregulatory RNAs to be identified.
Combinatorial control by cis-acting regulatory RNAs
Enough riboswitch-based regulatory mechanisms have been discovered that it is worthwhile to begin to compare transcriptional and posttranscriptional strategies as genetic regulatory solutions. For transcription initiation-based mechanisms, complex regulatory outcomes can be easily achieved using only a few accessory protein factors, such as sigma factors and DNA-binding transcription factors (Figure 1). Combinations of these components allow for control of expression in response to combinatorial signals and with variations in individual parameters of the regulatory response (e.g., cooperativity, robustness, rapidity, dynamic range, etc.). In contrast, most metabolite-sensing riboswitches respond to a single metabolic cue and function through simple feedback inhibition. However, several unique riboswitch arrangements have been uncovered recently that hint at greater complexity. For example, tandem arrangements of cis-acting regulatory RNAs have been identified for multiple classes of metabolite- and tRNA-sensing RNA elements (Figure 1b) [24–28]. The sequential arrangement of fully intact riboswitches can alter the dynamic range of the regulatory response such that the regulatory response has digital-like character or a modest increase in ligand sensitivity [24,25,28]. In addition to shrinking the dynamic range required for maximal gene expression through tandem riboswitches that sense the same ligand, the sequential arrangement of riboswitches responding to different ligands can allow downstream genes to be independently regulated by two chemical input signals [25]. Moreover, it stands to reason that sequential or coordinated regulatory RNAs need not be comprised only of metabolite-sensing RNAs. Indeed, it has now become clear that multiple classes of posttranscriptional regulatory mechanisms can coordinate with one another to regulate gene expression. For example, recent data on an ethanolamine catabolic operon (eut) in Enterococcus, Listeria, and Clostridium species revealed coordinated regulation of the eut locus by multiple posttranscriptional mechanisms, including an AdoCbl-activated riboswitch and an ethanolamine-activated RNA-binding protein [30]. A nearly identical operon in Salmonella is also regulated in response to adenosylcobalamin and ethanolamine but instead through their association with a single DNA-binding transcription factor [31]. Therefore, these data together are beginning to demonstrate how transcriptional and posttranscriptional regulatory mechanisms offer equivalent solutions in different organisms for common sets of regulatory tasks.
An increasingly diverse repertoire of riboswitch genetic mechanisms
Riboswitches elicit control of gene expression primarily through the use of two mechanisms: transcription attenuation and translation inhibition (Figure 2). For the former, association of riboswitch ligands is coupled with a change in secondary structure arrangement of the 5′ UTR such that a transcription termination signal is either promoted or prevented, depending on whether the riboswitch activates or represses expression. Similarly, metabolite-induced conformational changes can influence the relative accessibility of the ribosome binding site in order to control translation initiation efficiency. However, in the past few years several riboswitch classes have been demonstrated to control gene expression through other mechanisms, broadening expectations for the functional roles of metabolite-sensing RNAs.
Control of bacterial mRNA stability
An alternate genetic mechanism was proposed for the glucosamine-6-phosphate (GlcN6P)-sensing RNA, located upstream of the glmS gene [29]. Prior studies revealed that GlcN6P promoted an autocatalytic, site-specific cleavage event near the 5′ terminus in vitro [8,32,33]. A reciprocal relationship was observed between ribozyme self-cleavage and gene expression in vivo, suggesting that the B. subtilis glmS ribozyme regulated gene expression through feedback repression. However, the underlying genetic mechanism was not initially intuitively obvious. The most straightforward expectation was that a rise in GlcN6P would induce self-cleavage by the glmS ribozyme, which would then impart a shortened half-life on the glmS transcript (Figure 2c). However, ribozyme self-cleavage results in a 5′ cleavage product containing a 2′, 3′-cyclic phosphate and a downstream 3′ cleavage product (corresponding to the glmS transcript) containing a 5′ hydroxyl group. In E. coli, mRNAs containing a 5′ hydroxyl group are degraded poorly. Therefore, based upon current models for mRNA degradation pathways in E. coli, the downstream glmS transcript should be stabilized rather than destabilized upon self-cleavage, in contrast to expectations for negative feedback-based control of mRNA stability. This conundrum was solved upon demonstration that an RNase enzyme present in B. subtilis but absent in E. coli is explicitly required for degradation of the 3′ cleavage product upon glmS self-cleavage. Destabilization of the 3′ cleavage product by the enzyme, RNase J, was dependent upon the presence of a 5′ hydroxyl; RNAs with a 5′ triphosphate group resulted in significant intracellular accumulation. This contrasts with mRNA degradation pathways in E. coli, which are mediated largely by the endoribonuclease RNase E. RNase E exhibits a preference for RNA substrates containing a 5′ monophosphate groups and degrades only poorly those with a 5′ hydroxyl or triphosphate group [34,35]. Together these data suggest that recognition of the 5′ terminus of bacterial transcripts is likely to be a rate-limiting step in mRNA turnover for bacteria in general, regardless of the identity of the central RNase that is involved. With this knowledge, it should be possible to hunt for more examples of natural signal-responsive genetic ribozymes and begin the practical design of synthetic versions.
