Abstract
Differentiation of the Drosophila retina occurs as a morphogenetic furrow sweeps anteriorly across the eye imaginal disc, driven by Hedgehog secretion from photoreceptor precursors differentiating behind the furrow. A BTB protein, Roadkill, is expressed posterior to the furrow and targets the Hedgehog signal transduction component Cubitus interruptus for degradation by Cullin-3 and the proteosome. Clonal analysis and conditional mutant studies establish that roadkill transcription is activated by the EGF receptor and Ras pathway in most differentiating retinal cells, and by both EGF receptor/Ras and by Hedgehog signaling in cells that remain unspecified. These findings outline a circuit by which Hedgehog signal transduction is modified as Hedgehog signaling initiates retinal differentiation. A model is presented for regulation of the Cullin-3 and Cullin-1 pathways that modifies Hedgehog signaling as the morphogenetic furrow moves and the responses of retinal cells change.
Keywords: Drosophila eye, Signal transduction, Morphogenetic furrow, Hedgehog, Cubitus Interruptus, EGF receptor
Introduction
Drosophila eye development is associated with a morphogenetic furrow that moves across the eye primordium over a 2-day period, initiating retinal differentiation as it travels. The first individual cell fate specifications occur within the morphogenetic furrow. By the time the furrow has passed, most cells are differentiating, and they continue to mature further while the furrow advances more anteriorly (Wolff and Ready, 1993).
Hedgehog (Hh) is the principal signal that drives the wave of differentiation across the eye disc. Hh is secreted from behind the furrow and recruits cells ahead of the furrow to withdraw from the cell cycle and begin specifying retinal fates. As each new region of the eye disc differentiates, it begins secreting Hh that drives the furrow still further anteriorly across the eye disc (Ma et al., 1993). Some of the functions of Hh are direct, and some are mediated by Hh-dependent secondary signals, including Dpp (Heberlein et al., 1993; Lee and Treisman, 2002). The relationship of Hh expression to the wave of eye differentiation is illustrated in Figs. 1A–C.
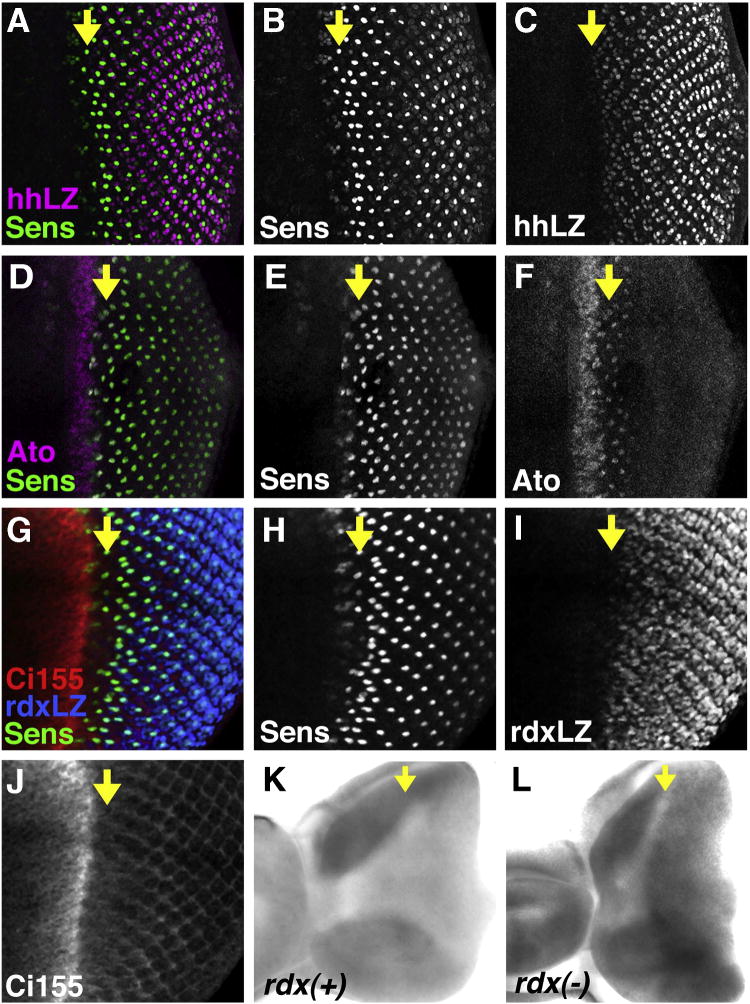
Fig. 1.
Expression and activity of Hedgehog in the Drosophila eye. All panels show eye imaginal discs with anterior to the left. Yellow arrows indicate the position of ommatidial column 0 at the base of the morphogenetic furrow. (A) Merge of Senseless expression (an R8 cell marker) in green and hh-LacZ enhancer trap in magenta. hh-LacZ is first detected around column 1–2, after R8 cell specification, although hh-LacZ might appear slightly delayed with respect to hh transcript (Ma et al., 1993). (B) Sens channel. (C) hh-LacZ channel. (D) Merge of Senseless expression in green and Atonal protein in magenta. Atonal expression initiates in a stripe ahead of the furrow that depends largely on Hh signaling, (although it can also be induced by Dpp after a delay) (Dominguez, 1999; Greenwood and Struhl, 1999; Curtiss and Mlodzik, 2000). The stripe of Atonal expression resolves to single R8 cells before the stage that hhLacZ is detected (compare panel (A)). (E) Ato channel. (F) Sens Channel. (G) Merge of Sens expression in green, Ci155 protein in red, and rdx-LacZ enhancer trap in blue. Ci155 peaks ahead of the furrow as Atonal expression is induced, dropping abruptly at the anterior of the morphogenetic furrow exactly as Sens expression begins, R8 specification begins, and egfr-dependent MAPK phosphorylation is observed (Kumar et al., 1998; Spencer et al., 1998; Chen and Chien, 1999; Greenwood and Struhl, 1999) (Lesokhin et al., 1999). Ci155 remains detectable at lower levels in undifferentiated unspecified cells. rdxLacZ is first detected around column 1. It is not known whether rdxLacZ lags behind endogenous Rdx protein expression, although it does not seem to lag endogenous rdx RNA (see panel L below). (H) Ci155 channel. (I) Sens channel. (J) rdxLacZ channel. (K) in situ hybridization with sense (negative control) probe for rdx transcripts. Arrow indicates the morphogenetic furrow. (L) in situ hybridization with antisense probe for rdx transcripts. Transcripts are detected posterior to the morphogenetic furrow (arrow), corresponding to the Rdx-LacZ pattern.
One feature of this moving differentiation wave is that cellular properties change once the furrow has passed. This includes responses to signaling pathways. For example, one morphogenetic furrow target is the proneural transcription factor Atonal, which specifies the R8 photoreceptor cell class. Each R8 cell founds an ommatidium as the source of EGFR-dependent Ras activation that recruits differentiation of additional photoreceptor neurons and has many other effects in the differentiating retina (Roignant and Treisman, 2009). Hh initiates atonal expression in a transient stripe as the furrow approaches, but ato is not expressed posterior to the furrow (Jarman et al., 1994) (Figs. 1D–F). The passage of the furrow must change the competence of the eye cells so that they do not continue to make the same responses to Hh (and Dpp). The mechanisms by which this occurs are of interest in understanding the progression of development.
