Abstract
Background
The three-dimensional shape of grain, measured as grain length, width, and thickness (GL, GW, and GT), is one of the most important components of grain appearance in rice. Determining the genetic basis of variations in grain shape could facilitate efficient improvements in grain appearance. In this study, an F7:8 recombinant inbred line population (RIL) derived from a cross between indica and japonica cultivars (Nanyangzhan and Chuan7) contrasting in grain size was used for quantitative trait locus (QTL) mapping. A genetic linkage map was constructed with 164 simple sequence repeat (SSR) markers. The major aim of this study was to detect a QTL for grain shape and to fine map a minor QTL, qGL7.
Results
Four QTLs for GL were detected on chromosomes 3 and 7, and 10 QTLs for GW and 9 QTLs for GT were identified on chromosomes 2, 3, 5, 7, 9 and 10, respectively. A total of 28 QTLs were identified, of which several are reported for the first time; four major QTLs and six minor QTLs for grain shape were also commonly detected in both years. The minor QTL, qGL7, exhibited pleiotropic effects on GL, GW, GT, 1000-grain weight (TGW), and spikelets per panicle (SPP) and was further validated in a near isogenic F2 population (NIL-F2). Finally, qGL7 was narrowed down to an interval between InDel marker RID711 and SSR marker RM6389, covering a 258-kb region in the Nipponbare genome, and cosegregated with InDel markers RID710 and RID76.
Conclusion
Materials with very different phenotypes were used to develop mapping populations to detect QTLs because of their complex genetic background. Progeny tests proved that the minor QTL, qGL7, could display a single mendelian characteristic. Therefore, we suggested that minor QTLs for traits with high heritability could be isolated using a map-based cloning strategy in a large NIL-F2 population. In addition, combinations of different QTLs produced diverse grain shapes, which provide the ability to breed more varieties of rice to satisfy consumer preferences.
Background
Rice is one of the most important cereal crops and staple foods in Asia. According to its grain shape, rice is primarily classified into long, medium, and short categories, by which the ratios of grain length (GL) to grain width (GW) are more than 3.0, between 2.1 and 2.9, and smaller than 2.0, respectively http://www.ams.usda.gov/AMSv1.0/getfile?dDocName=STELDEV3003761. People from North and South America, Southern China, and Europe usually prefer rice with long and slender grains, whereas those from Japan, Northern China, and North and South Korea prefer rice with short and round grains [1,2]. Juliano and Villareal reviewed grain quality of world rice types and reported that the correlations between GW and cooked rice hardness were significant in 18 countries/locations [3]. In addition, Yang et al. and Luo et al. also mentioned that cooking quality is associated with grain shape [4,5]. However, difference in cooking quality among the three shape categories is mainly determined by the chemical components and texture of their grains. In most cases, long grain rice has a high grain amylose content and after cooking, it is often firm and fluffy (not sticky); medium grain rice has a low amylose content and after cooking, it is often soft, moist, and sticky in texture. The cooking quality and amylose content in short grain rice are similar to those of rice in the medium grain category.
Grain size is usually evaluated by 1000-grain weight (TGW), which is one of the three key components, along with grain number per panicle (GPP) and panicle number per plant, contributing to rice grain yield and is positively correlated with grain shape traits including GL, GW, and grain thickness (GT) [6,7]. Grain shape has attracted significant attention in rice breeding programs due to its contributions to rice yield and quality [8].
Grain shape has been widely accepted as a complex trait controlled by multiple genes with small effects. Extensive efforts to determine the genetic basis of grain shape have used forward and reverse genetic strategies. Initially, studies focused on characterizing mutants and the expression of major genes associated with grain size, for example, the Lk-f gene, which confers long kernel size [9], or Mi, which confers short kernel size [10]. However, finding mutants of most grain shape genes in nature is not easy. Alternatively, quantitative trait locus (QTL) analysis based on genome wide mapping is a good strategy that has been widely used for mapping rice grain shape genes during the last 20 years. Previous studies used different populations, such as F2, F3, backcross, double-haploids, and recombinant inbred lines (RILs) http://www.gramene.org/qtl. Among these, the RIL population has particular advantages including repeatability, which favors the genetic analysis of quantitative traits because experiments can be replicated over years and locations. Combined with several fine genetic maps developed in recent years, many QTLs for grain shape have been identified. Tan et al., using F2:3 and RIL populations, detected two major QTLs for GL and GW on chromosomes 3 and 5, respectively. Several minor QTLs were also detected [11]. The two major QTLs for grain shape were also commonly detected in the same intervals on chromosomes 3 and 5, respectively, across different populations [7,11-15].