Control of eukaryotic splicing
One of the major differences in genetic organization between eubacteria and eukaryotes is the presence of introns within pre-mRNAs, which must be excised in order to yield functional transcripts. Alternative splicing is the process in which introns are excised and exons are separated and rejoined in different combinations to produce alternative mRNA transcripts [36]. In most instances regulation of splicing is dependent on the interaction of RNA-binding proteins and pre-mRNA sequences proximal to splice sites. However, for certain fungal, plant, and algal species, alternative splicing of genes involved in thiamine biosynthesis is directly controlled by TPP riboswitches located within intronic regions (Figure 2d, e). For Neurospora crassa, a total of three genes are regulated thusly; two are repressed in response to TPP, while expression of the third gene is activated [37–40]. Binding of TPP to these riboswitches alters availability of a nucleotide tract within the riboswitch that could otherwise form base-pairing interactions with a riboswitch-proximal splice site or branch point components. In this manner, availability of TPP controls whether or not an upstream open reading frame (uORF) is formed, the presence of which decreases expression of the downstream, main ORF. TPP also controls formation of an uORF for regulation of thiamine biosynthesis genes in the algal species, Chlamydomonas reinhardti and Volvox carteri [40]. Finally, TPP riboswitches have also been found to control alternative splicing of plant biosynthesis genes (Figure 2e) [38,39]. However, these TPP riboswitches are located within the 3′ UTR of plant thiamine synthesis genes. Under low thiamine conditions a short oligonucleotide stretch within the riboswitch is capable of base-pairing to a complementary sequence that overlaps the 5′ splice site, thereby preventing splicing at that site. This particular splicing pattern then results in inclusion of an RNA processing site that leads to a shortened 3′ UTR, conditions that lead to mRNA stabilization and increased translation of the target gene. Upon an increase in TPP, the riboswitch oligonucleotide tract cannot pair with the 5′ splice site and the altered splicing pattern leads to a loss of the 3′ UTR mRNA processing site and, ultimately, destabilization of the overall transcript. At the heart of these mechanisms is the ligand-mediated influence on mutually exclusive base-paired regions, conceptually common amongst all riboswitches. Therefore, discovery of these mechanisms would suggest there is no reason not to anticipate that other riboswitches could control eukaryotic alternative splicing and stability.
An increasingly diverse repertoire of riboswitch functional roles
Feedback inhibition
Most but not all riboswitches have been found to control gene expression through feedback inhibition of biosynthesis or transport genes (Figure 3). However, increasingly diverse functional roles have been observed for a subset of riboswitches as greater numbers of individual examples and new classes are identified. Even novel variations of simple feedback inhibition mechanisms have been discovered in the past several years. For example, rare representatives of a subclass of the well-characterized purine riboswitch structural element were recently found upstream of ribonucleotide reductase genes in Mollicute species [41]. These RNAs resembled classical purine riboswitches except for carrying certain key nucleotide substitutions within the ligand-binding core. Biochemical tests of this riboswitch variant revealed that it bound specifically the nucleoside deoxyguanosine. Therefore, it would appear that for these microorganisms the purine riboswitch, which is usually employed for feedback inhibition of purine biosynthesis, evolved the capacity to recognize a structurally related but distinct metabolite ligand for pseudo-feedback regulation of ribonucleotide reductase activity. This discovery highlights the evolutionary malleability of riboswitch aptamer domains and genetic mechanisms.