The distribution of the Ci155 protein shows that Hh signal transduction itself is reduced posterior to the furrow (Motzny and Holmgren, 1995; Strutt and Mlodzik, 1996) (Figs. 1G–J). Ci is a zinc-finger protein gene that regulates transcription of Hh target genes. Hh signaling converts Ci from a repressor to an activator, both by inhibiting the processing of full-length Ci155 protein to the truncated, Ci75 repressor form, and through additional modifications that activate Ci155 (Kalderon, 2005; Jiang and Hui, 2008). An antibody that detects Ci155 but not Ci75 tracks Hh signaling by identifying cells where Ci155 is stabilized (Motzny and Holmgren, 1995; Aza-Blanc et al., 1997). In the eye disc, Ci155 accumulates in increasing amounts as the furrow approaches and Hh signaling rises. As soon as retinal differentiation begins with the specification of R8 photoreceptor precursor cells that defines column 0, Ci155 drops to low levels in all cells. While more cell types are progressively recruited to ommatidial fates, Ci155 drops further in these differentiating ommatidial cells and becomes undetectable by immunohistochemistry (Figs. 1G–J).
The reduction in Ci155 levels is associated with transcriptional induction of the roadkill gene (rdx) and a change in the proteolysis of Ci (Kent et al., 2006; Zhang et al., 2006a) (Figs. 1G–J). Ci155 processing is mediated by the proteosome and requires a ubiquitin-ligase complex that targets Ci155 through the F-box protein Cullin1 (Cul1). Cul1 is required for Ci155 processing in most regions of the body but is dispensable posterior to the morphogenetic furrow. Accordingly, cul1 mutant cells accumulate Ci155 in the anterior eye and in most body regions regulated by Hh, but not posterior to the morphogenetic furrow. By contrast, posterior to the furrow Ci155 accumulates in cells mutant for the related gene cullin3 (cul3), but Cul3 does not affect Ci155 levels in most other tissues (Ou et al., 2002; Mistry et al., 2004). Cul3 mediates the complete degradation of Ci155, rather than partial degradation to the Ci75 repressor protein (Ou et al., 2002). Both cul1 and cul3 genes are transcribed uniformly, but rdx encodes a BTB protein that couples Ci to Cul3 posterior to the morphogenetic furrow (Kent et al., 2006; Zhang et al., 2006a).
In order to understand how the morphogenetic furrow moves, we sought to understand how rdx transcription is induced posterior to the furrow, and whether this is sufficient to explain the changes in Hh signaling. The fact that Hh is the primary driver of furrow movement also raises the question of how rdx transcription could be maintained by Hh signaling if Rdx protein shuts down Hh signal transduction. Some previous studies have suggested complex models in which Hh has multiple, opposite roles in Ci regulation, or in which Cul3-mediated Ci proteolysis reflects a second mode of Hh signal transduction, parallel to the regulation of Cul1-mediated Ci processing (Dominguez, 1999; Ou et al., 2002; Kent et al., 2006; Zhang et al., 2006a). It could also be that rdx transcription was maintained epigenetically.
We have investigated the regulation of rdx transcription and of Cul3-dependent Ci degradation as the morphogenetic furrow moves. It was already known that rdx-LacZ reporter expression was lost from smo mutant cells that cannot respond to Hh (Zhang et al., 2006a). This dependence of rdx on Hh signaling could be quite indirect, however, since almost every event in the posterior eye is ultimately downstream of Hh. We report here that rdx transcription is induced by Hh signaling and also by Ras signaling. Ras signaling acts as part of an indirect feedback loop that helps keep Ci low after Hh signaling defines the R8 cells that are the source of signals that activate the EGFR. Our data suggest Cul1-dependent Ci155 processing may also be affected as the furrow moves. In our model of morphogenetic furrow movement, Hh and Ras signals induce rdx and couple Ci to Cul3-dependent degradation, and Dpp contributes to an uncoupling of Ci155 from Cul1-dependent processing. These coordinated steps together change the sensitivity of cells to Hh as the retina differentiates, and keep the morphogenetic furrow moving only forward.
Materials and methods
Drosophila strains and husbandry
All flies were raised at 25 °C except where otherwise specified. FLP/FRT-mediated mitotic recombination was used to obtain mosaic clones (Golic, 1991; Xu and Rubin, 1993). The arm-LacZ transgene was used to identify recombinant cells (Vincent et al., 1994). The smo egfr mutant clones were obtained after heat shock of the genotype:
hsFlp; smo3 FRT42 egfrf24 / smoD16 FRT42 P{smo+} arm-LacZ M(2) 56i; GMRp35/+.
The following other transgenic and mutant strains were used:
rdx1 (also known as P{PZ}mei-P1903477 and rdxLacZ) (Kent et al., 2006),
hhts2 (Ma et al., 1993),
hh-LacZ (Lee et al., 1992),
hsNintra (Struhl et al., 1993),
egfrf24 (also known as topCO) and egfrtop18A (Price et al., 1989),
egfrts1a (Kumar et al., 1998),
hs-RasV12 (Miller and Cagan, 1998),
smo3 and smoD16 (Chen and Struhl, 1998),
P{smo+} (Methot and Basler, 1999),
mago3 (Boswell et al., 1991),
GMRp35 (Hay et al., 1994),
pnt 88 (Morimoto et al., 1996).
Antibody labeling and in situ hybridization
Antibodies and immunochemistry have been described previously (Fu and Baker, 2003; Firth et al., 2006). The in situ hybridization was performed as described (Firth and Baker, 2007), with hybridization at 55 °C. An in situ probe was based on four coding exons common to all rdx splice variants. Total RNA was isolated from 3rd instar eye-antennal discs using TriZol (Invitrogen), and total cDNA synthesized using oligo(dT)20 primer and the SuperScript III First-Strand Synthesis System (Invitrogen). Regions of rdx transcript were then amplified from this cDNA using the following primers:
Forward Primer 5′ GGCCGCGGGAAGCGTGAGGAAACCAAAG 3′;
Reverse Primer 5′ CCCGGGGCTCCTCGCACATCACTTTCAG 3′.
Control (sense) and experimental (antisense) probes were then synthesized as described (Firth and Baker, 2007).
Results
Cell-autonomous and non-autonomous regulation of rdx by Hh signaling
Transcription of rdx is detected using an enhancer-trap P-element insertion in the rdx1 mutant allele, henceforth referred to as rdx-LacZ (Figs. 1G–J). Rdx-lacZ is detected in all eye disc cells posterior to the morphogenetic furrow, beginning at column 0, where the first R8 precursor cells are specified and Ci155 levels drop (Figs. 1G–J) (Kent et al., 2006; Zhang et al., 2006a). In situ hybridization confirms that endogenous rdx transcripts are expressed in a similar pattern (Figs. 1K, L) (Zhang et al., 2006a). Because the P{PZ} element inserted in Rdx-LacZ encodes a Nuclear Localization Signal, beta-galactosidase accumulates in the nuclei of expressing cells (Mlodzik and Hiromi, 1992). In the posterior eye disc, this includes the differentiating retinal cells, whose nuclei move apically in the eye disc epithelium, and the unspecified cells, whose nuclei remain basally located (Tomlinson, 1985; Kent et al., 2006) (Fig. 2).
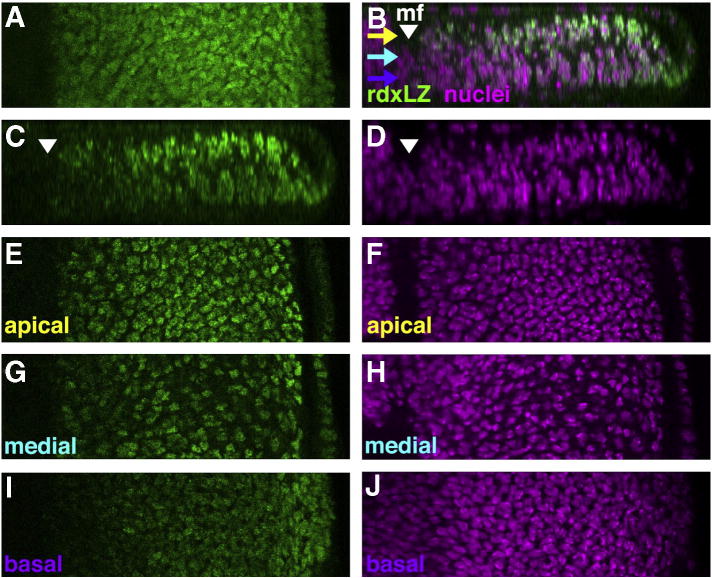
Fig. 2.
