Abstract
Upon transmission, human immunodeficiency virus type 1 (HIV-1) establishes infection of the lymphatic reservoir, leading to profound depletion of the memory CD4+ T cell population, despite the induction of the adaptive immune response. The rapid evolution and association of viral variants having distinct characteristics with different stages of infection, the level of viral burden, and rate of disease progression suggest a role for viral variants in this process. Here, we review the literature on HIV-1 variants and disease and discuss the importance of viral fitness for transmission and disease.
Keywords: HIV, SIV, fitness, CD4, T cells, macrophages, AIDS
Transmission and selection of variants
Transmission of HIV-1 can occur via sexual, parenteral, or vertical routes of infection (Lamers et al., 1993; Mulder-Kampinga et al., 1993; Pang et al., 1992; Scarlatti et al., 1993; Wolfs et al., 1992; Wolinsky et al., 1992; Zhang et al., 1993; Zhu et al., 1993). Each of these represents a distinct environment and therefore a distinct set of factors affecting selection of viral variants. Research focusing on factors affecting HIV-1 selection has addressed a multitude of issues ranging from stochastic versus selective models of transmission, single versus multiple variant transmissions, cell-free versus cell-associated virus transmission, selection criteria for restricting variant transmission and compartmentalization of variants within hosts biasing variant selection during transmission.
In the context of sexual transmission, the type of sexual encounter, gender of the transmitter and recipient, nature of the mucosal surface and presence of other genital tract infections can all play an integral role in selection during transmission. In order to distinguish the presence or absence of viral variants, the highly variable sequences of HIV-1 envelope (env) have been used as markers of diversity, especially in the V1–V2 and V3 regions of env which are important for immune recognition (Clerici et al., 1991; Javaherian et al., 1990; LaRosa et al., 1991), replication efficiency (Takeuchi et al., 1991) and cellular tropism (Chesebro et al., 1991; de Jong et al., 1992; Hwang et al., 1991; O’Brien et al., 1990; Shioda et al., 1991; Westervelt et al., 1991; Westervelt et al., 1992) and are under constant selection, resulting in a high rate of variation (Delwart et al., 1994; Delwart et al., 1993; Holmes et al., 1992; Javaherian et al., 1990; Kuiken et al., 1993; Wang et al., 1995; Zhu et al., 1993). Due to the increased variability, these regions have been used extensively to characterize the level of heterogeneity in donor/recipient HIV-1 transmission events. Transmission of variants found in low abundance (minor variants) would argue against a stochastic transmission model and many reports find that minor variants are often the predominant variants in the newly infected host (Zhu et al., 1996; Zhu et al., 1993; Wolfs et al., 1992; Wolinsky et al., 1992; Zhang et al., 1993). The majority of reports find that HIV-1 env sequences in newly infected individuals are relatively homogenous, despite the fact that the transmitters harbor heterogeneous genotypes (Delwart et al., 2002; Derdeyn et al., 2004; Learn et al., 2002; Wolinsky et al., 1992; Wolfs et al., 1992; Zhang et al., 1993; Zhu et al., 1993; Zhu et al., 1996). This sequence homogenization relative to the sequence heterogeneity of the transmitter could be a result of selection based on low inoculum levels (i.e. a founder effect), selective penetration of virus from donor to recipient and/or selective amplification of particular viral variants within the newly infected recipient. Factors such as cell-free or cell-associated virion transmission are also thought to affect virus transmission (Ludlam et al., 1985; Zhu et al., 1996). For instance, cell-free virions could be more likely to result in one or a few viral variants being transmitted, whereas cell-associated transmission could increase the likelihood of a multiple variant transmission event. Recent data support the complexity of transmission events in that some infections were initiated from a single virus while other infections resulted from a multi-virus transmission event (Keele et al., 2008). Gender differences also play a role in determining variant transmission profiles (Grobler et al., 2004; Haase, 2005; Long et al., 2000; Long et al., 2002; Ritola et al., 2004; Poss et al., 1995), potentially due to the type and duration of exposure to mucosal surfaces which may favor single or multiple viral variant transfers (Pilcher et al., 2004a; Pilcher et al., 2004b; Pope and Haase, 2003; Sagar et al., 2004; Vernazza et al., 1999; Wawer et al., 2005). It remains to be determined whether these differences are based on mucosal tissue morphology or on more complex biological factors.
Vertical transmission or mother-to child transmission (MTCT) has also been extensively studied and research has attempted to address similar questions as with sexual transmission. However, with MTCT, an additional layer of factors regarding the temporal nature of the transmission event arises. Specifically, does transmission occur prepartum (in utero), intrapartum (at delivery) and/or postpartum (via breast feeding) and what, if any, effect does this have on variant transmission profiles? Studies have found that transmission of virus can occur during any of these phases of pregnancy, but the effect on variant transmission remains elusive (Courgnaud et al., 1991; Ehrnst et al., 1991; De Rossi et al., 1992; Lepage et al., 1987; Soeiro et al., 1992; Ziegler et al., 1985). Similar to sexual transmission, major, minor and multiple variant transmission events are also seen in MTCT cases (Dickover et al., 2001; Kliks et al., 1994; Lamers et al., 1994; Narwa et al., 1996; Nowak et al., 2002; Briant et al., 1995; Pasquier et al., 1998; Wade et al., 1998; van’t Wout et al., 1994). However, most studies have found single variant transmission events and greater homogeneity within the newly infected child when compared to the mother, suggesting selective pressures during transmission (Dickover et al., 2001; Becquart et al., 2002; Nowak et al., 2002; Wike et al., 1992; Wolinsky et al., 1992; Mulder-Kampinga et al., 1993; Ahmad et al., 1995; Roth et al., 1996; Scarlatti et al., 1993). Whether these selective criteria are the same or similar to those involved in sexual transmission remains unknown.
Quasi-species evolution post transmission
Once a successful transmission event has occurred, mutations in the viral genome, host immune pressures and target cell availability drive HIV-1 diversification and evolution, eventually resulting in viral variants that differ from the founding parent viruses. While there may be a slight loss in replicative capacity initially, these new variants are capable of evading host immune defenses, persisting, and eventually driving CD4+ T cell depletion and progression to AIDS.
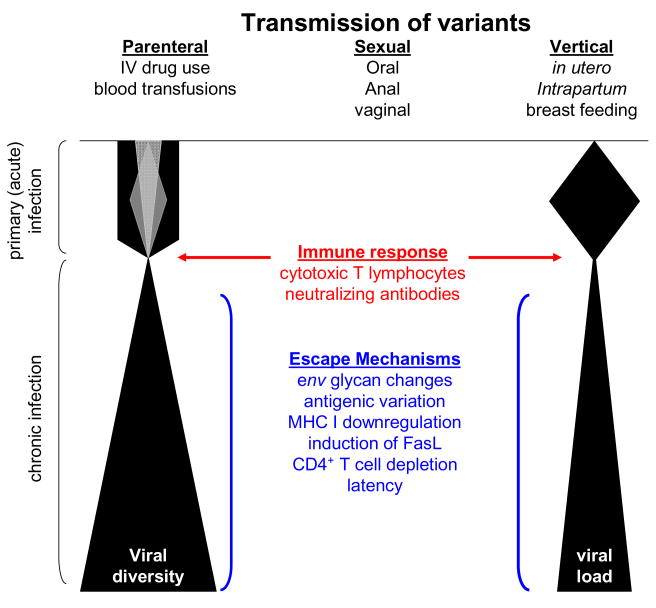
HIV-1 evolution is rapid due to a high viral replication rate, an error-prone replication of reverse transcriptase (Coffin, 1995; Mansky and Temin, 1995; Preston et al., 1988; Roberts et al., 1988), transcription by host RNA polymerase II (Laakso and Sutton, 2006), recombination events between co-infecting HIV-1 viral variants (Charpentier et al., 2006; Coffin, 1995; Jung et al., 2002; Kemal et al., 2003; Levy et al., 2004; Philpott et al., 2005; van Rij et al., 2003; Zhuang et al., 2002) and rapid immune system-mediated selection of viral variants (Jung et al., 2002; Williamson et al., 2005). The combination of these events continually drives HIV-1 diversity in infected hosts (Figure 1). However constraints are placed on the mutations that can be incorporated into the viral genome due to structural and functional requirements of the encoded proteins involved in viral replication (Draenert et al., 2006).
Figure 1.
Schematic diagram representing changes in viral diversity and viral load throughout the course of infection. The amount of viral diversity at or shortly following transmission is indicated by the different shadings during primary infection. Factors that may influence viral diversity, selection, and persistence are shown.
During the initial stages of infection, selected mutations appear to favor immune escape rather than enhanced viral replication (Martinez-Picado et al., 2006; Leslie et al., 2004; Goepfert et al., 2008). However, late during the asymptomatic chronic stage of infection, when immune pressures have been ablated by destruction of immune cells or exhaustion of immune responses, mutations increasing viral replication begin appearing or reappearing within the virus population (Mild et al., 2007). These data suggest that early during infection mutations are selected based on immune evasion rather than enhanced viral replication, whereas during later stages of infection more pathogenic, but less immune-evasive viruses appear to drive disease progression. This hypothesis is supported by studies of SIV in macaques, where there is a fitness cost to escape from CTL responses early after infection (Friedrich et al., 2004). However, with time, variants having increased replicative capacity emerge and eventually drive disease progression (Kimata et al., 1999; Rudensey et al., 1995).
While most basic and clinical studies seem to agree that variants isolated early during infection appear to be less pathogenic and have lower replicative capacity compared to late stage isolates, mathematical modeling of within-host virus evolution suggests that over the course of host infection viral variants move toward reduced replicative fitness (Wodarz and Levy, 2007; Ball et al., 2007). This apparent discrepancy could result from the fact that mathematical modeling has not accounted for the plethora of complex factors that play roles in viral fitness in vivo. However, one would be remiss to disregard key aspects of these mathematical models. Indeed, the overall conclusion from mathematical modeling is that the most replicatively fit HIV-1 viral variant would not be able to sustain infection, especially as is seen in chronic HIV-1 infections, due to the rapid destruction and depletion of the essential target CD4+ T cell population. Thus, an evolutionary ceiling is placed above viral replicative fitness. By contrast, studies examining replicative fitness do indicate that as infection progresses more pathogenic viral variants with higher replicative capacities appear at late stages of infection (Kimata et al., 1999; Kimata, 2006). Interestingly, both the mathematical models and clinical studies demonstrate that, even in the presence of these pathogenic and replicatively fit viral variants, less pathogenic and replicatively robust viral variants are still found in circulation (Ball et al., 2007; Wodarz and Levy, 2007; Mild et al., 2007; Mansky and Temin, 1995; Gali et al., 2007).