Figure 3.
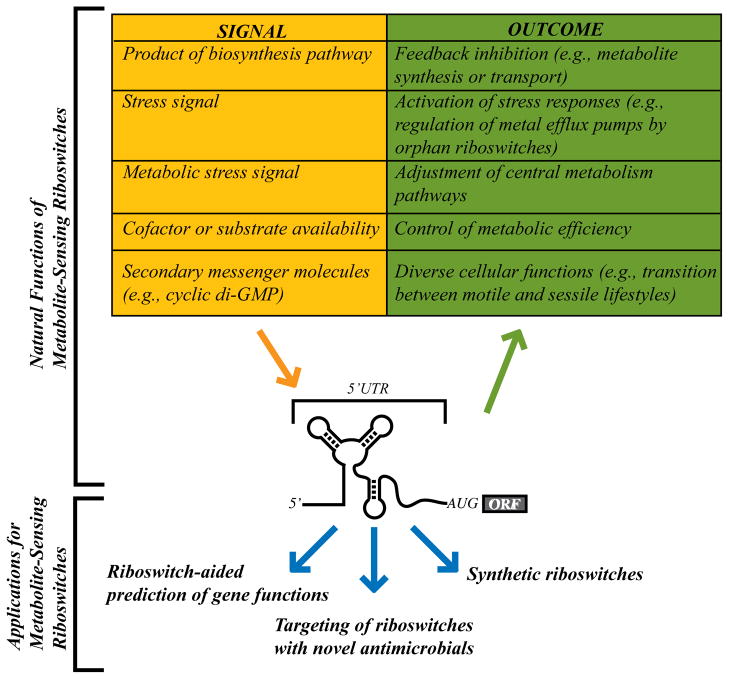
Applications and biological roles of riboswitch RNAs. Metabolite-sensing riboswitches have been discovered that respond to a variety of metabolites. Most riboswitches are used in feedback loops for negative regulation of biosynthesis or transport genes. However, certain riboswitch classes respond to stress signals or the availability of a key enzymatic cofactor or substrate. A particular riboswitch class responds to the second messenger, cyclic di-GMP, for control of numerous fundamental cellular processes. In addition to these natural roles, riboswitches are also increasingly viewed as useful tools in assisting the functional analyses of riboswitch-associated genes, as drug targets, and as scaffolds for construction of artificial or reengineered riboswitches [44–46].
Sensing of key cofactors or metabolic substrates
A substantial proportion of proteins expressed within the cell rely upon cofactors for catalysis. Many riboswitch examples function as sensors of enzymatic cofactors. Yet, the majority of these riboswitches are used solely for feedback regulation of cofactor biosynthesis or transport genes. However, recent examples reveal riboswitches that regulate expression of enzymes based upon availability of the cofactor to which they bind. In general, these latter riboswitches are utilized for maximizing metabolic efficiency by limiting expression of inefficient enzymes until concentrations of key cofactors are sufficient. One such example can be found with Bacillus clausii, which expresses two different proteins designated metE and metH for conversion of homocysteine to methionine and a third gene, metK, for conversion of methionine to SAM [25]. Expression for all three genes is repressed by a SAM-sensing riboswitch. Interestingly, in addition to the SAM riboswitch the metE 5′ UTR also contains an AdoCbl riboswitch. This extra regulatory mechanism allows the cell to repress metE under conditions of plentiful AdoCbl, instead favoring expression of metK, which is more efficient but is strictly dependent upon AdoCbl for catalysis [25]. Similarly, certain Moco-dependent enzymes are also subject to regulation by the Moco riboswitch [15]. Another example of a riboswitch that regulates gene expression in response to enzyme cofactor availability is that of ribonucleotide reductase (RNR) in Streptomyces coelicolor [41]. This organism possesses two RNRs (termed class Ia and class II), although either enzyme is sufficient for vegetative growth. The class Ia RNR requires oxygen for activity while the class II does not require oxygen but is dependent on AdoCbl. An AdoCbl riboswitch present in the 5′ UTR of the class Ia RNR is responsible for signaling to the cell when AdoCbl levels are sufficient that it should switch to the more efficient class II enzyme.