To date, thousands of QTLs have been detected in primary rice mapping populations. For fine mapping and cloning these QTLs, it is vital to make a population in which the targeted QTLs behave as the characters of a single mendelian factor. Therefore, an advanced population such as a near isogenic line population (NIL), which minimizes the noise of genetic background, is a prerequisite for QTL map-based cloning. Recently, increasing numbers of NILs have been used for QTL fine mapping and cloning. In terms of grain shape, Li et al. reported fine mapping of gw3.1 in the pericentromeric region of chromosome 3 using NILs, which is derived from a tropical japonica cultivar as the recurrent parent backcrossed to a wild rice [16]. Wan et al. found QTL qGL-3 for GL and qGW-5 for GW using chromosomal segment substitution lines (CSSLs) derived from the cross Asominori × IR24 [14]. Interestingly, these two QTLs were repeatedly identified in eight different environments. Fan et al. finely mapped the GS3 locus, a major QTL for GL and GW, and determined its candidate gene using advanced populations [17]. Based on these related reports, Zhang suggested that GL is mostly controlled by the GS3 locus on chromosome 3 and GW is largely conditioned by GS5 on chromosome 5 [18]. So far, one GL QTL, GS3 (encoding a transmembrane protein), two GW QTLs, GW2 (encoding RING-type E3 ubiquitin ligase) and qSW5/GW5 (whose biochemical function remains unclear), have been cloned [17,19-21]. In addition, Xie et al. finely mapped two TGW QTLs, gw8.1 and gw9.1, and concluded that there were significant correlations between TGW and grain shape [22,23].
Consecutive backcrossing is the conventional strategy for developing NILs and has been extensively used in mapping QTLs. Although molecular marker-assisted selection (MAS) can aim to any genome region, advanced backcross QTL-NIL development is both laborious and time consuming. An efficient method for a rapid NIL development is to seek the heterogeneous inbred family (HIF) at a QTL region [24]. The basic work flow for HIF-based NIL (HIF-NIL) development is first to screen the RIL heterozygous in the target QTL (high generation, eg, F5 or F6), second to evaluate the variation of target trait phenotype, and finally, to get the NILs (seeds harvested from the heterozygotes at the targeted genome region). Therefore, HIF-NIL development is much more efficient and more rapid for either major or minor QTLs as compared to the conventional strategies.
In this study, an RIL population derived from a cross between two rice cultivars, Nanyangzhan and Chuan7, was used for QTL mapping for grain size and grain shape. An NIL-F2 population of minor QTLs was constructed. The objectives of this study were (1) to detect QTLs for GL, GW, and GT, (2) to evaluate the power of QTL detection for grain shape as compared with other reports, and (3) to fine map a minor QTL, qGL7, with pleiotropic effects on GL, TGW, and spikelets per panicle (SPP).
Methods
Mapping population and field experiment
A total of 185 RILs were derived from the cross between japonica rice Nanyangzhan and indica rice Chuan7 by single-seed descent. Nanyangzhan has large grains, whereas Chuan7 has small grains. The RILs F7 and F8 were planted in a bird-net-equipped field on the experimental farm of Huazhong Agricultural University in the 2006 and 2007 rice-growing seasons in Wuhan, China. Field trials were carried out following the randomized complete block design with two replications within each year. At about 25 days old, 10 seedlings of each line were transplanted into one row in the main field with a distance of 16.5 cm between the plants within a row and 26.4 cm between rows. Eight plants in the middle of each row were harvested individually to score the traits.
Based on QTL mapping results from 2006, 185 RILs were screened for heterozygotes at the markers flanking the QTLs. In RIL76, the interval between RM22065 and RM5720 exhibited heterozygosity, but was homozygous in other genetic backgrounds. Selfing of RIL76 produced progeny segregating for qGL7 population in a near isogenic background. The qGL7 NIL-F2 population of 201 individuals was planted in Wuhan during the summer and their F3 families of 16 plants each were grown in Hainan during the winter of 2007.
Trait measurement
Twenty full-filled rice grains were chosen randomly from each plant for trait measurement. GL was estimated by placing 20 grains one by one in a straight line along a ruler. These 20 seeds were individually measured for GW and GT using an electronic digital caliper (Guanglu Measuring Instrument Co. Ltd, China) with a precision of 0.1 mm. The averaged GL, GW, and GT of 20 grains as the trait values of that line were used for data analysis. The grain length-to-width ratio (LWR) is equal to GL divided by its GW. SPP was measured as the total number of spikelets per plant divided by its panicle number. TGW was calculated as the grain weight per plant divided by its grain number multiplied by 1000.
DNA makers and linkage map construction
Fresh leaves were collected from 185 RILs (F7) individually for DNA extraction. DNA was extracted using a microisolation method as described by Cho et al. with minor modifications [25]. Polymorphic simple sequence repeat (SSR) markers between the parents Nanyangzhan and Chuan7 were used to genotype the population. All polymorphic markers were used for mapping the RIL population. The SSR assay was carried out essentially as described by Wu and Tanksley [26]. The genetic linkage map with RILs was constructed by using MAPMAKER/EXP version 3.0b [27]. The local linkage map based on the NIL-F2 population was developed. Genetic distance was calculated in Haldane function [28]. According to the sequence alignment between japonica cultivar Nipponbare and indica cultivar 93-11, three primer pairs for three InDel markers were designed with software "Primer3" for fine mapping qGL7 (Table 1).