Additional correlates of AIDS disease progression include slower rate of synonymous substitution rates, indicative of general, non-selective mutation rates, (Lemey et al., 2007; Stilianakis and Schenzle, 2006), increased viral replication (Kimata et al., 1999; Birch et al., 2001; Dyer et al., 1999; Kirchhoff et al., 1995; Learmont et al., 1999), persistent immune activation (Bofill et al., 1996; Grossman et al., 2006; Giorgi et al., 1999; Sousa et al., 2002), broad-range CTL responses (Karlsson et al., 2007; Fernandez et al., 2007) and specific host human leukocyte antigen (HLA) class I alleles (Carrington et al., 1999; Trachtenberg et al., 2003).
Correlates of pathogenicity: phenotypic changes during viral infection
Phenotypic characteristics of HIV-1 that have been extensively studied for correlation to disease progression include replicative capacity (also commonly referred to as replicative fitness), which is generally classified as rapid/high or slow/low in relation to replication and production of virus (De Rossi et al., 2005; De Rossi et al., 1993; Connor et al., 1993), syncytium induction, classified as non-syncytium inducing (NSI) or syncytium inducing (SI) virus (Koot et al., 1992; Jurriaans et al., 1994), co-receptor usage with the vast majority of viral variants being classified as CCR5-using (R5), CXCR4-using (X4) or dual-tropic (R5X4) viruses (Littman, 1998; Doms and Peiper, 1997) and macrophage-tropic (M-tropic) or T cell-tropic (T-tropic) variants. Previously, it was believed that these phenotypic characteristics were intimately linked such that rapid/high viral variants were also SI, X4, T-tropic variants and slow/low viruses were NSI, R5, M-tropic variants (Alkhatib et al., 1996a; Tersmette et al., 1988; Tersmette et al., 1989). However, while there is a correlation with co-receptor usage, SI ability and tropism, these are separable phenotypic features and therefore should be individually tested for when characterizing viral isolates (Aquino-de Jesus et al., 2000; Peters et al., 2006).
Following transmission, R5-tropic viruses typically predominate early stages of infections (Connor et al., 1997). However, it remains unclear as to whether R5 viruses are the only viruses transmitted or whether both R5 and X4 viruses can be transmitted but that X4 viruses are less fit, resulting in only R5 variants being detected during the early stages of infection. These virus isolation studies also show that X4 and dual-tropic viruses generally are not detected until very late in infections at the juncture of transition from asymptomatic infection to AIDS. Morever, X4 variants are only found in 50% of cases (Berger et al., 1999), demonstrating that X4 variants are not required to drive progression to AIDS (Campbell et al., 2003; Kimata et al., 1999; Kwa et al., 2003; Koot et al., 1993; Tersmette et al., 1989).
There is experimental evidence that X4 variants may be more susceptible to control by CD8+ cytotoxic T cells than R5 viruses (Harouse et al., 2003). Indeed, X4 variants may be rapidly selected against during primary infection, allowing R5 variants to emerge and predominate in the infection. Thus, X4 viruses appearing late in infection may not necessarily drive disease progression, but rather serve as indicators of an exhausted and dysfunctional immune system, which allows an unchecked replication of viruses and destruction of the remaining CD4+ T cell population.
Cell tropism has also been extensively studied in vivo and in vitro. A current model is that M-tropic viruses predominate during early stages of transmission, since it is believed that tissue-resident macrophages, monocytes and dendritic cells are initial cell targets during the actual transmission event. While infection of these cell types remain important throughout the course of disease progression in regard to latently infected cell populations (Aquino-de Jesus et al., 2000), a shift in cell tropism occurs early and rapidly as virus is trafficked from the site of infection to lymphatic tissues where robust replication can take place in CD4+ T cells.
Lastly, there appears to be selection of NSI variants during transmission (van’t Wout et al., 1994; Zhu et al., 1993; Tersmette et al., 1988; Keet et al., 1993), but whether the phenotypic change from the transmitted NSI virus to a SI variant is important for progression to AIDS requires further exploration. Virological studies indicate that the switch from NSI to SI phenotype is not required for AIDS progression (Fitzgibbon et al., 1998; Spencer et al., 1994), but may increase the rate of AIDS progression (Fauci, 1996; Glushakova et al., 1998).
A simple explanation of the appearance of each of these phenotypic characteristics during infection is that as viral diversity increases the rate of AIDS progression increases, thus the diversity, as indicated by the appearance of these phenotypes, rather than the functions of the respective phenotypes drives AIDS progression (Sagar et al., 2003). Therefore it may not be the presence per se of X4 and SI viral variants, but rather the weakening of selective pressures from the host immune response on generalized viral replication, allowing an outgrowth of previously immune response-targeted phenotypes which serve simply as indicators of the weakened and dysfunctional immune response (Troyer et al., 2005).
Finally, changes in N-linked glycosylation and length of Env variable regions V1 and V2 have been reported to occur with infection. Initially, it was observed in SIV-infected macaques that variants with limited N-linked glycans in the V1/V2 region of Env dominated the early stages of infection (Overbaugh and Rudensey, 1992). As the animals progressed to disease, additional N-linked glycosylation sites appeared and the V1/V2 region lengthened. Some of these changes correlated with protection from neutralizing antibodies and loss of macrophage tropism (Rudensey et al., 1995; Rudensey et al., 1998). More recently several groups have reported similar glycosylation changes to occur with HIV-1 (Chohan et al., 2005; Derdeyn et al., 2004; Sagar et al., 2006; Wu et al., 2006) following sexual transmission in discordant couples and vertically from mother to child. However, while less glycosylated Env proteins from early stages of infection were associated with sensitivity to neutralizing antibodies after sexual transmission, this was not observed with vertically transmitted mother to child variants. These data raise questions about the functional changes in Env conferred by additional glycans. Recent studies demonstrate that GAG-specific CTL responses have a more profound impact on pathogenicity and viral load than do the ENV-specific CTL response (Kiepiela et al., 2007; Peut and Kent, 2007), lending support to the hypothesis that env-mediated phenotypic changes serve as indicators of dampened immune selection rather than functional mediators of pathogenicity.
Viral determinants altering phenotype
Phenotypic differences among viral variants have been studied and mapped to specific regions of the viral genome, including env, pol and nef. The env gene is a major determinant in viral replicative fitness as its protein products, gp120 and gp41, mediate cell binding via the receptor and co-receptors and fusion of the cellular plasma membrane and the viral membranous envelope (Baribaud and Doms, 2001; Berger et al., 1999; Poignard et al., 2001). Numerous studies have documented that env sequences influence viral transmission (Hsu et al., 2003; Tersmette et al., 1988), cell tropism (Berger, 1997; Hoffman and Doms, 1999; Alkhatib et al., 1996b; Choe et al., 1996; Deng et al., 1996) and are major targets of the host immune response (Levy, 1993; Richman et al., 2003; Wei et al., 2003b), including both CTL and neutralizing antibody responses (Jones et al., 2004; Borrow et al., 1997; Geels et al., 2003). Furthermore, env sequences appear to have the greatest impact on competitive viral replicative fitness in vitro in comparison to other regions of the viral genome (Ball et al., 2003).
The phenotypic change in co-receptor usage by viral variants has been mapped to env, specifically the variable regions 1, 2 and 3 (V1/V2 and V3). Mutations in V3 have been extensively studied and found to directly control tropism usage (Cocchi et al., 1996; Harrowe and Cheng-Mayer, 1995). However, mutations responsible for the switch from CCR5- to CXCR4-usage appear to confer a replication fitness disadvantage to the resulting virus (Wagner et al., 1999; Kelleher et al., 2001), either due to decreases in evasion of host CTL responses, enhanced sensitivity to antiviral drugs or decreased avidity for receptor/co-receptor molecules (Marozsan et al., 2005; Lobritz et al., 2007; Derdeyn et al., 2000; Labrosse et al., 2003; Strizki et al., 2001; Torre et al., 2000; Trkola et al., 1998). Mutations in the V1/V2 region also play a role in co-receptor usage (Groenink et al., 1993; Koito et al., 1994; Koito et al., 1995; Ogert et al., 2001; Ross and Cullen, 1998; Sullivan et al., 1993; Toohey et al., 1995; Wyatt et al., 1995; Yoshimura et al., 1996) with an apparent ability to compensate for the loss-of-fitness mutations in the V3 region, thus allowing the co-receptor switch to occur (Pastore et al., 2006). It is interesting to note that relatively few mutations are required for the R5/X4 co-receptor switch, but that this phenotypic change does not occur for several years after initial infection (Schuitemaker et al., 1992; Shankarappa et al., 1999). It is hypothesized that this delay in emergence of X4 variants is in part due to a very limited number of viable mutational pathways of transitional viral variants that maintain a minimally competitive replication fitness and evasion from host CTL responses (Pastore et al., 2006; Fernandez et al., 2005; Peyerl et al., 2004). Env mutations are also responsible for the NSI/SI phenotypic changes. Mutations responsible for this phenotypic switch have not been studied as extensively; however, due to the high degree of correlation between NSI/SI and R5/X4 phenotypic switching it is likely that mutations reside in the same sequences of env (e.g. V1/V2 and V3).
Functional reverse transcriptase (RT) is a heterodimer composed of subunits, p66 and p51, with p66 containing active RT and RNase H activity (Veronese et al., 1986). The majority of RT mutations and variants that affect functionality have been identified during studies with antiviral drugs designed to target RT-specific catalytic steps in the viral lifecycle. These drugs include nucleoside reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitors (NNRTIs) that inhibit RT activity by acting as DNA chain terminators or steric inhibitors of nucleoside binding, respectively. While it is clear that during highly active antiretroviral therapy (HAART) specific mutations are selected within RT that decrease sensitivity to prescribed drug regimens (Gu et al., 1992; Gao et al., 1993; Tisdale et al., 1997; Walter et al., 2002), some of these mutations can have positive effects on virus replication and fitness (Hu et al., 2007) while others have deleterious effects on replication (Wakefield et al., 1992; Larder et al., 1995; Olivares et al., 2004; White et al., 2002). The majority of these mutations appear to affect RT affinity for dNTPs which in turn alter RT DNA polymerase fidelity and/or processivity (White et al., 2002; Jonckheere et al., 1998; Bebenek et al., 1995; Back et al., 1996; Wainberg et al., 1996; Hsu et al., 1997; Feng and Anderson, 1999). Since deleterious mutations are more frequent than advantageous mutations within any genome, extreme RT infidelity would quickly result in non-functional HIV-1 genomes (Furió et al., 2005; Kimura, 1967). Conversely, increased RT fidelity would impact the intrinsic ability of HIV-1 to evade the host immune response and escape from HAART, thus RT fidelity is an intricate balance of genomic diversity and stability (Furió et al., 2007). Several studies have identified available dNTP levels as a factor that can affect RT fidelity (Back and Berkhout, 1997; Vartanian et al., 1997), yet very little research has been performed with HIV-1 RT fidelity mutations that occur naturally within infected hosts in the absence of HAART, despite the fact that cell types known to be targets for HIV-1 infection (e.g. macrophages, resting T cells, activated T cells) have variable dNTP pools (Traut, 1994; Diamond et al., 2004; Hauschka, 1973; Fuller et al., 1982; Skoog and Bjursell, 1974; Yao et al., 2003). A recent study with SIV RT variants isolated from infected macaques demonstrated rapid selection against an RT variant with higher replication fidelity (Biesinger et al., 2008). These data suggest that RT’s ability to misincorporate dNTPs in settings with limited resource availability or the indirect effect misincorporation has on processivity allows for more rapid genomic reverse transcription and integration and is an important aspect of replication fitness. Regardless, it is clear that intrinsic RT properties (e.g. fidelity and processivity) are important factors in determining viral replicative fitness in the context of host cell usage and overall viral production.