Yet another example for how availability of cofactors can be a driving force behind riboswitch evolution is the discovery of a unique AdoCbl subclass for control of ethanolamine catabolism (eut locus) in Enterococcus, Listeria, and Clostridium species [30]. Unlike the classical AdoCbl riboswitch the eut AdoCbl switch activates downstream eut expression in response to sufficient AdoCbl, a key cofactor for the rate-limiting step in ethanolamine catabolism.
Sensing of stress signals
Another theme in riboswitch-mediated regulation that will undoubtedly continue to emerge is the sensing of metabolites for control of different stress responses. For example, glycine riboswitches are most often utilized by bacteria for activation of genes encoding for glycine catabolism or efflux upon encountering toxic leves of glycine. [24]. Most recently, this regulatory prowess was shown to extend to control of central metabolism pathways as well [42–43]. Specifically, a glycine riboswitch was recently found to control expression of malate synthase in the marine bacterium, Candidatus Pelagibacter ubique. This arrangement presumably allows the organism to appropriately balance flux through the TCA cycle with the glyoxylate cycle, with glycine serving as an indicator of extracellular nutrient status.
S-adenosylhomocysteine (SAH) is another metabolite that is toxic if allowed to accumulate intracellularly, due to inhibition of SAM-dependent methyltransferases. SAM riboswitches preferentially recognize SAM over SAH. Recently, an RNA element was reported that selectively binds SAH and upregulates genes involved in the conversion of SAH to methionine [17]. This motif is predominantly found in β and γ-proteobacteria and in nearly all cases resides in front of a four gene operon encoding enzymes required for SAH detoxification. The SAH element does not exhibit any sequence or secondary structural similarities to the various SAM aptamers and is able to discriminate SAH over SAM by more than 1,000 fold [17], a remarkable feat when one considers that SAM and SAH differ by only one methyl group and a positively charged sulfur atom. More riboswitches dedicated to stress response regulation are very likely to be identified. Indeed, multiple classes of orphan riboswitches also appear to control stress response genes (e.g., metal efflux genes) in response to unknown signals.
Sensing of second messenger metabolites
Arguably the most exciting new functional role for metabolite-sensing riboswitches is the demonstration of a riboswitch responding to a bacterial second messenger molecule. Second messenger molecules are diffusible agents affecting gene expression or protein activity for many cellular responses by virtue of their intracellular concentration, which is established by controlling the activity of enzymes responsible for synthesis and degradation of the messenger molecule. Bacteria employ an array of second messengers, including cyclic di-guanosine monophosphate (cyclic di-GMP). Many different physiological and developmental processes, including regulation of virulence factors, motility, DNA uptake factors, and biofilm formation, are controlled by the synthesis and breakdown of cyclic di-GMP. The mechanisms for coupling cyclic di-GMP to these cellular responses are largely unresolved. However, a recent report identified a riboswitch class, known as the GEMM motif (for Genes for the Environment, for Membranes and for Motility), as a high affinity sensor for cyclic-di-GMP [18]. Biochemical analysis found the riboswitch to contain a single, saturable binding site, exhibiting a dissociation constant (KD) of approximately 1 nM. In concordance with a role in cyclic di-GMP signaling the GEMM riboswitch is predicted to regulate many different genes, including those encoding for flagellar assembly and regulation, competence factors, virulence factors, membrane transporters, cyclic di-GMP synthesis (GGDEF domain proteins), cyclic di-GMP degradation (EAL domain proteins), and many proteins of unknown function. Therefore, considering the exceptional ability of riboswitches in aiding prediction of function for unknown genes, the GEMM motif is likely to aid functional analyses of genes involved in the transition between motile and sessile lifestyles and in virulence gene regulation. Interestingly, the GEMM riboswitch is unusual in its application as both an “on” switch and “off” switch. While this is rarely observed with other riboswitches, the GEMM riboswitch appears to be freely interchangeable for metabolite-induced activation or repression of downstream expression, even within the same host cell. Remarkably, one organism, Geobacter uraniumreducens, contains 25 separate transcriptional units individually regulated by this riboswitch class. Of these, five are preceded by a tandem arrangement of cyclic di-GMP aptamers, raising the intriguing, albeit speculative hypothesis that not all intracellular cyclic di-GMP riboswitches will provoke cellular responses at the same ligand concentration, or with use of a similar dynamic range for the regulatory response. Clearly, identification of GEMM riboswitches in different bacterial species is going to continue to provide unique insights into the study of cyclic di-GMP regulation. However, a next challenge will be in deciphering how these RNA elements are carefully coordinated and what mechanisms are utilized for organisms that appear to lack the GEMM motif but still utilize cyclic di-GMP signaling pathways.