Rdx-LacZ expression in the nuclei of eye disc cells. (A) Z-projection of high resolution confocal data shows Rdx-LacZ expression in cells posterior to the morphogenetic furrow (green). (B) Y-axis reprojection of a slice from panel (A), double-labeled with propidium iodide to highlight nuclei of all cells (magenta). Posterior to the morphogenetic furrow (mf), nuclei of cells recruited to ommatidial fates rise apically in the epithelium while nuclei from cells that remain unspecified sink basally (Tomlinson, 1985). Rdx-LacZ overlaps with both apical nuclei and basal nuclei. Rdx-LacZ is reduced in the nucleoli that stain very brightly with propidium iodide. Rdx-LacZ is not detected in the disc epithelium anterior to the morphogenetic furrow, or anterior to the morphogenetic furrow. Yellow, turquoise and violet arrows indicate apical, medial, and basal planes that are shown separately in panels E–J. (C) Rdx-LacZ channel from panel (B). (D) Propidium Iodide channel from panel (B). (E) Rdx-LacZ in a single apical Z-plane, corresponding to the yellow arrow in panel (B). Nuclear localization in the differentiating cells is confirmed by comparison with propidium iodide labeling in panel (F). (G) Rdx-LacZ in a single medial Z-plane, corresponding to the turquoise arrow in panel (B). Nuclear localization is confirmed by comparison with propidium iodide labeling in panel (H). These nuclei correspond to cells early in the process of recruitment to ommatidial fates, and to differentiating R8 cells whose nuclei sink to a medial location (Tomlinson, 1985). (I) Rdx-LacZ in a single basal Z-plane, corresponding to the violet arrow in panel (B). Comparison with propidium iodide labeling in panel (J) confirms that basal Rdx-LacZ corresponds to the nuclei of unspecified cells.
Rdx-LacZ was examined in smo mutant cells to examine how rdx expression depends on Hh signaling (Fig. 3). Rdx-LacZ was absent from smo clones in the morphogenetic furrow, consistent with induction by Hh signaling (Figs. 3A–D) (Zhang et al., 2006a). More posteriorly, however, Rdx-LacZ expression occurred in smo mutant cells close to posterior clone margins (Figs. 3A–H). Such Rdx-LacZ expression was closely associated with the delayed cell differentiation that occurs in smo mutant clones (Figs. 3E–H). Thus, rdx can be transcribed in the absence of Hh signaling, although not in a timely fashion. Such delayed expression suggests a requirement for Hh signaling to activate a secondary non-autonomous signal that activates rdx. This signal can spread into smo mutant clones from cells beyond their posterior boundary, and activate rdx within smo mutant cells.
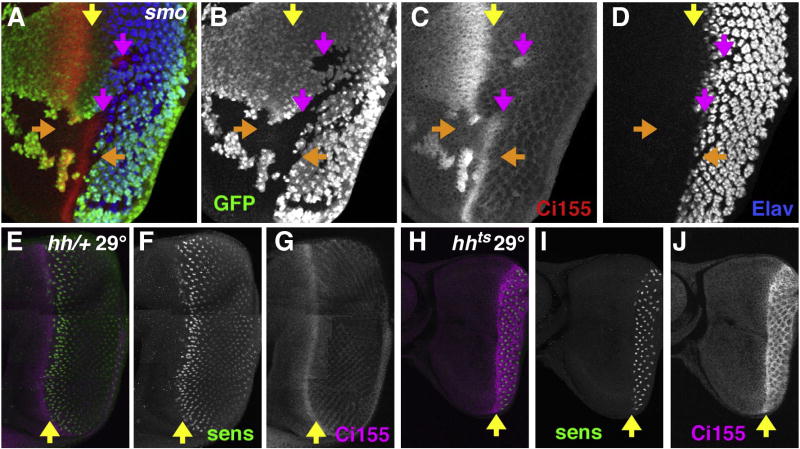
Fig. 3.
Hh signaling and rdx expression. Hh signaling and rdx expression. (A) Clones mutant for smo3 identified by the absence of GFP (green). Neural differentiation (Elav protein in blue) and rdxLacZ expression (red) are absent from much of the clones but spread into the mutant areas at their posterior boundaries (yellow arrows) (Strutt and Mlodzik, 1997; Greenwood and Struhl, 1999). (B) GFP channel. (C) Elav channel. (D) rdxLacZ channel. (E) Higher magnification of the disc shown in panel A. Yellow arrows highlight the spread of rdxLacZ (red) and Elav (blue) into the smo mutant clone. Although the expression of rdxLacZ and Elav generally correspond, some cells express one but not the other. (F) GFP channel. (G) Elav channel. (H) rdxLacZ channel. (I) control eye disc (hhts2/TM6B) labeled for Sens protein (green) and rdxLacZ expression (magenta) at 29 °C for 10 h. Orange arrow indicates ommatidial column 0 in the morphogenetic furrow. (J) Sens channel. (K) rdxLacZ channel. (L) Y-axis reprojection of a slice from panel (I), apical disc surface uppermost. R8 nuclei labeled for Sens (green) lie within the apical half of the disc epithelium; more basally, many nuclei from undifferentiated cells also express rdxLacZ (magenta). Lower panel shows a tracing of the image with Sens-expressing nuclei superimposed. R8 cells have the most basal nuclei among the differentiating ommatidia, but the nuclei of many undifferentiated cells are more basal still (Tomlinson and Ready, 1987). A dotted line traces the limit of the apical differentiating nuclei, including the R8 cells. (M) hhts2 eye disc labeled for Sens protein (green) and rdxLacZ expression (magenta) after 10 h at 29 °C (the restrictive temperature). Morphogenetic furrow progression has stopped, so that no Sens expression is beginning ahead of the first differentiating ommatidia (orange arrow). rdxLacZ expression is limited to clusters of nuclei. These correspond to the differentiating ommatidial cells. The intervening undifferentiated cells are unlabeled, so that the ommatidial clusters appear distinctly separated, by contrast to panel I. (N) Sens channel. (O) rdxLacZ channel. Note the clear reduction of labeling between the differentiating ommatidial clusters. (P) Y-axis reprojection of a slice from panel (M), apical disc surface uppermost. R8 nuclei labeled for Sens (green) lie within the apical half of the disc epithelium; more basal nuclei from undifferentiated cells lack most rdxLacZ expression (magenta). Lower panel shows a tracing of the image with Sens-expressing nuclei superimposed on the apical and basal nuclear layers. A dotted line traces the limit of the apical differentiating nuclei, including the R8 cells. The R8 nuclei are the most basal nuclei expressing significant levels of rdxLacZ in this genotype, as rdxLacZ drops to low or undetectable levels in the basal nuclei of unspecified cells.