Nef is a multifunctional viral protein with key roles in the viral life cycle which include downregulation of CD4, CD28 and class I major histocompatibility complex (MHC-I), alteration of CD4+ T cell activation, enhancement of viral infectivity and replication and sensitization to apoptotic pathways (Laforge et al., 2007; Anderson and Hope, 2004; Collins and Baltimore, 1999; Johnson and Desrosiers, 2002; Renkema and Saksela, 2000; Roeth and Collins, 2006; Wei et al., 2003a). The essential nature of Nef activity for viral replication in vivo has been well-documented. However, the details of how Nef is able to exert its pleiotropic effects remain poorly described. Nef functionality in vivo exerts a dramatic effect on viral fitness and progression to AIDS. However, which function of Nef provides this enhancement is still unclear. Several possibilities, although not mutually exclusive, have been presented in the literature, including CTL escape via MHC-I downregulation (Ali et al., 2003), modulation of T cell activation via interactions with cellular factors such as PAK2 (Lu et al., 1996) and increased host cell apoptosis through unknown cellular interactions (Laforge et al., 2007). Finally, SIV-macaque studies suggest that the major function of Nef may be to enhance virion infectivity, as nef mutants that fail to enhance viral infectivity replicated poorly in vivo (Iafrate et al., 2000; Patel et al., 2002). Despite the fact that nef is essential for robust HIV-1 replication in vivo and evasion from the host immune response, it has recently been reported that highly attenuated nef-deleted viral infections still resulted in HIV-associated diseases and loss of CD4+ T cell levels (Gorry et al., 2007), suggesting that Nef functions are dispensable in context of AIDS progression over extended periods of time.
Model systems to measure viral phenotype and fitness
Correlations of viral phenotype and replicative fitness are found throughout the literature. However, most studies to determine phenotype and phenotypic changes (e.g. tropism, infectivity and ability to induce syncytia) are generally performed in in vitro single virus replication assays. The classical definition of fitness has necessarily required some aspect of direct competition for limited resources; therefore single-virus infections and assays are unable to be applied to directly answer whether one viral variant is more fit in a given environment versus another variant in the same environment. Relatively few studies have combined studies of viral phenotype with dual-virus replication assays to determine viral variant fitness.
In vivo studies to determine fitness of HIV-1 in the presence of immune responses have been hampered by the lack of an animal model to validate observations, and collection of samples from co-infections or super-infections within human cohorts are limited (Piantadosi et al., 2007; Kozaczynska et al., 2007; Gottlieb et al., 2007). In vitro competition assays have been used to characterize general viral fitness in regards to infectivity, replication and cytopathicity. These types of experiments suggest increasing fitness trends in the overall HIV-1 population over time despite the ‘reset’ of viral phenotype that occurs during each transmission event (Gali et al., 2007). Additionally, competition assays demonstrated that R5 and X4 phenotype do not necessarily correlate with lower or higher replicative fitness, respectively. One study found that R5 and X4 clones were equally fit in terms of viral replication in mitogen-activated T cells, but that X4 viruses did appear to have a selective advantage in DC-T cell co-cultures (Arien et al., 2006), while another study found that some R5 clones were more fit than X4 clones (Quinones-Mateu et al., 2000). These observations could help explain why the shift from R5 to X4 is not essential for progression to AIDS, since mass activation of the host CD4+ T cell population is characteristic of late stages of HIV-1 infection allowing outgrowth of either R5 or X4 viruses, but that moderate increases in viral spread which is correlated with X4 clones, would shorten the time to AIDS onset. Specific mutations in env have now been correlated with changes in fitness and entry inhibitor resistance (Lobritz et al., 2007; Rangel et al., 2003). Competition assays have also been used to explore the effects of environmental factors on viral fitness, such as the availability of permissive cell populations (Goodenow et al., 2003). Finally, the analysis of HIV-1 superinfection in infected individuals provides a rare glimpse into viral fitness in vivo (Gottlieb et al., 2007; Blish et al., 2007; Kozaczynska et al., 2007). However, research is limited by the rarity of these occurrences and knowledge of the properties of the viruses.
Because of the limitations for studying HIV-1 fitness in vivo, infection of macaques with SIV has remained a critical model for exploring questions about HIV-1 fitness and pathogenesis. The model system allows incorporation of viral fitness scores in the presence of shifting target cell populations, immune response and host variability and in context of infection with a related lentivirus. It also enables fitness to be examined with variants of known in vitro phenotype and in vivo pathogenicity. Indeed, a recent study showed that a SIV variant with increased replicative capacity and pathogenicity demonstrated higher competitive viral replication fitness in vitro compared to the slower-replicating and minimally pathogenic parental clone (Voronin et al., 2005). These types of studies provide important experimental evidence in support of the hypothesis that viral fitness impacts HIV-1 pathogenesis.
The macaque model system has also been used to study effects of dual virus infections in the host. These studies have mainly used R5- and X4-tropic SHIV chimeric constructs (Burke et al., 2006; Wolinsky et al., 2004; Otten et al., 1999). These dual virus infection studies are vital to understanding viral fitness in the full context of an infection where both viral and host factors are present and influence fitness outcomes of variants. However, few studies have been performed using dual virus infections to specifically study the relative fitness of variants and which determinants are able to confer fitness advantages at various stages of infection and disease states (Harouse et al., 2003). Furthermore, these studies have been limited to competitions with R5-tropic and X4-tropic viruses. Additional studies with different variants, including R5-tropic viruses with distinct phenotypes, would further enhance our understanding of viral determinants influencing for transmission, persistence and disease.
CONCLUSION
Route of HIV-1 transmission appears to dictate to some extent the type and number of viral variants that comprise the founder population of viruses in a newly infected host. Particularly interesting, as discussed above, are the aspects controlling selection during transmission where different factors, ranging from time of contact between viruses trapped within the vaginal cavity versus rectal tissues to cell-free virions versus cell-associated viruses to the glycosylation patterns found on gp120, appear to affect the number and type of transmitted viruses. Additionally, host immune responses and the corresponding counter-defenses of the infecting variants that enable evasion of the immune response represent an environment of attrition. While the strength of the immune response may initially determine viral set-points and rates of CD4+ T cell loss, latency and high mutability enable HIV-1 to subvert some host immune responses and evade other immune responses, leading to profound depletion of CD4+ T cell populations and onset of AIDS.
Correlating viral phenotypes with pathogenicity has resulted in identification of regions of interest within the viral genome that impact replication and persistence. Added studies of competitive replication fitness will be necessary to further shed light on the significance of these changes for viral fitness at early and late stages of infection. In this regard, the infection of macaques with SIV variants will be needed to provide invaluable results for deciphering fitness determinants for transmission and pathogenesis.
Acknowledgments
J. T. K. is supported by NIH grant R01 AI047725 and T. B. is supported by an NIH training grant in Molecular Virology (T32-AI07471).
Reference List
- Ahmad N, Baroudy BM, Baker RC, Chappey C. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J Virol. 1995;69:1001–1012. doi: 10.1128/jvi.69.2.1001-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Pillai S, Ng H, Lubong R, Richman DD, Jamieson BD, Ding Y, McElrath MJ, Guatelli JC, Yang OO. Broadly Increased Sensitivity to Cytotoxic T Lymphocytes Resulting from Nef Epitope Escape Mutations. J Immuno. 2003;171:3999–4005. doi: 10.4049/jimmunol.171.8.3999. [DOI] [PubMed] [Google Scholar]
- Alkhatib G, Broder CC, Berger EA. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996a;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: A RANTES, MIP-1alpha, MIP-1beta Receptor as a Fusion Cofactor for Macrophage-Tropic HIV-1. Science. 1996b;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Anderson J, Hope T. HIV accessory proteins and surviving the host cell. Current HIV/AIDS Reports. 2004;1:47–53. doi: 10.1007/s11904-004-0007-x. [DOI] [PubMed] [Google Scholar]
- Aquino-de Jesus MJ, Anders C, Miller G, Sleasman JW, Goodenow MM, Andiman WA. Genetically and epidemionlogically related “non-syncytium-inducing” isolates of HIV-1 display heterogeneous growth patterns in macrophages. Journal of Medical Virology. 2000;61:171–180. doi: 10.1002/(sici)1096-9071(200006)61:2<171::aid-jmv1>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Arien KK, Gali Y, El Abdellati A, Heyndrickx L, Janssens W, Vanham G. Replicative fitness of CCR5-using and CXCR4-using human immunodeficiency virus type 1 biological clones. Virology. 2006;347:65–74. doi: 10.1016/j.virol.2005.11.045. [DOI] [PubMed] [Google Scholar]