Conclusion
A first generation of riboswitch analysis resulted in the discovery of several riboswitch classes and established their general importance in regulation of important biochemical pathways. This was rapidly followed by biochemical advancements and atomic-resolution structures of the molecular mechanisms used by riboswitches for ligand discrimination and genetic control. Most of these riboswitches were found to sense metabolic products for feedback control of biosynthesis genes or transporters. Recent discoveries have built upon all of these prior findings by revealing that riboswitches are also used to sense molecules outside of metabolic pathways, such as second messenger metabolites or intracellular metals. Recent discoveries have also broken boundaries by demonstrating that certain riboswitches can employ genetic mechanisms other than transcription attenuation or translation inhibition. What will be most exciting in the future will not simply be more discoveries of metabolite-binding RNAs, indeed that has become an expectation, but instead the investigation of their functional equivalency with protein-based mechanisms and unrelated posttranscriptional mechanisms, and the full range of their capabilities in controlling metabolic pathways.
Acknowledgments
Work on RNA-mediated regulation in the Winkler lab has been supported by the Searle Scholars Program, Welch Foundation (I-1643), National Institutes of Health (RO1GM081882, R21AI078104), and The University of Texas Southwestern Medical Center Endowed Scholars Program.
References
- 1.Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 2.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Storz G, Opdyke JA, Wassarman KM. Regulating bacterial transcription with small RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:269–273. doi: 10.1101/sqb.2006.71.033. [DOI] [PubMed] [Google Scholar]
- 5.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irnov, Kertsburg A, Winkler WC. Genetic control by cis-acting regulatory RNAs in Bacillus subtilis: general principles and prospects for discovery. Cold Spring Harb Symp Quant Biol. 2006;71:239–249. doi: 10.1101/sqb.2006.71.021. [DOI] [PubMed] [Google Scholar]
- 7.Narberhaus F, Waldminghaus T, Chowdhury S. RNA thermometers. FEMS Microbiol Rev. 2006;30:3–16. doi: 10.1111/j.1574-6976.2005.004.x. [DOI] [PubMed] [Google Scholar]
- 8.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 9.Edwards TE, Klein DJ, Ferre-D’Amare AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 11.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 14.Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci U S A. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regulski EE, Moy RH, Weinberg Z, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol Microbiol. 2008;68:918–932. doi: 10.1111/j.1365-2958.2008.06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA. 2008;14:685–695. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell. 2008;29:691–702. doi: 10.1016/j.molcel.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borsuk P, Przykorska A, Blachnio K, Koper M, Pawlowicz JM, Pekala M, Weglenski P. L-arginine influences the structure and function of arginase mRNA in Aspergillus nidulans. Biol Chem. 2007;388:135–144. doi: 10.1515/BC.2007.015. [DOI] [PubMed] [Google Scholar]
- 20•.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. Atomic resolution structure of a S-adenosylmethionine-sensing riboswitch revealed how a common architecture (H-type pseudoknot) can be adapted for riboswitch function. [DOI] [PubMed] [Google Scholar]
- 21.Baugh PE, Collison D, Garner CD, Joule JA. Molybdenum metalloenzymes. In: Sinnott M, editor. ComprehensiveBiological Catalysis. III Academic Press; 1998. pp. 377–400. [Google Scholar]
- 22•.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. This manuscript provides the first experimental evidence suggesting that riboswitches may function as sensors of metal ions instead of small organic molecules. [DOI] [PubMed] [Google Scholar]
- 23••.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. The manuscript describes the detailed analysis of a metal-sensing regulatory RNA and revealed many similarities and some differences between metalloregulatory RNAs and metabolite-sensing riboswitches. [DOI] [PubMed] [Google Scholar]