To test when Hh signaling contributes to Rdx expression, Hh signaling was reduced using the temperature-sensitive allele hhts2. After 10 h at the restrictive temperature Hh function has been lost and the morphogenetic furrow stopped (Ma et al., 1993) (Figs. 3I–P). Rdx-LacZ levels dropped in the basal nuclei of undifferentiated cells everywhere from the morphogenetic furrow to the posterior margin of the eye disc, becoming undetectable or low in these cells (Figs. 3I–P). By contrast, rdx-LacZ remained unchanged in nuclei of ommatidial cells that had already begun differentiating before Hh function was interrupted (Figs. 3I–P). These data indicated that Hh signaling was required to initiate and maintain rdx transcription in unspecified cells. By contrast, Hh signaling was not required to maintain rdx transcription in differentiating ommatidial cells.
EGFR/Ras signaling is required cell-autonomously for rdx expression
A pathway or pathways other than Hh signaling must be sufficient to maintain rdx transcription in differentiating ommatidial cells. One candidate was Notch signaling, because it was reported previously that clones of cells null for the receptor protein Notch accumulated Ci155 posterior to the morphogenetic furrow (Baker and Yu, 1997). However, Notch signaling is thought not to occur in some of the ommatidial cells that express rdx (Baker, 2002). We also find that ectopic N signaling does not lead to Ci degradation anterior to the furrow (data not shown), and that high Ci155 levels were noted in posterior cells that had been prevented from differentiating by ectopic N signaling (Supplementary Fig. 1). Elevated Ci155 levels in both high and low N signaling suggests that Ci processing is affected indirectly by N, through its effects on ommatidial differentiation, which is prevented by both ectopic N signaling and by complete absence of N.
The differentiation of most retinal cells depends on activation of the Ras pathway by the EGF receptor. The isolated R8 cells that found each ommatidium are a primary response to Hh and the morphogenetic furrow signals, and secrete the EGFR ligands that recruit many other ommatidial cell types (Freeman, 1997; Roignant and Treisman, 2009). To test whether the EGFR/Ras pathway was required for rdx expression, clones of egfr mutant cells were examined. Fig. 4 shows a cell-autonomous failure to express rdx-LacZ in egfr mutant clones posterior to the morphogenetic furrow. A small number of scattered cells contained rdx-LacZ at the posterior edge of the morphogenetic furrow (Figs. 4A–B). These scattered cells also label for Elav (Figs. 4D–G), showing that these must be R8 cell precursors, the only cells to be specified and express Elav in the absence of EGFR signaling (Dominguez et al., 1998; Baker and Yu, 2001; Yang and Baker, 2001). R8 cells can also express rdx without egfr signaling (Fig. 4). These data indicate that EGFR signaling was required for rdx expression in most cells, both differentiating and unspecified. Undifferentiated cells required both EGFR and Hh signaling for rdx expression, whereas differentiated cells except for R8 require EGFR but do not directly require Hh.
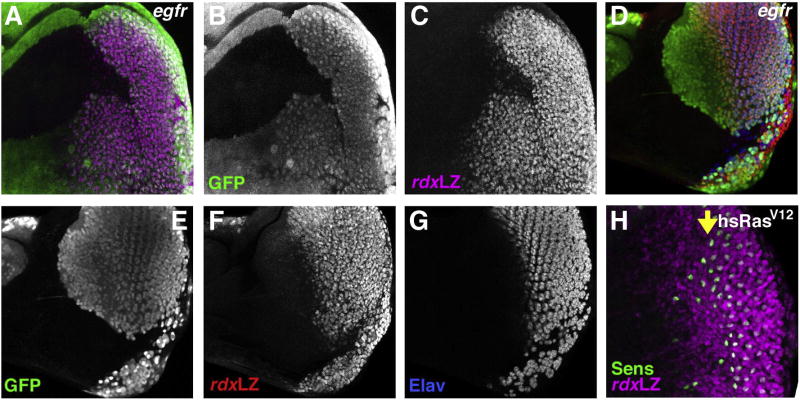
Fig. 4.
EGFR signaling and rdx expression. (A) Clone mutant for egfrf24 identified by the absence of GFP (green). Expression of rdxLacZ expression (magenta) is almost absent from the clones. A few scattered cells show some expression but this occurs throughout the clone and is not non-autonomous rescue near the clone boundaries. The GMRp35 transgene was used to suppress cell death in egfrf24 clones posterior to the furrow. (B) GFP channel. (C) rdxLacZ channel. (D) Clone mutant for egfrf24 identified by the absence of GFP (green). A few scattered cells that show some rdxLacZ expression (red) are also positive for the neural differentiation marker Elav (blue). Previous studies show that only R8 cells express Elav in the egfr mutant clones (Yang and Baker, 2001). The GMRp35 transgene was used to suppress cell death in egfrf24 clones posterior to the furrow. (E) GFP channel. (F) rdxLacZ channel. (G) Elav channel. (H) Effect of ectopic Ras activity in a hsRasV12 eye disc, 10 h after heat shock at 37 °C for 10 min. Sens labeling in green. Yellow arrow shows ommatidial column 0 in the morphogenetic furrow. Some ectopic R8 photoreceptors differentiate anterior to the furrow. rdxLacZ expression is induced anterior to the morphogenetic furrow (magenta).
To test whether Ras activity is sufficient to induce rdx expression, RasV12 was expressed uniformly using heat shock induction. Transcription of rdx was induced anterior to the furrow by ectopic Ras activity (Fig. 4H). The ectopic Rdx-LacZ expression was patchy. Although more cells expressed rdx-LacZ close to the morphogenetic furrow, there seemed to be no region of the eye disc where rdx-LacZ expression could not occur, and expression did not appear to be in a gradient, as would be expected if Hh activity was also required. Ci155 levels were greatly reduced wherever rdx was expressed ahead of the furrow (data not shown). These findings confirm that Ras activity is able to induce rdx transcription in the eye disc.
These findings document the following scheme for rdx regulation. Anterior to the morphogenetic furrow, neither Hh nor Ras signaling levels are high enough to activate rdx transcription. In the morphogenetic furrow, high Hh and Ras activity together turn rdx on in all cells. In the absence of Ras signaling, Hh activity is only sufficient to turn on rdx in the R8 precursor cells. In the absence of Hh signaling, Ras can still turn on rdx in other differentiating ommatidial cells. An explanation for the delayed activation of rdx observed in smo clones can now be suggested: where delayed differentiation spreads into the clones from the posterior under the influence of Dpp and N signaling, the EGFR activity that accompanies differentiation turns on rdx.
Activity of the Rdx/Cul3 pathway is independent of Hh
The regulatory pathway of rdx transcription leaves some observations unexplained, such as how Ci155 protein accumulates in some smo mutant cells posterior to the furrow (Dominguez, 1999) (Figs. 5A–D). It is easy to see that mutating smo can prevent Ci155 degradation through the Cul3 pathway, because of the direct and indirect effects of Hh signaling on rdx expression. It is harder to see why Ci155 should not be processed by the Cul1 pathway in smo clones. Because Ci155 only accumulates in smo mutant cells, not wild type cells posterior to the furrow, it has been suggested that this posterior accumulation has something to do with Hh signaling (Dominguez, 1999). However, posterior Ci155 accumulation cannot be explained by Hh inhibiting Cul1-dependent Ci processing in the normal manner that requires smo function, hence the suggestion that Hh and Smo might stimulate the activity of Cul3 (Ou et al., 2002).
Fig. 5.