- Back NK, Berkhout B. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1997;41:2484–2491. doi: 10.1128/aac.41.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back NK, Nijhuis M, Keulen W, Boucher CA, Oude Essink BO, van Kuilenburg AB, van Gennip AH, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- Ball C, Gilchrist M, Coombs D. Modeling Within-Host Evolution of HIV: Mutation, Competition and Strain Replacement. Bulletin of Mathematical Biology. 2007;69:2361–2385. doi: 10.1007/s11538-007-9223-z. [DOI] [PubMed] [Google Scholar]
- Ball SC, Abraha A, Collins KR, Marozsan AJ, Baird H, Quinones-Mateu ME, Penn-Nicholson A, Murray M, Richard N, Lobritz M, Zimmerman PA, Kawamura T, Blauvelt A, Arts EJ. Comparing the Ex Vivo Fitness of CCR5-Tropic Human Immunodeficiency Virus Type 1 Isolates of Subtypes B and C. J Virol. 2003;77:1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baribaud F, Doms RW. The impact of chemokine receptor conformational heterogeneity on HIV infection. Cellular and Molecular Biology. 2001;47:653–660. [PubMed] [Google Scholar]
- Bebenek K, Beard WA, Casas-Finet JR, Kim HR, Darden TA, Wilson SH, Kunkel TA. Reduced Frameshift Fidelity and Processivity of HIV-1 Reverse Transcriptase Mutants Containing Alanine Substitutions in Helix H of the Thumb Subdomain. Journal of Biological Chemistry. 1995;270:19516–19523. doi: 10.1074/jbc.270.33.19516. [DOI] [PubMed] [Google Scholar]
- Becquart P, Chomont N, Roques P, Ayouba A, Kazatchkine MD, Belec L, Hocini H. Compartmentalization of HIV-1 between Breast Milk and Blood of HIV-Infected Mothers. Virology. 2002;300:109–117. doi: 10.1006/viro.2002.1537. [DOI] [PubMed] [Google Scholar]
- Berger EA. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- Berger EA, Murphy PM, Farber JM. CHEMOKINE RECEPTORS AS HIV-1 CORECEPTORS: Roles in Viral Entry, Tropism, and Disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Biesinger T, Yu Kimata MT, Kimata JT. Changes in simian immunodeficiency virus reverse transcriptase alleles that appear during infection of macaques enhance infectivity and replication in CD4+ T cells. Virology. 2008;370:184–193. doi: 10.1016/j.virol.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch MR, Learmont JC, Dyer WB, Deacon NJ, Zaunders JJ, Saksena N, Cunningham AL, Mills J, Sullivan JS. An examination of signs of disease progression in survivors of the Sydney Blood Bank Cohort (SBBC) Journal of Clinical Virology. 2001;22:263–270. doi: 10.1016/s1386-6532(01)00198-6. [DOI] [PubMed] [Google Scholar]
- Blish CA, Blay WM, Haigwood NL, Overbaugh J. Transmission of HIV-1 in the face of neutralizing antibodies. Current HIV Research. 2007;5:578–587. doi: 10.2174/157016207782418461. [DOI] [PubMed] [Google Scholar]
- Bofill M, Mocroft A, Lipman M, Medina E, Borthwick NJ, Sabin CA, Timms A, Winter M, Baptista L, Johnson MA, Lee CA, Phillips AN, Janossy G. Increased numbers of primed activated CD8+ CD38+ CD45RO+ T cells predict the decline of CD4+ T cells in HIV-1-infected patients. AIDS. 1996;10:827–834. doi: 10.1097/00002030-199607000-00005. [DOI] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MBA, Shaw GM. Antiviral pressure exerted by HIV-l-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- Briant L, Wade CM, Puel J, Brown AJ, Guyader M. Analysis of envelope sequence variants suggests multiple mechanisms of mother-to-child transmission of human immunodeficiency virus type 1. J Virol. 1995;69:3778–3788. doi: 10.1128/jvi.69.6.3778-3788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Derby NR, Kraft Z, Saunders CJ, Dai C, Llewellyn N, Zharkikh I, Vojtech L, Zhu T, Srivastava IK, Barnett SW, Stamatatos L. Viral evolution in macaques coinfected with CCR5- and CXCR4-tropic SHIVs in the presence or absence of vaccine-elicited anti-CCR5 SHIV neutralizing antibodies. Virology. 2006;355:138–151. doi: 10.1016/j.virol.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Campbell TB, Schneider K, Wrin T, Petropoulos CJ, Connick E. Relationship between In Vitro Human Immunodeficiency Virus Type 1 Replication Rate and Virus Load in Plasma. J Virol. 2003;77:12105–12112. doi: 10.1128/JVI.77.22.12105-12112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O’Brien SJ. HLA and HIV-1: Heterozygote Advantage and B*35-Cw*04 Disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- Charpentier C, Nora T, Tenaillon O, Clavel F, Hance AJ. Extensive Recombination among Human Immunodeficiency Virus Type 1 Quasispecies Makes an Important Contribution to Viral Diversity in Individual Patients. J Virol. 2006;80:2472–2482. doi: 10.1128/JVI.80.5.2472-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Nishio J, Perryman S, Cann A, O’Brien W, Chen IS, Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J Virol. 1991;65:5782–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-Chemokine Receptors CCR3 and CCR5 Facilitate Infection by Primary HIV-1 Isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for Human Immunodeficiency Virus Type 1 Envelope Glycosylation Variants with Shorter V1–V2 Loop Sequences Occurs during Transmission of Certain Genetic Subtypes and May Impact Viral RNA Levels. J Virol. 2005;79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Lucey DR, Zajac RA, Boswell RN, Gebel HM, Takahashi H, Berzofsky JA, Shearer GM. Detection of cytotoxic T lymphocytes specific for synthetic peptides of gp160 in HIV-seropositive individuals. J Immuno. 1991;146:2214–2219. [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- Coffin JM. HIV Population Dynamics in Vivo: Implications for Genetic Variation, Pathogenesis, and Therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Collins KL, Baltimore D. HIV’s evasion of the cellular immune response. Immuno Rev. 1999;168:65–74. doi: 10.1111/j.1600-065x.1999.tb01283.x. [DOI] [PubMed] [Google Scholar]
- Connor RI, Mohri H, Cao Y, Ho DD. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in Coreceptor Use Correlates with Disease Progression in HIV-1-Infected Individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Laure F, Brossard A, Bignozzi C, Goudeau A, Barin F, Brechot C. Frequent and early in utero HIV-1 infection. AIDS Research and Human Retroviruses. 1991;7:337–341. doi: 10.1089/aid.1991.7.337. [DOI] [PubMed] [Google Scholar]
- de Jong JJ, Goudsmit J, Keulen W, Klaver B, Krone W, Tersmette M, de Ronde A. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rossi A, Giaquinto C, Ometto L, Mammano F, Zanotto C, Dunn D, Chieco-Bianchi L. Replication and tropism of human immunodeficiency virus type 1 as predictors of disease outcome in infants with vertically acquired infection. Journal of Pediatrics. 1993;123:929–936. doi: 10.1016/s0022-3476(05)80389-0. [DOI] [PubMed] [Google Scholar]
- De Rossi A, Ometto L, Mammano F, Zanotto C, Giaquinto C, Chieco-Bianchi L. Vertical transmission of HIV-1: lack of detectable virus in peripheral blood cells of infected children at birth. AIDS. 1992;6:1117–1120. [PubMed] [Google Scholar]
- De Rossi A, Pasti M, Mammano F, Ometto L, Giaquinto C, Chieco-Bianchi L. Perinatal infection by human immunodeficiency virus type 1 (HIV-1): Relationship between proviral copy number in vivo, viral properties in vitro, and clinical outcome. Journal of Medical Virology. 2005;35:283–289. doi: 10.1002/jmv.1890350414. [DOI] [PubMed] [Google Scholar]
- Delwart EL, Sheppard HW, Walker BD, Goudsmit J, Mullins JI. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart E, Magierowska M, Royz M, Foley B, Peddada L, Smith R, Heldebrant C, Conrad A, Busch M. Homogeneous quasispecies in 16 out of 17 individuals during very early HIV-1 primary infection. [Miscellaneous Article] AIDS. 2002;16:189–195. doi: 10.1097/00002030-200201250-00007. [DOI] [PubMed] [Google Scholar]
- Delwart EL, Shpaer EG, Louwagie J, McCutchan FE, Grez M, Rubsamen-Waigmann H, Mullins JI. Genetic Relationships Determined by a DNA Heteroduplex Mobility Assay: Analysis of HIV-1 env Genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio PD, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. Envelope-Constrained Neutralization-Sensitive HIV-1 After Heterosexual Transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of Human Immunodeficiency Virus Type 1 to the Fusion Inhibitor T-20 Is Modulated by Coreceptor Specificity Defined by the V3 Loop of gp120. J Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. Macrophage Tropism of HIV-1 Depends on Efficient Cellular dNTP Utilization by Reverse Transcriptase. Journal of Biological Chemistry. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickover RE, Garratty EM, Plaeger S, Bryson YJ. Perinatal Transmission of Major, Minor, and Multiple Maternal Human Immunodeficiency Virus Type 1 Variants In Utero and Intrapartum. J Virol. 2001;75:2194–2203. doi: 10.1128/JVI.75.5.2194-2203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms RW, Peiper SC. Unwelcomed Guests with Master Keys: How HIV Uses Chemokine Receptors for Cellular Entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- Draenert R, Allen TM, Liu Y, Wrin T, Chappey C, Verrill CL, Sirera G, Eldridge RL, Lahaie MP, Ruiz L, Clotet B, Petropoulos CJ, Walker BD, Martinez-Picado J. Constraints on HIV-1 evolution and immunodominance revealed in monozygotic adult twins infected with the same virus. J Exp Med. 2006;203:529–539. doi: 10.1084/jem.20052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer WB, Ogg GS, Demoitie MA, Jin X, Geczy AF, Rowland-Jones SL, McMichael AJ, Nixon DF, Sullivan JS. Strong Human Immunodeficiency Virus (HIV)-Specific Cytotoxic T-Lymphocyte Activity in Sydney Blood Bank Cohort Patients Infected with nef-Defective HIV Type 1. J Virol. 1999;73:436–443. doi: 10.1128/jvi.73.1.436-443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnst A, Lindgren S, Dictor M, Johansson B, Sonnerborg A, Czajkowski J, Sundin G, Bohlin AB. HIV in pregnant women and their offspring: evidence for late transmission. Lancet. 1991;338:203–207. doi: 10.1016/0140-6736(91)90347-r. [DOI] [PubMed] [Google Scholar]
- Fauci AS. Host factors in the pathogenesis of HIV disease. Antibiotics and Chemotherapy. 1996;48:4–12. doi: 10.1159/000425151. [DOI] [PubMed] [Google Scholar]
- Feng JY, Anderson KS. Mechanistic Studies Examining the Efficiency and Fidelity of DNA Synthesis by the 3TC-Resistant Mutant (184V) of HIV-1 Reverse Transcriptase. Biochemistry. 1999;38:9440–9448. doi: 10.1021/bi990709m. [DOI] [PubMed] [Google Scholar]