- 24.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 25•.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. The authors provide a demonstration that riboswitches can function in a combinatorial manner for complex control of gene expression. [DOI] [PubMed] [Google Scholar]
- 26••.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. This manuscript provides the first description of a riboswitch that responds to a second messenger molecule. These data demonstrate that riboswitches can be used to control complex cellular behavior, including organelle synthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. A thorough bioinformatics-based analysis of a class of regulatory RNAs, T box RNAs, that is widely used amongst Fimicutes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 30.Fox KARA, Stearns JE, Bourgogne A, Reyes A, Winkler WC, Garsin DA. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilizationEnterococcus faecalis. Proceedings of the National Academy of Sciences; 2008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roof DM, Roth JR. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J Bacteriol. 1992;174:6634–6643. doi: 10.1128/jb.174.20.6634-6643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein DJ, Ferre-D’Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 33.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Condon C. Maturation and degradation of RNA in bacteria. Curr Opin Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 37••.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. This is one of two manuscripts that reveal how riboswitches are used to control fungal or algal alternative splicing for control of an upstream ORF and inhibition of a downstream coding sequence. Similar to bacterial riboswitches, ligand-mediated control of mutually exclusive secondary structure elements are at the heart of the mechanism. [DOI] [PubMed] [Google Scholar]
- 38•.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. This is one of two manuscripts that reveal how a riboswitch is used in plants for control of mRNA stability, which is in turn controlled by alternative spliicig. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. The second manuscript that reveals how a riboswitch is used in plants for control of mRNA stability, which is in turn controlled by alternative spliicig. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci U S A. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. This is the second manuscript that reveals how riboswitches are used to control fungal or algal alternative splicing for control of an upstream ORF and inhibition of a downstream coding sequence. Similar to bacterial riboswitches, ligand-mediated control of mutually exclusive secondary structure elements are at the heart of the mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borovok I, Gorovitz B, Schreiber R, Aharonowitz Y, Cohen G. Coenzyme B12 controls transcription of the Streptomyces class Ia ribonucleotide reductase nrdABS operon via a riboswitch mechanism. J Bacteriol. 2006;188:2512–2520. doi: 10.1128/JB.188.7.2512-2520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tripp HJ, Schwalbach MS, Meyer MM, Kitner JB, Breaker RR, Giovannoni SJ. Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ Microbiol. 2009;11:230–238. doi: 10.1111/j.1462-2920.2008.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kazanov MD, Vitreschak AG, Gelfand MS. Abundance and functional diversity of riboswitches in microbial communities. BMC Genomics. 2007;8:347. doi: 10.1186/1471-2164-8-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallivan JP. Toward reprogramming bacteria with small molecules and RNA. Curr Opin Chem Biol. 2007;11:612–619. doi: 10.1016/j.cbpa.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Sharma V, Nomura Y, Yokobayashi Y. Engineering Complex Riboswitch Regulation by Dual Genetic Selection. J Am Chem Soc. 2008 doi: 10.1021/ja805203w. The latest in a line of manuscripts describing efforts at engineering synthetic riboswitch. In this case, the authors demonstrate a unique in vivo pathway for reengineering of riboswitch genetic mechanisms. [DOI] [PubMed] [Google Scholar]
- 46.Suess B, Weigand JE. Engineered riboswitches: overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]