Hh signaling and Ci155 accumulation. (A) Clone mutant for smo3 identified by the absence of GFP (green). Ci155 (red) is undetectable at positions ahead of the furrow where Ci155 accumulates in wild type cells or close to the posterior margins of smo clones, where delayed ommatidial differentiation is occurring. Neuronal Elav expression is in blue. Orange arrows indicate regions anterior and posterior to the furrow where Ci155 is undetectable. Yellow arrow shows ommatidial column 0 in the morphogenetic furrow in wild type cells. In smo clones, Ci155 is detected only in a stripe of cells located posterior to the normal location of the morphogenetic furrow (magenta arrows), and separated from the posterior clone boundaries by the region where differentiation is creeping into the clone. (B) GFP channel. (C) Ci155 channel. (D) Elav channel. (E) hhts2/TM6B disc showing normal differentiation and Ci155 accumulation after 10 h at 29 °C. R8 differentiation (green) and Ci155 protein (magenta) are shown. Yellow arrow shows ommatidial column 0 in the morphogenetic furrow. (F) Sens channel. (G) Ci155 channel. (H) hhts2 eye disc labeled for Sens protein (green) and Ci155 protein (magenta) after 10 h at 29 °C (the restrictive temperature). Morphogenetic furrow progression has stopped, so that no Sens expression is beginning ahead of the first differentiating ommatidia (yellow arrow). There is no anterior gradient of Ci155 accumulation, reflecting the loss of Hh signaling. Ci155 is not detected in differentiating cells posterior to the furrow. In undifferentiated cells posterior to the furrow, Ci155 accumulates to higher levels than in controls (compare to panels (E), (G); these images were recorded and processed identically and in parallel). (I) Sens channel. (J) Ci155 channel.
We first examined the relationship between the regulation of rdx expression and of Ci155 accumulation more closely. The smo mutant cells lack Ci155 at the normal location of the morphogenetic furrow, accumulate Ci155 more posteriorly, and lose Ci155 again close to the posterior clone margins (Figs. 5A–D). rdx is also expressed in only some smo mutant cells, but rdx was induced near posterior boundaries of smo clones, where ommatidial differentiation was occurring (Figs. 3A–H), after Ci155 had disappeared again (Figs. 5A–D). Ci155 accumulation in smo clones was therefore unlikely to be related to the Cul3 pathway, since rdx has yet to be transcribed in the cells where Ci155 accumulated.
The hhts2 mutation was used to examine how Ci155 depends on Hh in another way (Figs. 5E–I). After Hh signaling was inhibited, Ci155 accumulated to higher levels in the unspecified cells that separate the ommatidia (Figs. 5H–J). These unspecified cells also lost rdx-LacZ expression (Figs. 3N–P). Because the differentiating ommatidial cells that retained rdx-LacZ expression still lacked Ci155 (Figs. 5H–J), degradation of Ci155 by Rdx and Cul3 in the differentiating ommatidial cells appeared not to depend on Hh signaling.
Ci155 accumulation even in the absence of Hh signaling is surprising given the expression of Cul1 in these cells. The hhts2 mutation was sufficient to prevent Ci155 accumulation ahead of the furrow in the same experiments (Figs. 5E–J), so even in the absence of Hh, Ci155 seemed a better substrate for the Cul1 pathway ahead of the furrow than in the posterior undifferentiated cells.
Cul1-mediated processing of Ci in the posterior eye disc
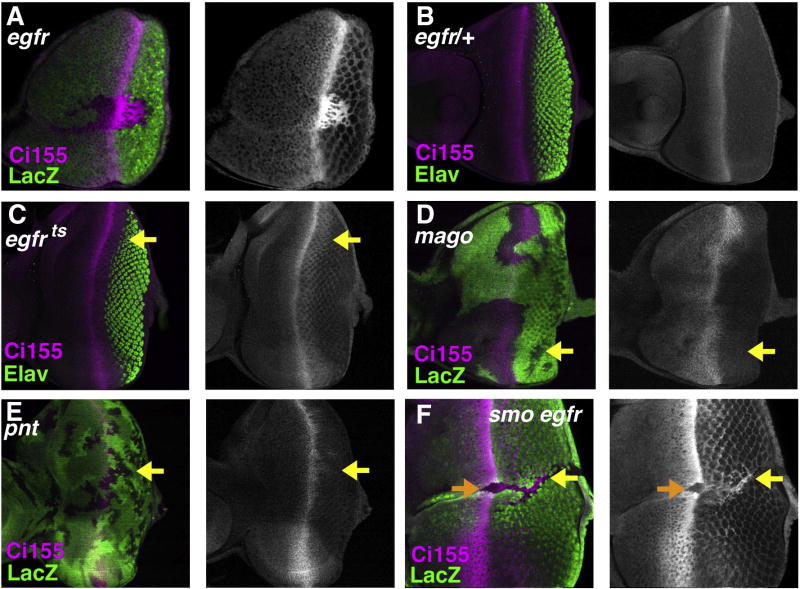
Ci155 should not be coupled to the Cul3 pathway in egfr mutant cells that lacked rdx expression. Accordingly, Ci155 accumulated cell autonomously in egfr mutant clones posterior to the morphogenetic furrow (Fig. 6A). Ci155 remained uniformly high as posterior as column 14, but levels often diminished in more posterior mutant cells (data not shown). The late reduction was not due to apoptosis; we prevented cell death in these experiments by expression of baculovirus p35 posterior to the morphogenetic furrow (see Methods). In another experiment, EGFR activity was interrupted using the conditional allelic combination egfrts2/egfrtop18a. Within 4 h at the restrictive temperature, Ci155 levels increased as far posterior as column 15, but were little changed more posteriorly (Figs. 6B–C). Downstream components of the EGFR pathway were also examined. We found that Ci155 accumulated in posterior clones mutant for mago (Fig. 6D). The requirement for mago has been positioned between raf and MAPK in this signaling pathway (Roignant and Treisman, personal communication). Much weaker effects were seen in clones of cells mutant for pnt, a transcription factor that is required for many Ras signaling outcomes. Little Ci155 accumulated in pnt clones, and accumulation was only detected quite close to the furrow (Fig. 6E). Although many functions of the Ras pathway are mediated by Pnt, not all are; Ras can induce mitosis in the eye disc without Pnt, for example (Yang and Baker, 2003). These results confirm that the failure to induce rdx in the absence of EGFR signaling protects Ci155 in posterior eye cells. The experiments again suggest that Ci155 is a poor Cul1 target in the posterior eye, at least in egfr mutant cells.
Fig. 6.
EGFR signaling and Ci155 accumulation. (A) Clone mutant for egfrf24 identified by the absence of LacZ (green). Ci155 (magenta) is maintained posterior to the furrow except for the few R8 cells differentiating within the clones. The GMRp35 transgene was used to suppress cell death in egfrf24 clones posterior to the furrow. (B) Clone mutant for mago3 identified by the absence of LacZ (green). Ci155 (magenta) is maintained until column 15. Clones of mago cells in more posterior regions lose Ci155 protein, however (yellow arrow). (C) Clone mutant for pnt 88 identified by the absence of LacZ (green). Ci155 (magenta) is somewhat elevated in pnt mutant cells, but not to the same levels or for as long as in mago or egfr mutant cells (yellow arrow). (D) egfr18a/+ eye disc after 4 h at 29 °C. Ci155 protein (magenta) and neural differentiation (Elav protein in green) remain distributed as in wild type. (E) egfr18a/egfrts eye disc after 4 h at 29 °C (restrictive temperature). Ci155 (magenta) accumulates to higher levels than in the control, from the furrow until about column 14 (yellow arrow). (F) Clone mutant for smo and egfrf24 identified by the absence of LacZ (green). The GMRp35 transgene was used to suppress cell death in egfrf24 clones posterior to the furrow. Ci155 (magenta) is maintained posterior to the furrow (yellow arrow), similar to egfr clones (panel A), and different from smo clones (compare Fig. 4A). Like smo clones, smo egfr clones prevent Ci155 accumulation ahead of the furrow (orange arrow).