- Fernandez CS, Smith MZ, Batten CJ, De Rose R, Reece JC, Rollman E, Venturi V, Davenport MP, Kent SJ. Vaccine-Induced T Cells Control Reversion of AIDS Virus Immune Escape Mutants. J Virol. 2007;81:4137–4144. doi: 10.1128/JVI.02193-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CS, Stratov I, De Rose R, Walsh K, Dale CJ, Smith MZ, Agy MB, Hu Sl, Krebs K, Watkins DI, O’Connor DH, Davenport MP, Kent SJ. Rapid Viral Escape at an Immunodominant Simian-Human Immunodeficiency Virus Cytotoxic T-Lymphocyte Epitope Exacts a Dramatic Fitness Cost. J Virol. 2005;79:5721–5731. doi: 10.1128/JVI.79.9.5721-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon JE, Gaur S, Gavai M, Gregory P, Frenkel LD, John JF., Jr Effect of the HIV-1 syncytium-inducing phenotype on disease stage in vertically-infected children. Journal of Medical Virology. 1998;55:56–63. doi: 10.1002/(sici)1096-9071(199805)55:1<56::aid-jmv10>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, Cullen C, Evans DT, Desrosiers RC, Mothe BR, Sidney J, Sette A, Kuntsman K, Wolinsky S, Piatak M, Lifson J, Hughes AL, Wilson N, O’Conner DH, Watkins DI. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nature Medicine. 2004;10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- Fuller SA, Hutton JJ, Meier J, Coleman MS. Deoxynucleotide-interconverting enzymes and the quantification of deoxynucleoside triphosphates in mammalian cells. Biochemical Journal. 1982;206:131–138. doi: 10.1042/bj2060131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furió V, Moya A, Sanjuán R. The cost of replication fidelity in an RNA virus. Proc Natl Acad Sci. 2005;102:10233–10237. doi: 10.1073/pnas.0501062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furió V, Moya A, Sanjuán R. The cost of replication fidelity in human immunodeficiency virus type 1. Proceedings of the Royal Society B: Biological Sciences. 2007;274:225–230. doi: 10.1098/rspb.2006.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gali Y, Berkhout B, Vanham G, Bakker M, Back NKT, Arien KK. Survey of the temporal changes in HIV-1 replicative fitness in the Amsterdam Cohort. Virology. 2007;364:140–146. doi: 10.1016/j.virol.2007.02.021. [DOI] [PubMed] [Google Scholar]
- Gao Q, Gu Z, Parniak MA, Cameron J, Cammack N, Boucher C, Wainberg MA. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geels MJ, Cornelissen M, Schuitemaker H, Anderson K, Kwa D, Maas J, Dekker JT, Baan E, Zorgdrager F, van den Burg R, van Beelen M, Lukashov VV, Fu TM, Paxton WA, van der Hoek L, Dubey SA, Shiver JW, Goudsmit J. Identification of Sequential Viral Escape Mutants Associated with Altered T-Cell Responses in a Human Immunodeficiency Virus Type 1-Infected Individual. J Virol. 2003;77:12430–12440. doi: 10.1128/JVI.77.23.12430-12440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi J, Hultin L, McKeating J, Johnson T, Owens B, Jacobson L, Shih R, Lewis J, Wiley D, Phair J, Wolinsky S, Detels R. Shorter Survival in Advanced Human Immunodeficiency Virus Type 1 Infection Is More Closely Associated with T Lymphocyte Activation than with Plasma Virus Burden or Virus Chemokine Coreceptor Usage. The Journal of Infectious Diseases. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- Glushakova S, Grivel JC, Fitzgerald W, Sylwester A, Zimmerberg J. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat Med. 1998;4:346–349. doi: 10.1038/nm0398-346. [DOI] [PubMed] [Google Scholar]
- Goepfert PA, Lumm W, Farmer P, Matthews P, Prendergast A, Carlson JM, Derdeyn CA, Tang J, Kaslow RA, Bansal A, Yusim K, Heckerman D, Mulenga J, Allen S, Goulder PJR, Hunter E. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205:1009–1017. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenow MM, Rose SL, Tuttle DL, Sleasman JW. HIV-1 fitness and macrophages. J Leuk Biol. 2003;74:657–666. doi: 10.1189/jlb.0403186. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Churchill M, Learmont J, Cherry C, Dyer WB, Wesselingh SL, Sullivan JS. Replication-Dependent Pathogenicity of Attenuated nef-Deleted HIV-1 In Vivo. Journal of Acquired Immune Deficiency Syndrome. 2007;46:390–394. doi: 10.1097/QAI.0b013e31815aba08. [DOI] [PubMed] [Google Scholar]
- Gottlieb G, Nickle D, Jensen M, Wong K, Kaslow R, Shepherd J, Margolick J, Mullins J. HIV Type 1 Superinfection with a Dual Tropic Virus and Rapid Progression to AIDS: A Case Report. Clinical Infectious Diseases. 2007;45:501–509. doi: 10.1086/520024. [DOI] [PubMed] [Google Scholar]
- Grobler J, Gray C, Rademeyer C, Seoighe C, Ramjee G, Karim S, Morris L, Williamson C. Incidence of HIV-1 Dual Infection and Its Association with Increased Viral Load Set Point in a Cohort of HIV-1 Subtype C-Infected Female Sex Workers. The Journal of Infectious Diseases. 2004;190:1355–1359. doi: 10.1086/423940. [DOI] [PubMed] [Google Scholar]
- Groenink M, Fouchier RA, Broersen S, Baker CH, Koot M, van’t Wout AB, Huisman HG, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- Grossman Z, Meier-Schellersheim M, Paul WE, Picker LJ. Pathogenesis of HIV infection: what the virus spares is as important as what it destroys. Nat Med. 2006;12:289–295. doi: 10.1038/nm1380. [DOI] [PubMed] [Google Scholar]
- Gu Z, Gao Q, Li X, Parniak MA, Wainberg MA. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J Virol. 1992;66:7128–7135. doi: 10.1128/jvi.66.12.7128-7135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AT. Perils At Mucosal Front Lines For HIV And SIV And Their Hosts. Nat Rev Immunol. 2005;5:783–792. doi: 10.1038/nri1706. [DOI] [PubMed] [Google Scholar]
- Harouse JM, Buckner C, Gettie A, Fuller R, Bohm R, Blanchard J, Cheng-Mayer C. CD8+ T cell-mediated CXC chemokine receptor 4-simian/human immunodeficiency virus suppression in dually infected rhesus macaques. Proc Natl Acad Sci. 2003;100:10977–10982. doi: 10.1073/pnas.1933268100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrowe G, Cheng-Mayer C. Amino Acid Substitutions in the V3 Loop Are Responsible for Adaptation to Growth in Transformed T-Cell Lines of a Primary Human Immunodeficiency Virus Type 1. Virology. 1995;210:490–494. doi: 10.1006/viro.1995.1367. [DOI] [PubMed] [Google Scholar]
- Hauschka PV. Analysis of nucleotide pools in animal cells. Methods in Cell Biology. 1973;7:361–462. doi: 10.1016/s0091-679x(08)61787-2. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Doms RW. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Molecular Membrane Biology. 1999;16:57–65. doi: 10.1080/096876899294760. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Zhang LQ, Simmonds P, Ludlam CA, Brown AJL. Convergent and Divergent Sequence Evolution in the Surface Envelope Glycoprotein of Human Immunodeficiency Virus Type 1 within a Single Infected Patient. Proc Natl Acad Sci. 1992;89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M, Inouye P, Rezende L, Richard N, Li Z, Prasad VR, Wainberg MA. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant than wild-type reverse transcriptase of human immunodeficiency virus type 1. Nucl Acids Res. 1997;25:4532–4536. doi: 10.1093/nar/25.22.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M, Harouse JM, Gettie A, Buckner C, Blanchard J, Cheng-Mayer C. Increased Mucosal Transmission but Not Enhanced Pathogenicity of the CCR5-Tropic, Simian AIDS-Inducing Simian/Human Immunodeficiency Virus SHIVSF162P3 Maps to Envelope gp120. J Virol. 2003;77:989–998. doi: 10.1128/JVI.77.2.989-998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Hatano H, Hammond SP, Smith D, Wild M, Gupta S, Whitcomb J, Kalayjian RC, Gripshover B, Kuritzkes DR. Virologic Characterization of HIV Type 1 With a Codon 70 Deletion in Reverse Transcriptase. J Acquir Immune Defic Syndr. 2007;45:494–500. doi: 10.1097/QAI.0b013e31806ada48. [DOI] [PubMed] [Google Scholar]
- Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Carl S, Bronson S, Stahl-Hennig C, Swigut T, Skowronski J, Kirchhoff F. Disrupting Surfaces of Nef Required for Downregulation of CD4 and for Enhancement of Virion Infectivity Attenuates Simian Immunodeficiency Virus Replication In Vivo. J Virol. 2000;74:9836–9844. doi: 10.1128/jvi.74.21.9836-9844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaherian K, Langlois AJ, LaRosa GJ, Profy AT, Bolognesi DP, Herlihy WC, Putney SD, Matthews TJ. Broadly Neutralizing Antibodies Elicited by the Hypervariable Neutralizing Determinant of HIV-1. Science. 1990;250:1590–1593. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Desrosiers RC. VIRAL PERSISTENCE: HIV’s Strategies of Immune System Evasion. Annual Review of Medicine. 2002;53:499–518. doi: 10.1146/annurev.med.53.082901.104053. [DOI] [PubMed] [Google Scholar]
- Jonckheere H, Witvrouw M, De Clercq E, Anné J. Lamivudine resistance of HIV type 1 does not delay development of resistance to nonnucleoside HIV type 1-specific reverse transcriptase inhibitors as compared with wild-type HIV type 1. AIDS Research and Human Retroviruses. 1998;14:249–253. doi: 10.1089/aid.1998.14.249. [DOI] [PubMed] [Google Scholar]
- Jones NA, Wei X, Flower DR, Wong M, Michor F, Saag MS, Hahn BH, Nowak MA, Shaw GM, Borrow P. Determinants of Human Immunodeficiency Virus Type 1 Escape from the Primary CD8+ Cytotoxic T Lymphocyte Response. J Exp Med. 2004;200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A, Maier R, Vartanian JP, Bocharov G, Jung V, Fischer U, Meese E, Wain-Hobson S, Meyerhans A. Multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- Jurriaans S, Van Gemen B, Weverling GJ, Van Strup D, Nara P, Coutinho R, Koot M, Schuitemaker H, Goudsmit J. The Natural History of HIV-1 Infection: Virus Load and Virus Phenotype Independent Determinants of Clinical Course? Virology. 1994;204:223–233. doi: 10.1006/viro.1994.1526. [DOI] [PubMed] [Google Scholar]
- Karlsson AC, Hecht FM, Nixon DF. Sequential Broadening of CTL Responses in Early HIV-1 Infection Is Associated with Viral Escape. PLoS ONE. 2007;2:e225. doi: 10.1371/journal.pone.0000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keet IP, Krihnen P, Koot M, Lange JM, Miedema F, Goudsmit J, Coutinho RA. Predictors of rapid progression to AIDS in HIV-1 seroconverters. AIDS. 1993;7:51–57. doi: 10.1097/00002030-199301000-00008. [DOI] [PubMed] [Google Scholar]
- Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. Clustered Mutations in HIV-1 gag Are Consistently Required for Escape from HLA-B27-restricted Cytotoxic T Lymphocyte Responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal KS, Foley B, Burger H, Anastos K, Minkoff H, Kitchen C, Philpott SM, Gao W, Robison E, Holman S, Dehner C, Beck S, Meyer WA, III, Landay A, Kovacs A, Bremer J, Weiser B. HIV-1 in genital tract and plasma of women: Compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci. 2003;100:12972–12977. doi: 10.1073/pnas.2134064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- Kimata JT. HIV-1 Fitness and Disease Progression: Insights from the SIV-Macaque Model. Current HIV Research. 2006;4:65–77. doi: 10.2174/157016206775197628. [DOI] [PubMed] [Google Scholar]
- Kimata JT, Kuller L, Anderson DB, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nature Medicine. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- Kimura M. On the evolutionary adjustment of spontaneous mutation rates. Genetic Research. 1967;9:23–34. [Google Scholar]
- Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Absence of Intact nef Sequences in a Long-Term Survivor with Nonprogressive HIV-1 Infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- Kliks SC, Wara DW, Landers DV, Levy JA. Features of HIV-1 that could influence maternal-child transmission. JAMA: The Journal of the American Medical Association. 1994;272:467–474. [PubMed] [Google Scholar]
- Koito A, Harrowe G, Levy JA, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koito A, Stamatatos L, Cheng-Mayer C. Small Amino Acid Sequence Changes within the V2 Domain Can Affect the Function of a T-Cell Line-Tropic Human Immunodeficiency Virus Type 1 Envelope gp120. Virology. 1995;206:878–884. doi: 10.1006/viro.1995.1010. [DOI] [PubMed] [Google Scholar]
- Koot M, Vos AH, Keet RP, de Goede RE, Dercksen MW, Terpstra FG, Coutinho RA, Miedema F, Tersmette M. HIV-1 biological phenotype in long-term infected individuals evaluated with an MT-2 cocultivation assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- Koot M, Keet IPM, Vos AHV, de Goede REY, Roos MT, Coutinho RA, Miedema F, Schellekens PT, Tersmette M. Prognostic Value of HIV-1 Syncytium-inducing Phenotype for Rate of CD4+ Cell Depletion and Progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- Kozaczynska K, Cornelissen M, Reiss P, Zorgdrager F, van der Kuyl AC. HIV-1 sequence evolution in vivo after superinfection with three viral strains. Retrovirology. 2007;4:59–72. doi: 10.1186/1742-4690-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken CL, Zwart G, Baan E, Coutinho RA, Hoek JA, Goudsmit J. Increasing Antigenic and Genetic Diversity of the V3 Variable Domain of the Human Immunodeficiency Virus Envelope Protein in the Course of the AIDS Epidemic. Proc Natl Acad Sci. 1993;90:9061–9065. doi: 10.1073/pnas.90.19.9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa D, Vingerhoed J, Boeser B, Schuitemaker H. Increased In Vitro Cytopathicity of CC Chemokine Receptor 5□ÇôRestricted Human Immunodeficiency Virus Type 1 Primary Isolates Correlates with a Progressive Clinical Course of Infection. The Journal of Infectious Diseases. 2003;187:1397–1403. doi: 10.1086/374650. [DOI] [PubMed] [Google Scholar]
- Laakso MM, Sutton RE. Replicative fidelity of lentiviral vectors produced by transient transfection. Virology. 2006;348:406–417. doi: 10.1016/j.virol.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Labrosse B, Labernardiere JL, Dam E, Trouplin V, Skrabal K, Clavel F, Mammano F. Baseline Susceptibility of Primary Human Immunodeficiency Virus Type 1 to Entry Inhibitors. J Virol. 2003;77:1610–1613. doi: 10.1128/JVI.77.2.1610-1613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforge M, Petit F, Estaquier J, Senik A. Commitment to Apoptosis in CD4+ T Lymphocytes Productively Infected with Human Immunodeficiency Virus Type 1 Is Initiated by Lysosomal Membrane Permeabilization, Itself Induced by the Isolated Expression of the Viral Protein Nef. J Virol. 2007;81:11426–11440. doi: 10.1128/JVI.00597-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Sleasman JW, She JX, Barrie KA, Pomeroy SM, Barrett DJ, Goodenow MM. Independent variation and positive selection in env V1 and V2 domains within maternal-infant strains of human immunodeficiency virus type 1 in vivo. J Virol. 1993;67:3951–3960. doi: 10.1128/jvi.67.7.3951-3960.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers SL, Sleasman JW, She JX, Barrie KA, Pomeroy SM, Barrett DJ, Goodenow MM. Persistence of multiple maternal genotypes of human immunodeficiency virus type I in infants infected by vertical transmission. J Clin Invest. 1994;93:380–390. doi: 10.1172/JCI116970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder BA, Kemp SD, Harrigan PR. Potential Mechanism for Sustained Antiretroviral Efficacy of AZT-3TC Combination Therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- LaRosa GJ, Davide JP, Weinhold K, Waterbury JA, Profy AT, Lewis JA, Langlois AJ, Dreesman GR, Boswell RN, Shadduck P, Holley LH, Karplus M, Bolognesi DP, Matthews TJ, Emini EA, Putney SD. Conserved Sequence and Structural Elements in the HIV-1 Principal Neutralizing Determinant: Corrections and Clarifications. Science. 1991;251:811. doi: 10.1126/science.1990444. [DOI] [PubMed] [Google Scholar]
- Learmont JC, Geczy AF, Mills J, Ashton LJ, Raynes-Greenow CH, Garsia RJ, Dyer WB, McIntyre L, Oelrichs RB, Rhodes DI, Deacon NJ, Sullivan JS, McPhee DA, Crowe S, Solomon AE, Chatfield C, Blasdall S, Kuipers H The Sydney Blood Bank Cohort Research Group. Immunologic and Virologc Status after 14 to 18 Years of Infection with an Attenuated Strain of HIV-1 -- A Report from the Sydney Blood Bank Cohort. N Engl J Med. 1999;340:1715–1722. doi: 10.1056/NEJM199906033402203. [DOI] [PubMed] [Google Scholar]
- Learn GH, Muthui D, Brodie SJ, Zhu T, Diem K, Mullins JI, Corey L. Virus Population Homogenization following Acute Human Immunodeficiency Virus Type 1 Infection. J Virol. 2002;76:11953–11959. doi: 10.1128/JVI.76.23.11953-11959.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Kosakovsky Pond SL, Drummond AJ, Pybus OG, Shapiro B, Barroso H, Taveira N, Rambaut A. Synonymous Substitution Rates Predict HIV Disease Progression as a Result of Underlying Replication Dynamics. PLoS Computational Biology. 2007;3:e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P, Van de Perre PHIL, Carael M, Nsengumuremyi F, Nkurunziza J, Butzler JP, Sprecher S. Postnatal transmission of HIV from mother to child. Lancet. 1987;2:400. doi: 10.1016/s0140-6736(87)92423-8. [DOI] [PubMed] [Google Scholar]
- Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, John AS, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJR. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci. 2004;101:4204–4209. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JA. Pathogenesis of human immunodeficiency virus infection. Microbiol Mol Biol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman DR. Chemokine Receptors: Keys to AIDS Pathogenesis? Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- Lobritz MA, Marozsan AJ, Troyer RM, Arts EJ. Natural Variation in the V3 Crown of Human Immunodeficiency Virus Type 1 Affects Replicative Fitness and Entry Inhibitor Sensitivity. J Virol. 2007;81:8258–8269. doi: 10.1128/JVI.02739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EM, Martin HL, Kreiss JK, Rainwater SMJ, Lavreys L, Jackson DJ, Rakwar J, Mandaliya K, Overbaugh J. Gender differences in HIV-1 diversity at time of infection. Nat Med. 2000;6:71–75. doi: 10.1038/71563. [DOI] [PubMed] [Google Scholar]
- Long EM, Rainwater SMJ, Lavreys L, Mandaliya K, Overbaugh J. HIV Type 1 Variants Transmitted to Women in Kenya Require the CCR5 Coreceptor for Entry, Regardless of the Genetic Complexity of the Infecting Virus. AIDS Research and Human Retroviruses. 2002;18:567–576. doi: 10.1089/088922202753747914. [DOI] [PubMed] [Google Scholar]
- Lu X, Wu X, Plemenitas A, Yu H, Sawai ET, Abo A, Peterlin BM. CDC42 and Rac1 are implicated in the activation of the Nef-associated kinase and replication of HIV-1. Current Biology. 1996;6:1677–1684. doi: 10.1016/s0960-9822(02)70792-6. [DOI] [PubMed] [Google Scholar]
- Ludlam CA, Tucker J, Steel CM, Tedder RS, Cheingsong-Popov R, Weiss RA, McClelland DB, Philp I, Prescott RJ. Human T-lymphotropic virus type III (HTLV-III) infection in seronegative haemophiliacs after transfusion of factor VIII. Lancet. 1985;ii:233–236. doi: 10.1016/s0140-6736(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozsan AJ, Moore DM, Lobritz MA, Fraundorf E, Abraha A, Reeves JD, Arts EJ. Differences in the Fitness of Two Diverse Wild-Type Human Immunodeficiency Virus Type 1 Isolates Are Related to the Efficiency of Cell Binding and Entry. J Virol. 2005;79:7121–7134. doi: 10.1128/JVI.79.11.7121-7134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. Fitness Cost of Escape Mutations in p24 Gag in Association with Control of Human Immunodeficiency Virus Type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mild M, Esbjornsson J, Fenyo EM, Medstrand P. Frequent Intrapatient Recombination between Human Immunodeficiency Virus Type 1 R5 and X4 Envelopes: Implications for Coreceptor Switch. J Virol. 2007;81:3369–3376. doi: 10.1128/JVI.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder-Kampinga GA, Kuiken C, Dekker J, Scherpbier HJ, Boer K, Goudsmit J. Genomic human immunodeficiency virus type 1 RNA variation in mother and child following intra-uterine virus transmission. J Gen Virol. 1993;74:1747–1756. doi: 10.1099/0022-1317-74-9-1747. [DOI] [PubMed] [Google Scholar]
- Narwa R, Roques P, Courpotin C, Parnet-Mathieu F, Boussin F, Roane A, Marce D, Lasfargues G, Dormont D. Characterization of human immunodeficiency virus type 1 p17 matrix protein motifs associated with mother-to-child transmission. J Virol. 1996;70:4474–4483. doi: 10.1128/jvi.70.7.4474-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak P, Karlsson AC, Naver L, Bohlin AB, Piasek A, Sonnerborg A. The selection and evolution of viral quasispecies in HIV-1 infected children. HIV Medicine. 2002;3:1–11. doi: 10.1046/j.1464-2662.2001.00097.x. [DOI] [PubMed] [Google Scholar]