One obvious explanation for Ci155 being stable in egfr clones, apparently not degraded by either Cullin, would be if the Cul1 pathway was inhibited by its known inhibitor Hh. If Hh was responsible, we would expect Ci155 accumulation in egfr mutant cells to depend on smo. Contrary to this expectation, Ci155 accumulated in smo egfr mutant cells (Fig. 6F). Ci155 accumulation began posterior to the furrow in smo egfr clones just as in smo clones, but Ci155 accumulated right to the clone boundaries, and persisted in much more posterior regions of the eye disc (Fig. 6F). This is consistent with the finding that EGFR activity associated with differentiation is responsible for rdx transcription near the posterior of smo clones. Ci155 levels have not been compared quantitatively between egfr and smo egfr clones to determine whether any Hh dependence is measurable, but the persistence of Ci155 in smo egfr clones shows that the Cul1 pathway is not sufficient to degrade Ci155 in the posterior eye in the absence of Rdx/Cul3 activity, even in cells that cannot respond to Hh.
Discussion
As the morphogenetic furrow crosses the eye disc, Ci155 accumulates most highly just anterior to the morphogenetic furrow, even though Hh is secreted posterior to the morphogenetic furrow. The sharp reduction in Ci155 as the furrow passes is associated with a switch from Cul1-dependent processing to Cul3-dependent degradation (Ou et al., 2002). The posterior eye expresses rdx, encoding a BTB protein that couples Ci 155 to the Cul3 pathway (Kent et al., 2006) (Zhang et al., 2006a). Here, the signals that induce rdx and process Ci155 in the posterior eye are identified. A model for Ci155 processing in the eye is shown in Fig. 7A, our results are summarized schematically in Figs. 7B–M, and the regulatory network proposed to tie Ci155 processing to the progression of the morphogenetic furrow illustrated in Fig. 7N.
Fig. 7.
Data summary and models. (A) Cullin activity in the eye disc. Cullin-1 is required for Hh-sensitive Ci155 processing anterior to the furrow. Cullin-3 is required for Ci155 degradation posterior to the furrow. Note that the hypothesized lack of Cul1-mediated Ci155 processing posterior to the furrow is not contradicted by the continuous requirement for Hh signaling to maintain rdx transcription in unspecified cells, as this could be mediated by Hh-dependent activation of Ci155. There may be a zone within the morphogenetic furrow where both Cul-1 and Cul-3 pathways are active, because in smo clones, Ci155 starts accumulating just posterior to the location of the morphogenetic furrow in wild type cells, slightly later than the Hh-dependent Ci155 accumulation in the furrow of wild type regions (Figs. 2, 6). This indicates when Ci155 has uncoupled from the Cul1 pathway, and this is after Ci155 has already become unstable and cul3-dependent in nearby wild type cells. However, it could be that the timing of uncoupling is affected in smo cells, and that the precise timing of uncoupling in wild type cells is different. (B) Ci155 accumulation in the wild type. Ci155 accumulates in a gradient decreasing anteriorly from the morphogenetic furrow. Posterior to the furrow, Ci155 accumulates at lower levels in between the differentiating ommatidia. (C–M) Panels C–M summarize the results of clonal analysis as the Ci155 levels within hypothetical clones staddling the morphogenetic furrow. (C) Clones mutant for cul1 prevent Ci155 processing ahead of the furrow, so that Ci155 accumulates, but have no effect posterior to the furrow (Ou et al., 2002; Mistry et al., 2004). (D) Clones mutant for cul3 prevent Ci155 degradation posterior to the furrow, but have no effect anterior to the furrow (Ou et al., 2002; Mistry et al., 2004). (E) Clones mutant for smo prevent Ci155 accumulation ahead of the furrow (Dominguez, 1999). Posterior to the furrow, smo cells accumulate Ci155 like cul3 clones at first, but then lose Ci155 close to the posterior clone boundaries (Dominguez, 1999; Fu and Baker, 2003) (See Fig. 4A). (F) At the restrictive temperature, hhts prevents Ci155 accumulation ahead of the furrow. Posterior to the furrow, Ci155 accumulates only in the cells between the ommatidia (see Fig. 4H). (G) Clones mutant for egfr maintain Ci155 posterior to the furrow, but have no effect anterior to the furrow (see Fig. 5A). (H) Activating Ras through inducible expression of RasV12 leads to widespread induction of rdx, and loss of Ci155, both anterior and posterior to the furrow (see Fig. 3H and data not shown). (I) Clones mutant for mago maintain Ci155 posterior to the furrow, but have no effect anterior to the furrow (see Fig. 5B). (J) Clones mutant for pnt reduce Ci155 degradation transiently posterior to the furrow, and have no effect anterior to the furrow (see Fig. 5C). (K) At the restrictive temperature, egfrts maintain Ci155 posterior to the furrow, but had no effect anterior to the furrow (see Fig. 5E). (L) Clones mutant for both smo and egfr prevent Ci155 accumulation anterior to the furrow, but accumulate Ci155 posterior to the furrow (see Fig. 5F). Because the egfr phenotype is epistatic to smo posterior to the furrow, loss of Ci155 from posterior smo clones requires egfr activity. (M) Clones mutant for smo and tkv prevent Ci155 accumulation both anterior and posterior to the furrow (Fu and Baker, 2003). Therefore, Ci155 accumulation in smo clones requires Dpp signaling. (N) Model of the genetic network regulating Ci155 processing to Ci75 through Cul1 and degradation through Cul3. Hh is proposed to stabilize Ci155 through two mechanisms. First is the well-known inhibition of Ci155 processing by Hh and Smo, which affects the Cul1 pathway. Second is uncoupling Ci155 from Cul1-mediated processing, which is achieved through or in conjunction with Dpp signaling. This implies Dpp inhibition of a factor (Factor X) coupling Ci155 to Cul1, or alternatively induction of an antagonist of Cul1 coupling (not shown). These dual effects on the Cul1 and Cul3 pathways mean that Ci155 is nowhere stable in smo tkv mutant cells (see panel M) (Fu and Baker, 2003). Hh is further proposed to promote Ci155 degradation through the direct and indirect (through Ras) induction of rdx transcription. The Rdx protein couples Ci155 to degradation via the Cul3 pathway (Kent et al., 2006; Zhang et al., 2006a). Such degradation is constitutive once Rdx is expressed, and ensures low Ci155 levels posterior to the furrow. Ras-dependent induction of rdx is delayed in smo clones, delaying the degradation of Ci155 through Cul3, and cannot occur in smo egfr clones, preventing the degradation of Ci155 through Cul3. Ci155 can accumulate in smo or smo egfr clones posterior to the furrow because Dpp signaling is not delayed (or not delayed as much), so the Cul1 pathway is inactivated also.
Regulation of rdx transcription
The induction of rdx transcription couples Ci155 processing to Cul3 (Kent et al., 2006; Zhang et al., 2006a). We found that rdx transcription was regulated by both Hh signaling and Ras signaling, and that there were distinctions between cell types (Figs. 3, 4). The smo mosaic and hhts2 experiments show that Hh signaling is continuously required for rdx transcription in unspecified cells with basal nuclei. In the absence of smo, EGFR-dependent rdx transcription occurs in differentiating photoreceptor cells only, not in unspecified cells. The egfr mosaics show that EGFR is essential for rdx transcription in all cells except the R8 photoreceptor class. Thus, EGFR-dependent differentiation was sufficient to induce rdx in photoreceptors even without Hh signaling, but Hh was not sufficient to induce rdx anywhere without EGFR signaling, except for the R8 cells. Undiffer-entiated cells might require both the Ras and Hh signaling pathways to induce rdx because the level of Ras signaling is lower in unspecified cells than in differentiating cells of the ommatidia (Yang and Baker, 2003). Alternatively, there may be a combinatorial requirement for both pathways in unspecified cells.