- O’Brien WA, Koyanagi Y, Namazie A, Zhao JQ, Diagne A, ldler K, Zack JA, Chen ISY. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- Ogert RA, Lee MK, Ross W, Buckler-White A, Martin MA, Cho MW. N-Linked Glycosylation Sites Adjacent to and within the V1/V2 and the V3 Loops of Dualtropic Human Immunodeficiency Virus Type 1 Isolate DH12 gp120 Affect Coreceptor Usage and Cellular Tropism. J Virol. 2001;75:5998–6006. doi: 10.1128/JVI.75.13.5998-6006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares I, Gutierrez-Rivas M, Lopez-Galindez C, Menendez-Arias L. Tryptophan scanning mutagenesis of aromatic residues within the polymerase domain of HIV-1 reverse transcriptase: critical role of Phe-130 for p51 function and second-site revertant restoring viral replication capacity. Virology. 2004;324:400–411. doi: 10.1016/j.virol.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Otten R, Ellenberger D, Adams D, Fridlund C, Jackson E, Pieniazek D, Rayfield M. Identification of a Window Period for Susceptibility to Dual Infection with Two Distinct Human Immunodeficiency Virus Type 2 Isolates in a Macaca nemestrina (Pig□ÇÉtailed Macaque) Model. The Journal of Infectious Diseases. 1999;180:673–684. doi: 10.1086/314968. [DOI] [PubMed] [Google Scholar]
- Overbaugh J, Rudensey LM. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol. 1992;66:5937–5948. doi: 10.1128/jvi.66.10.5937-5948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Schlesinger Y, Daar ES, Moudgil T, Ho DD, Chen IS. Rapid generation of sequence variation during primary HIV-1 infection. AIDS. 1992;6:452–460. doi: 10.1097/00002030-199205000-00003. [DOI] [PubMed] [Google Scholar]
- Pasquier C, Cayrou C, Blancher A, Tourne-Petheil C, Berrebi A, Tricoire J, Puel J, Izopet J. Molecular Evidence for Mother-to-Child Transmission of Multiple Variants by Analysis of RNA and DNA Sequences of Human Immunodeficiency Virus Type 1. J Virol. 1998;72:8493–8501. doi: 10.1128/jvi.72.11.8493-8501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore C, Nedellec R, Ramos A, Pontow S, Ratner L, Mosier DE. Human Immunodeficiency Virus Type 1 Coreceptor Switching: V1/V2 Gain-of-Fitness Mutations Compensate for V3 Loss-of-Fitness Mutations. J Virol. 2006;80:750–758. doi: 10.1128/JVI.80.2.750-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PG, Yu Kimata MT, Biggins JE, Wilson JM, Kimata JT. Highly Pathogenic Simian Immunodeficiency Virus mne Variants That Emerge during the Course of Infection Evolve Enhanced Infectivity and the Ability to Downregulate CD4 but Not Class I Major Histocompatibility Complex Antigens. J Virol. 2002;76:6425–6434. doi: 10.1128/JVI.76.13.6425-6434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Sullivan WM, Duenas-Decamp MJ, Bhattacharya J, Ankghuambom C, Brown R, Luzuriaga K, Bell J, Simmonds P, Ball J, Clapham PR. Non-Macrophage-Tropic Human Immunodeficiency Virus Type 1 R5 Envelopes Predominate in Blood, Lymph Nodes, and Semen: Implications for Transmission and Pathogenesis. J Virol. 2006;80:6324–6332. doi: 10.1128/JVI.02328-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peut V, Kent SJ. Utility of Human Immunodeficiency Virus Type 1 Envelope as a T-Cell Immunogen. J Virol. 2007;81:13125–13134. doi: 10.1128/JVI.01408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyerl FW, Bazick HS, Newberg MH, Barouch DH, Sodroski J, Letvin NL. Fitness Costs Limit Viral Escape from Cytotoxic T Lymphocytes at a Structurally Constrained Epitope. J Virol. 2004;78:13901–13910. doi: 10.1128/JVI.78.24.13901-13910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott S, Burger H, Tsoukas C, Foley B, Anastos K, Kitchen C, Weiser B. Human Immunodeficiency Virus Type 1 Genomic RNA Sequences in the Female Genital Tract and Blood: Compartmentalization and Intrapatient Recombination. J Virol. 2005;79:353–363. doi: 10.1128/JVI.79.1.353-363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantadosi A, Chohan B, Chohan V, McClelland RS, Overbaugh J. Chronic HIV-1 Infection Frequently Fails to Protect against Superinfection. PLos Pathogens. 2007;3:e177. doi: 10.1371/journal.ppat.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilcher CD, Price MA, Hoffman IF, Galvin S, Martinson FE, Kazembe PN, Eron JJ, Miller WC, Fiscus SA, Cohen MS. Frequent detection of acute primary HIV infection in men in Malawi. AIDS. 2004a;18:517–524. doi: 10.1097/00002030-200402200-00019. [DOI] [PubMed] [Google Scholar]
- Pilcher CD, Tien HC, Eron J, Vernazza PL, Leu SY, Stewart PW, Goh L, Cohen MS. Brief but Efficient: Acute HIV Infection and the Sexual Transmission of HIV. The Journal of Infectious Diseases. 2004b;189:1785–1792. doi: 10.1086/386333. [DOI] [PubMed] [Google Scholar]
- Poignard P, Saphire EO, Parren PW, Burton DR. GP120: Biologic Aspects of Structural Features. Annu Rev Immunol. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- Pope M, Haase AT. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat Med. 2003;9:847–852. doi: 10.1038/nm0703-847. [DOI] [PubMed] [Google Scholar]
- Poss M, Martin HL, Kreiss JK, Granville L, Chohan B, Nyange P, Mandaliya K, Overbaugh J. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol. 1995;69:8118–8122. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- Quinones-Mateu ME, Ball SC, Marozsan AJ, Torre VS, Albright JL, Vanham G, van der Groen G, Colebunders RL, Arts EJ. A Dual Infection/Competition Assay Shows a Correlation between Ex Vivo Human Immunodeficiency Virus Type 1 Fitness and Disease Progression. J Virol. 2000;74:9222–9233. doi: 10.1128/jvi.74.19.9222-9233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel HR, Weber J, Chakraborty B, Gutierrez A, Marotta ML, Mirza M, Kiser P, Martinez MA, Este JA, Quinones-Mateu ME. Role of the Human Immunodeficiency Virus Type 1 Envelope Gene in Viral Fitness. J Virol. 2003;77:9069–9073. doi: 10.1128/JVI.77.16.9069-9073.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkema GH, Saksela K. Interactions of HIV-1 Nef With Cellular Signal Transducing Proteins. Fron Biosci. 2000;5:268–283. doi: 10.2741/renkema. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritola K, Pilcher CD, Fiscus SA, Hoffman NG, Nelson JAE, Kitrinos KM, Hicks CB, Eron JJ, Jr, Swanstrom R. Multiple V1/V2 env Variants Are Frequently Present during Primary Infection with Human Immunodeficiency Virus Type 1. J Virol. 2004;78:11208–11218. doi: 10.1128/JVI.78.20.11208-11218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Roeth JF, Collins KL. Human Immunodeficiency Virus Type 1 Nef: Adapting to Intracellular Trafficking Pathways. Microbiol Mol Biol Rev. 2006;70:548–563. doi: 10.1128/MMBR.00042-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross TM, Cullen BR. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth WW, Zuberi JA, Stringer HG, Jr, Davidson SK, Bond VC. Examination of HIV type 1 variants in mother-child pairs. AIDS Research and Human Retroviruses. 1996;12:925–930. doi: 10.1089/aid.1996.12.925. [DOI] [PubMed] [Google Scholar]
- Rudensey LM, Kimata JT, Benveniste RE, Overbaugh J. Progression to AIDS in Macaques Is Associated with Changes in the Replication, Tropism, and Cytopathic Properties of the Simian Immunodeficiency Virus Variant Population. Virology. 1995;207:528–542. doi: 10.1006/viro.1995.1113. [DOI] [PubMed] [Google Scholar]
- Rudensey LM, Kimata JT, Long EM, Chackerian B, Overbaugh J. Changes in the Extracellular Envelope Glycoprotein of Variants That Evolve during the Course of Simian Immunodeficiency Virus SIVMne Infection Affect Neutralizing Antibody Recognition, Syncytium Formation, and Macrophage Tropism but Not Replication, Cytopathicity, or CCR-5 Coreceptor Recognition. J Virol. 1998;72:209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Kirkegaard E, Long EM, Celum C, Buchbinder S, Daar ES, Overbaugh J. Human Immunodeficiency Virus Type 1 (HIV-1) Diversity at Time of Infection Is Not Restricted to Certain Risk Groups or Specific HIV-1 Subtypes. J Virol. 2004;78:7279–7283. doi: 10.1128/JVI.78.13.7279-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Lavreys L, Baeten JM, Richardson BA, Mandaliya K, Chohan BH, Kreiss JK, Overbaugh J. Infection with Multiple Human Immunodeficiency Virus Type 1 Variants Is Associated with Faster Disease Progression. J Virol. 2003;77:12921–12926. doi: 10.1128/JVI.77.23.12921-12926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Wu X, Lee S, Overbaugh J. Human Immunodeficiency Virus Type 1 V1-V2 Envelope Loop Sequences Expand and Add Glycosylation Sites over the Course of Infection, and These Modifications Affect Antibody Neutralization Sensitivity. J Virol. 2006;80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti G, Leitner T, Halapi E, Wahlberg J, Marchisio P, Clerici-Schoeller MA, Wigzell H, Fenyo EM, Albert J, Uhlen M, Rossi P. Comparison of Variable Region 3 Sequences of Human Immunodeficiency Virus Type 1 from Infected Children with the RNA and DNA Sequences of the Virus Populations of Their Mothers. Proc Natl Acad Sci. 1993;90:1721–1725. doi: 10.1073/pnas.90.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, van Steenwijk RP, Lange JM, Schattenkerk JK, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent Viral Evolutionary Changes Associated with the Progression of Human Immunodeficiency Virus Type 1 Infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T, Levy JA, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp!20 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- Skoog L, Bjursell G. Nuclear and Cytoplasmic Pools of Deoxyribonucleoside Triphosphates in Chinese Hamster Ovary Cells. Journal of Biological Chemistry. 1974;249:6434–6438. [PubMed] [Google Scholar]
- Soeiro R, Rubinstein A, Rashbaum WK, Lyman WD. Maternofetal transmission of AIDS: frequency of human immunodeficiency virus type 1 nucleic acid sequences in human fetal DNA. Journal of Infectious Diseases. 1992;166:699–703. doi: 10.1093/infdis/166.4.699. [DOI] [PubMed] [Google Scholar]
- Sousa AE, Carneiro J, Meier-Schellersheim M, Grossman Z, Victorino RMM. CD4 T Cell Depletion Is Linked Directly to Immune Activation in the Pathogenesis of HIV-1 and HIV-2 but Only Indirectly to the Viral Load. J Immuno. 2002;169:3400–3406. doi: 10.4049/jimmunol.169.6.3400. [DOI] [PubMed] [Google Scholar]
- Spencer LT, Ogino MT, Dankner WM, Spector SA. Clinical significance of human immunodeficiency virus type 1 phenotypes in infected children. Journal of Infectious Diseases. 1994;169:491–495. doi: 10.1093/infdis/169.3.491. [DOI] [PubMed] [Google Scholar]
- Stilianakis NI, Schenzle D. On the intra-host dynamics of HIV-1 infections. Mathematical Biosciences. 2006;199:1–25. doi: 10.1016/j.mbs.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Strizki JM, Xu S, Wagner NE, Wojcik L, Liu J, Hou Y, Endres M, Palani A, Shapiro S, Clader JW, Greenlee WJ, Tagat JR, McCombie S, Cox K, Fawzi AB, Chou CC, Pugliese-Sivo C, Davies L, Moreno ME, Ho DD, Trkola A, Stoddart CA, Moore JP, Reyes GR, Baroudy BM. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc Natl Acad Sci. 2001;98:12718–12723. doi: 10.1073/pnas.221375398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N, Thali M, Furman C, Ho DD, Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J Virol. 1993;67:3674–3679. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Akutsu M, Murayama K, Shimizu N, Hoshino H. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol. 1991;65:1710–1718. doi: 10.1128/jvi.65.4.1710-1718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersmette M, de Goede RE, Al BJ, Winkel IN, Gruters RA, Cuypers HT, Huisman HG, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersmette M, Gruters RA, de Wolf F, de Goede RE, Lange JM, Schellekens PT, Goudsmit J, Huisman HG, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale M, Alnadaf T, Cousens D. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob Agents Chemother. 1997;41:1094–1098. doi: 10.1128/aac.41.5.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. Human Immunodeficiency Virus Envelope V1 and V2 Regions Influence Replication Efficiency in Macrophages by Affecting Virus Spread. Virology. 1995;213:70–79. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- Torre VS, Marozsan AJ, Albright JL, Collins KR, Hartley O, Offord RE, Quinones-Mateu ME, Arts EJ. Variable Sensitivity of CCR5-Tropic Human Immunodeficiency Virus Type 1 Isolates to Inhibition by RANTES Analogs. J Virol. 2000;74:4868–4876. doi: 10.1128/jvi.74.10.4868-4876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg E, Korber B, Sollars C, Kepler TB, Hraber PT, Hayes E, Funkhouser R, Fugate M, Theiler J, Hsu YS, Kunstman K, Wu S, Phair J, Erlich H, Wolinsky S. Advantage of rare HLA supertype in HIV disease progression. Nat Med. 2003;9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- Traut TW. Physiological concentrations of purines and pyrimidines. Molecular and Cellular Biochemistry. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Trkola A, Paxton WA, Monard SP, Hoxie JA, Siani MA, Thompson DA, Wu L, Mackay CR, Horuk R, Moore JP. Genetic Subtype-Independent Inhibition of Human Immunodeficiency Virus Type 1áReplication by CC and CXC Chemokines. J Virol. 1998;72:396–404. doi: 10.1128/jvi.72.1.396-404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, Toossi Z, Colebunders RL, Jensen MA, Mullins JI, Vanham G, Arts EJ. Changes in Human Immunodeficiency Virus Type 1 Fitness and Genetic Diversity during Disease Progression. J Virol. 2005;79:9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Worobey M, Visser JA, Schuitemaker H. Evolution of R5 and X4 human immunodeficiency virus type 1 gag sequences in vivo: evidence for recombination. Virology. 2003;314:451–459. doi: 10.1016/s0042-6822(03)00454-9. [DOI] [PubMed] [Google Scholar]
- van’t Wout AB, Kootstra NA, Mulder-Kampinga GA, Albrecht-van Lent N, Scherpbier HJ, Veenstra J, Boer K, Coutinho RA, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian JP, Plikat U, Henry M, Mahieux R, Guillemot L, Meyerhans A, Wain-Hobson S. HIV genetic variation is directed and restricted by DNA precursor availability. Journal of Molecular Biology. 1997;270:139–151. doi: 10.1006/jmbi.1997.1104. [DOI] [PubMed] [Google Scholar]
- Vernazza PL, Eron JJ, Fiscus SA, Cohen MS. Sexual transmission of HIV: infectiousness and prevention. AIDS. 1999;13:155–166. doi: 10.1097/00002030-199902040-00003. [DOI] [PubMed] [Google Scholar]
- Veronese FD, Copeland TD, DeVico AL, Rahman R, Oroszlan S, Gallo RC, Sarngadharan MG. Characterization of Highly Immunogenic p66/p51 as the Reverse Transcriptase of HTLV-III/LAV. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- Voronin Y, Overbaugh J, Emerman M. Simian Immunodeficiency Virus Variants That Differ in Pathogenicity Differ in Fitness under Rapid Cell Turnover Conditions. J Virol. 2005;79:15091–15098. doi: 10.1128/JVI.79.24.15091-15098.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Lobidel D, Brown AJ. Analysis of human immunodeficiency virus type 1 env and gag sequence variants derived from a mother and two vertically infected children provides evidence for the transmission of multiple sequence variants. J Gen Virol. 1998;79:1055–1068. doi: 10.1099/0022-1317-79-5-1055. [DOI] [PubMed] [Google Scholar]
- Wagner R, Leschonsky B, Harrer E, Paulus C, Weber C, Walker BD, Buchbinder S, Wolf H, Kalden JR, Harrer T. Molecular and Functional Analysis of a Conserved CTL Epitope in HIV-1 p24 Recognized from a Long-Term Nonprogressor: Constraints on Immune Escape Associated with Targeting a Sequence Essential for Viral Replication. J Immuno. 1999;162:3727–3734. [PubMed] [Google Scholar]
- Wainberg MA, Drosopoulos WC, Salomon H, Hsu M, Borkow G, Parniak MA, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad VR. Enhanced Fidelity of 3TC-Selected Mutant HIV-1 Reverse Transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- Wakefield JK, Jablonski SA, Morrow CD. In vitro enzymatic activity of human immunodeficiency virus type 1 reverse transcriptase mutants in the highly conserved YMDD amino acid motif correlates with the infectious potential of the proviral genome. J Virol. 1992;66:6806–6812. doi: 10.1128/jvi.66.11.6806-6812.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter H, Schmidt B, Werwein M, Schwingel E, Korn K. Prediction of Abacavir Resistance from Genotypic Data: Impact of Zidovudine and Lamivudine Resistance In Vitro and In Vivo. Antimicrob Agents Chemother. 2002;46:89–94. doi: 10.1128/AAC.46.1.89-94.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zhu T, Ho DD. Sequence diversity of V1 and V2 domains of gp120 from human immunodeficiency virus type 1: lack of correlation with viral phenotype. J Virol. 1995;69:2708–2715. doi: 10.1128/jvi.69.4.2708-2715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawer MJ, Gray RH, Sewankambo NK, Serwadda D, Li X, Laeyendecker O, Kiwanuka N, Kigozi G, Kiddugavu M, Lutalo T, Nalugoda F, Wabwire-Mangen F, Meehan M, Quinn T. Rates of HIV-1 Transmission per Coital Act, by Stage of HIV-1 Infection, in Rakai, Uganda. The Journal of Infectious Diseases. 2005;191:1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- Wei BL, Arora VK, Foster JL, Sodora DL, Garcia JV. In Vivo Analysis of Nef Function. Current HIV Research. 2003a;1:41–50. doi: 10.2174/1570162033352057. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003b;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Westervelt P, Gendelman HE, Ratner L. Identification of a Determinant within the Human Immunodeficiency Virus 1 Surface Envelope Glycoprotein Critical for Productive Infection of Primary Monocytes. Proc Natl Acad Sci. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westervelt P, Trowbridge DB, Epstein LG, Blumberg BM, Li Y, Hahn BH, Shaw GM, Price RW, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KL, Margot NA, Wrin T, Petropoulos CJ, Miller MD, Naeger LK. Molecular Mechanisms of Resistance to Human Immunodeficiency Virus Type 1 with Reverse Transcriptase Mutations K65R and K65R+M184V and Their Effects on Enzyme Function and Viral Replication Capacity. Antimicrob Agents Chemother. 2002;46:3437–3446. doi: 10.1128/AAC.46.11.3437-3446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wike CM, Korber BT, Daniels MR, Hutto C, Munoz J, Furtado M, Parks W, Saah A, Bulterys M, Kurawige JB. HIV-1 sequence variation between isolates from mother-to-infant transmission pairs. AIDS Research and Human Retroviruses. 1992;8:1297–1300. doi: 10.1089/aid.1992.8.1297. [DOI] [PubMed] [Google Scholar]
- Williamson S, Perry SM, Bustamante CD, Orive ME, Stearns MN, Kelly JK. A Statistical Characterization of Consistent Patterns of Human Immunodeficiency Virus Evolution Within Infected Patients. Mol Biol Evol. 2005;22:456–468. doi: 10.1093/molbev/msi029. [DOI] [PubMed] [Google Scholar]
- Wodarz D, Levy DN. Human immunodeficiency virus evolution towards reduced replicative fitness in vivo and the development of AIDS. Proceedings of the Royal Society B: Biological Sciences. 2007;274:2481–2490. doi: 10.1098/rspb.2007.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfs TFW, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- Wolinsky SM, Veazey RS, Kunstman KJ, Klasse PJ, Dufour J, Marozsan AJ, Springer MS, Moore JP. Effect of a CCR5 inhibitor on viral loads in macaques dual-infected with R5 and X4 primate immunodeficiency viruses. Virology. 2004;328:19–29. doi: 10.1016/j.virol.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Wolinsky SM, Wike CM, Korber BTM, Hutto C, Parks WP, Rosenblum LL, Kunstman KJ, Furtado MR, Munoz JL. Selective Transmission of Human Immunodeficiency Virus Type-1 Variants from Mothers to Infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SMJ, Overbaugh J. Neutralization Escape Variants of Human Immunodeficiency Virus Type 1 Are Transmitted from Mother to Infant. J Virol. 2006;80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci. 2003;100:6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Matsushita S, Hayashi A, Takatsuki K. Relationship of HIV-1 envelope V2 and V3 sequences of the primary isolates to the viral phenotype. Microbiology and Immunology. 1996;40:277–287. doi: 10.1111/j.1348-0421.1996.tb03347.x. [DOI] [PubMed] [Google Scholar]
- Zhang LQ, MacKenzie P, Cleland A, Holmes EC, Brown AJ, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Wang N, Carr A, Nam DS, Moor-Jankowski R, Cooper DA, Ho DD. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. Genotypic and Phenotypic Characterization of HIV-1 in Patients with Primary Infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Jetzt AE, Sun G, Yu H, Klarmann G, Ron Y, Preston BD, Dougherty JP. Human Immunodeficiency Virus Type 1 Recombination: Rate, Fidelity, and Putative Hot Spots. J Virol. 2002;76:11273–11282. doi: 10.1128/JVI.76.22.11273-11282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler JB, Cooper DA, Johnson RO, Gold J. Postnatal transmission of AIDS-associated retrovirus from mother to infant. Lancet. 1985;1:896–898. doi: 10.1016/s0140-6736(85)91673-3. [DOI] [PubMed] [Google Scholar]