Regulation of Rdx/Cul3 activity
There has been some discussion of whether proteolysis of Ci155 by Cul-3 is regulated directly by Hh, as is Cul-1 dependent Ci processing (Dominguez, 1999; Ou et al., 2002; Kent et al., 2006; Zhang et al., 2006a). Our studies provide no support for this idea. In all the genotypes we have examined, Ci proteolysis correlates with the expression of rdx, and the simplest explanation is that the only effect of Hh on the Cul-3 pathway is through rdx transcription, directly in unspecified cells, and indirectly via EGFR-mediated differentiation in most specified cells.
Regulatory changes behind the furrow
Two mechanisms, acting in different cells, appear to reduce Hh responses through Ci155 after the furrow passes. One also occurs in wing development, where rdx is transcribed only by cells experiencing high Hh signaling levels close to the source of Hh (Kent et al., 2006). In wing development, rdx and the Cul3-pathway modulate the amount of Ci155 available for Cul1-dependent processing, lowering the maximum level of Ci155 activity at high Hh levels. Rdx could lower Ci155 levels in unspecified eye cells posterior to the furrow by this mechanism, in which an equilibrium between Hh-dependent induction of rdx, and rdx- and Cul3-dependent degradation of Ci155, leads to a lower level of Ci155 protein than anterior to the furrow. By contrast, in the specified, differentiating eye cells, rdx transcription becomes independent of Hh signaling, and Ci155 is degraded more completely.
If there is Hh signaling posterior to the furrow, as our studies find maintains rdx transcription in unspecified retinal cells, why are genes such as atonal that are induced by Hh signaling ahead of the furrow not also expressed posterior to the furrow? There are at least three possible explanations. First, rdx may dampen Ci155 accumulation in unspecified cells such that the threshold necessary for ato expression is not achieved posterior to the furrow. This is unlikely to be the sole explanation, since mutating rdx or cul3 permits Ci155 accumulation but does not lead to ectopic R8 specification (Kent et al., 2006; Ou et al., 2007), but it could contribute in conjunction with other mechanisms. Secondly, other genes may interfere posterior to the furrow. This could include egfr induction of Bar gene expression, since Bar genes antagonize ato expression (Lim and Choi, 2004). There seem to be multiple respects in which EGFR-dependent differentiation renders cells unable to continue anterior responses to Hh, and we also envisage that egfr might play a role in further mechanisms that modulate the response to Dpp signaling posterior to the furrow, should such mechanisms exist. Finally, recent evidence suggests that induction of ato by Hh is not so simple as the induction of a target gene above a threshold in a morphogen gradient, but depends indirectly on Hh repressing Eyeless and activating Sine Oculis, so that these transcription factors are coexpressed and turn on ato only in a domain ahead of the furrow (Zhang et al., 2006b; Firth and Baker, 2009). In this case, persistent Hh signaling would not be expected to activate ato expression once Ey had been repressed.
Recently, Hh has been discovered to induce compensatory proliferation in response to eye disc cell death (Fan and Bergmann, 2008), a further example of post-furrow Hh function. Our results now suggest the model that loss of EGFR-dependent rdx expression elevates Ci155 locally to permit Hh responses when photoreceptor cells that secrete EGFR ligands are lost. Consistent with this idea, loss of rdx or cul3 also result in proliferation of eye disc cells (Ou et al., 2007).
Evidence that Ci155 becomes a poor substrate for the Cul1 pathway
The regulation of rdx expression and thus degradation of Ci by Cullin-3 may not be sufficient to explain Ci regulation posterior to the furrow. In order for Ci155 to be stable, as observed in cul3 mutant clones and egfr mutant clones, Ci155 must escape processing to Ci75 by Cul-1. Ahead of the furrow, and in most other tissues, rdx is not expressed, Ci is not coupled to Cul3, and Ci155 is stabilized wherever Hh inhibits Smo and the Cul1 pathway. The observation that Ci155 is stable in cul3 clones, or in the genotypes where rdx is not expressed, shows that Ci155 escapes processing by the Cul1 pathway in the posterior eye as well, but this is not due to Hh. Ci155 accumulates in smo egfr mutant clones that do not express rdx and cannot respond to Hh (Fig. 6F).
One model would be that once rdx is induced, Ci155 is sequestered and not available to be processed by Cul1. This model cannot explain why Ci155 accumulates in egfr clones that lack rdx expression, where Ci155 should be available for Cul1. Therefore Ci155 must escape Cul1-mediated processing in the posterior eye by a distinct mechanism (Figs. 7A, N). This could be explained by the induction of a component distinct from Rdx that inhibits the processing of Ci155 by Cul1, or sequesters Ci155. It is equally possible that a component essential for processing of Ci155 by Cul1 is repressed posterior to the morphogenetic furrow.
Previous studies show that Ci155 never accumulates in smo tkv clones that are unable to respond to either Hh or Dpp signaling. Clones of cells unable to respond to Dpp, but able to respond to Hh and Ras, only show a subtle change in Ci155 labeling (Fu and Baker, 2003). These previously published observations suggest that Ci155 remains a target of Cul1 in the absence of both Dpp and Hh signaling, perhaps through failure to transcribe or repress transcription of a gene that modulates Ci155 proteolysis by Cul1 posterior to the furrow.
We can now account for why smo clones affect Ci155 levels differently from cul3 clones, a previously puzzling observation. In cul3 clones, or egfr clones that do not express rdx, the Cul3 pathway cannot degrade Ci155 and the Cul1 pathway is inactivated posterior to the furrow exactly as in wild type discs, so Ci155 accumulates (Fig. 5A) (Ou et al., 2002; Mistry et al., 2004). In smo clones, Ci155 transiently accumulates in those cells in which processing by Cul1 has been lost but rdx not yet induced. In such cells, Ci155 is not coupled to any cullin, and is stable. Eventually, differentiation spreads into the posterior of smo clones, leading to rdx expression, and Cul3-dependent Ci degradation. If differentiation and rdx expression are prevented, as in smo egfr clones, then Ci155 remains stable. Because there is a delay in expressing rdx in smo clones compared to wildtype, Ci155 is not subject to Cul3-mediated processing as soon as in wild type, and there is a period when Ci155 has been uncoupled from Cul1-processing but not yet coupled to the Cul3 pathway. It is during this period that Ci155 accumulates in smo mutant cells.
Implications for development and cancer
Our findings help explain how a wave of differentiation moves across the eye disc uni-directionally. Hh, secreted from differentiating photoreceptor cells, must be present at highest concentrations posterior to the furrow. Indeed, ahead of the furrow Ci155 is stabilized in a decreasing posterior-to-anterior gradient, consistent with a gradient of Hh protein coming from a source posterior to the furrow. Yet, the cell-autonomous responses to Hh signaling that are seen ahead of the furrow, such as cell cycle arrest and atonal expression, do not occur posterior to the furrow, where Ci is rendered unstable by Rdx and Cul3, induced both directly by Hh itself, and indirectly by the photoreceptor differentiation that is largely induced by EGFR posterior to the furrow.
There are other examples where Hh-secreting tissues are not the targets of Hh signaling. For, example, in Drosophila wing development, anterior compartments respond to Hh secreted by posterior compartments, but posterior compartment cells do not respond because ci transcription is repressed by the posterior-specific protein Engrailed (Aza-Blanc et al., 1997). In vertebrate development, notochord cells express Shh but the responses seen in the nearby spinal cord are not seen in notochord. Such segregation of Hh-producing cells from fields competent to respond to Hh makes sense, if the purpose of Hh signaling in development is to pattern new body regions. Hh signaling is also deregulated in many tumors (Jiang and Hui, 2008). Whether any of these tumors activate Hh signaling by affecting GLI protein stability, or other normal down-regulatory mechanisms, remains to be seen. In any case, mechanisms that render cells unresponsive to Hh by coupling Ci155 to the proteosome might prove useful in the treatment of cancers that depend on Hh signaling.
Supplementary Material
Acknowledgments
We thank K. Basler, R. Cagan, B. Hay, K. Moses, T. Schupbach, G. Struhl, J. Treisman, and the Drosophila Stock Center at Bloomington, Indiana for genetic strains. We thank R. Holgren and H. Bellen for antibodies. mAb40-1 was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. We thank L. Yang, D. Kalderon and J. Treisman for comments on the manuscript, and J. Treisman and J.-Y. Roignant for generously sharing unpublished reagents and data. Confocal microscopy was performed in the Analytical Imaging Facility at the Albert Einstein College of Medicine. We thank Sung-Yun Yu and Chunhui Fang for technical assistance and Lihui Yang for earlier work. This work was supported by grant from the National Institutes of Health (GM47892).
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2009.09.008.
References
- Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu S. Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol. 1997;7:122–132. doi: 10.1016/s0960-9822(06)00056-x. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/s0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- Baker NE. Notch and the patterning of ommatidial founder cells in the developing Drosophila eye. In: Moses K, editor. Drosophila Eye Development. Vol. 37. 2002. pp. 35–58. [DOI] [PubMed] [Google Scholar]
- Boswell RE, Prout ME, Steichen JC. Mutations in a newly identified Drosophila melanogaster gene, mago nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development. 1991;113:373–384. doi: 10.1242/dev.113.1.373. [DOI] [PubMed] [Google Scholar]
- Chen CK, Chien CT. Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc Natl Acad Sci U S A. 1999;96:5055–5060. doi: 10.1073/pnas.96.9.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Struhl G. In vivo evidence that patched and smoothened constitute distinct binding and transducing components of a Hh receptor complex. Development. 1998;125:4943–4948. doi: 10.1242/dev.125.24.4943. [DOI] [PubMed] [Google Scholar]
- Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–1336. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Wassarman JD, Freeman M. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol. 1998;8:1039–1048. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- Dominguez M. Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development. 1999;126:2345–2353. doi: 10.1242/dev.126.11.2345. [DOI] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev Cell. 2008;14:399–410. doi: 10.1016/j.devcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth LC, Li W, Zhang H, Baker NE. Analyses of RAS regulation of eye development in Drosophila melanogaster. Methods Enzymol. 2006;407:711–721. doi: 10.1016/S0076-6879(05)07056-4. [DOI] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Spitz from the retina regulates genes transcribed in the second mitotic wave, peripodial epithelium, glia and plasmatocytes of the Drosophila eye imaginal disc. Dev Biol. 2007;307:521–538. doi: 10.1016/j.ydbio.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Retinal determination genes as targets and possible effectors of extracellular signals. Dev Biol. 2009;327:366–375. doi: 10.1016/j.ydbio.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- Fu W, Baker NE. Deciphering synergistic and redundant roles of Hh, Dpp and N that drive the wave of differentiation in Drosophila eye development. Development. 2003;130(5229):5239. doi: 10.1242/dev.00764. [DOI] [PubMed] [Google Scholar]
- Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- Greenwood S, Struhl G. Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development. 1999;126:5795–5808. doi: 10.1242/dev.126.24.5795. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Wolff T, Rubin GM. The TGF-β homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell. 1993;75:913–926. doi: 10.1016/0092-8674(93)90535-x. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D. The mechanism of hedgehog signal transduction. Biochem Soc Trans. 2005;33:1509–1512. doi: 10.1042/BST0331509. [DOI] [PubMed] [Google Scholar]
- Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–2010. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Seger R, Shilo BZ, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Lee JD, Treisman JE. Regulators of the morphogenetic furrow. Results Problems Cell Differ. 2002;37:21–33. doi: 10.1007/978-3-540-45398-7_3. [DOI] [PubMed] [Google Scholar]
- Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- Lesokhin A, Yu SY, Katz J, Baker NE. Several levels of EGF Receptor signalling during photoreceptor specification in Ellipse, wild type, and null mutant Drosophila. Dev Biol. 1999;205:129–144. doi: 10.1006/dbio.1998.9121. [DOI] [PubMed] [Google Scholar]
- Lim J, Choi KW. Induction and autoregulation of the anti-proneural gene Bar during retinal neurogenesis in Drosophila. Development. 2004;131:5573–5580. doi: 10.1242/dev.01426. [DOI] [PubMed] [Google Scholar]
- Ma C, Zhou Y, Beachy PA, Moses K. The segment gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell. 1993;75:927–938. doi: 10.1016/0092-8674(93)90536-y. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and transcriptional repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- Miller DT, Cagan RL. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 1998;125:2327–2335. doi: 10.1242/dev.125.12.2327. [DOI] [PubMed] [Google Scholar]
- Mistry H, Wilson BA, Roberts IJ, O'Kane CJ, Skeath JB. Cullin-3 regulates pattern formation, external sensory organ development and cell survival during Drosophila development. Mech Dev. 2004;121:1495–1507. doi: 10.1016/j.mod.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y. Enhancer trap method in Drosophila: it's application to neurobiology. In: Conn PM, editor. Gene Expression in Neural Tissues. 1992. [Google Scholar]
- Morimoto AM, Jordan KC, Tietze K, Britton JS, O'Neill EM, Ruohola-Baker H. Pointed, and ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Development. 1996;122:3745–3754. doi: 10.1242/dev.122.12.3745. [DOI] [PubMed] [Google Scholar]
- Motzny CK, Holmgren R. The Drosophila Cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- Ou CY, Lin YF, Chen DF, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CY, Wang CH, Jiang J, Chien CT. Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Dev Biol. 2007;308:106–119. doi: 10.1016/j.ydbio.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Price JV, Clifford RJ, Schupbach T. The maternal ventralizing locus torpedo is allelic to faint little ball, an embryonic lethal, and encodes the Drosophila EGF receptor homolog. Cell. 1989;56:1085–1092. doi: 10.1016/0092-8674(89)90641-7. [DOI] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development. 1998;125:4777–4790. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- Strutt DI, Mlodzik M. The regulation of hedgehog and decapentaplegic during Drosophila eye imaginal disc development. Mech Dev. 1996;58:39–50. doi: 10.1016/s0925-4773(96)00555-2. [DOI] [PubMed] [Google Scholar]
- Strutt DI, Mlodzik M. Hedgehog is an indirect regulator of morphogenetic furrow progression in the Drosophila eye. Development. 1997;124:3233–3240. doi: 10.1242/dev.124.17.3233. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J Embryol Exp Morphol. 1985;89:313–331. [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in the Drosophila ommatidium. Dev Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Vincent J, Girdham C, O'Farrell P. A cell-autonomous, ubiquitous marker for the analysis of Drosophila genetic mosaics. Dev Biol. 1994;164:328–331. doi: 10.1006/dbio.1994.1203. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Vol. 2. Cold Spring Harbor LAboratory Press; 1993. pp. 1277–1325. [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in the developing and adult Drosophila tissues. Development. 1993;117:1223–1236. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development. 2001;128:1183–1191. doi: 10.1242/dev.128.7.1183. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev Cell. 2003;4:359–369. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006a;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006b;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.