Table 1.
Basic information for three self-developed InDel markers
| Markers | Physical location | Prod. size* | Anneal temperature | Primers | |
|---|---|---|---|---|---|
| Forward (5'-3') | Reverse (5'-3') | ||||
| RID76 | 28.303 Mb | 149 | 58°C | caccgaagactgatcagcaa | tcacattcgagtggagcaac |
| RID710 | 28.420 Mb | 170 | 58°C | catgttatcgtgtgggcttt | tctgttgctgcagctgaact |
| RID711 | 28.482 Mb | 138 | 58°C | gcacatgcatgctaggacat | agccggtaaatttcttgcac |
*Production size in bp was predicted from the sequence of japonica variety Nipponbare.
Data analysis
Heritability for the traits GL, GW, and GT was calculated based on the experiments using the formula: 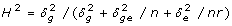 , where
, where 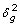 ,
, 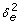 and
and 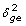 were the estimates of genetic, G by E and error variances derived from the mean square expectations of two-way ANOVA, n = 2 being the number of environments, and r = 2 being the number of replicates.
were the estimates of genetic, G by E and error variances derived from the mean square expectations of two-way ANOVA, n = 2 being the number of environments, and r = 2 being the number of replicates.
QTL analysis in RILs was performed by composite interval mapping (CIM) using the computer program Windows QTL cartographer 2.5 [29]. Window size was set as 10 cM and stepwise regression analysis was used to detect cofactors. The QTL main effects were estimated using the maximum-likelihood estimation method. The logarithm of odds (LOD) threshold at the experiment-wise significance level of 0.05 was determined by computing 1000 permutations of each morphologic character. The LOD threshold ranged from 2.8 to 3.1. In the qGL7 NIL-F2 population, MAPMAKER/QTL 1.1b software was used for QTL analysis. Walking speed for QTL scanning was 1 cM per step.
Two-way ANOVA was conducted to analyze digenic interaction between QTLs in the computer program Statistica 7.0 [30].
Results
Grain shape variation in the RIL population
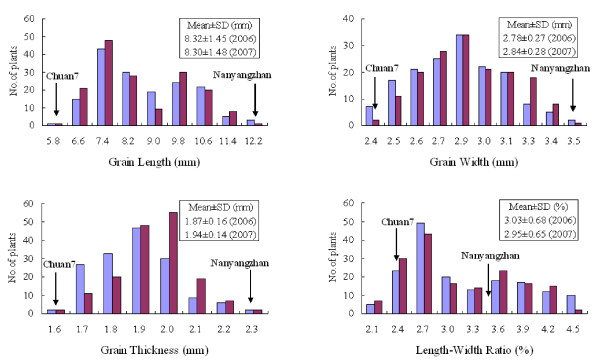
A highly significant difference for all four investigated traits was observed between the two parents, Nanyangzhan and Chuan7 (Figure 1). Nanyangzhan has a typical large grain size of 43.3 g/1000 grains with 12.2 mm in GL, 3.5 mm in GW, and 2.3 mm in GT. Chuan7 is a variety with an extremely small grain size of 11.4 g/1000 grains; it possesses low trait values of 5.8 mm in GL, 2.4 mm in GW, and 1.6 mm in GT. GL variation was wide, ranging from 5.8 to 12.2 mm; frequency distribution showed a bimodal pattern with GL 9.0 mm as the boundary for each year (Figure 2). GW and GT exhibited normal distribution patterns in the 2 years. No transgressive segregation was observed for any of the traits except LWR (Figure 2).
Figure 1.
Rice grains of both parents and two qGL7 homozygous NILs. qGL7 (NN) and qGL7 (CC) mean the NIL lines with Nanyangzhan and Chuan7 homozygous alleles at qGL7, respectively.
Figure 2.
Frequency distributions of grain shape traits in the RIL population. Blue bars and dark red bars indicate 2006 and 2007, respectively.
Heritability and correlation
GL, GW, and LWR showed high broad heritability of 97%, 91%, and 95%. GT was comparatively low, but its heritability still reached 78%. GL was positively correlated with GT and LWR in both years. GW was negatively correlated with GL and LWR, but positively correlated with GT (Table 2).
Table 2.
Correlation coefficients among the four traits
| Trait | GL | GW | GT | LWR |
|---|---|---|---|---|
| GL | -0.23** | 0.27** | 0.89** | |
| GW | -0.17* | 0.56** | -0.63** | |
| GT | 0.42** | 0.59** | -0.03 | |
| LWR | 0.89** | -0.59** | 0.08 |
The correlation coefficients in 2006 and 2007 are below and above the diagonal, respectively
*, **Significant at the level of α = 0.05 and 0.01, respectively
Genetic linkage map
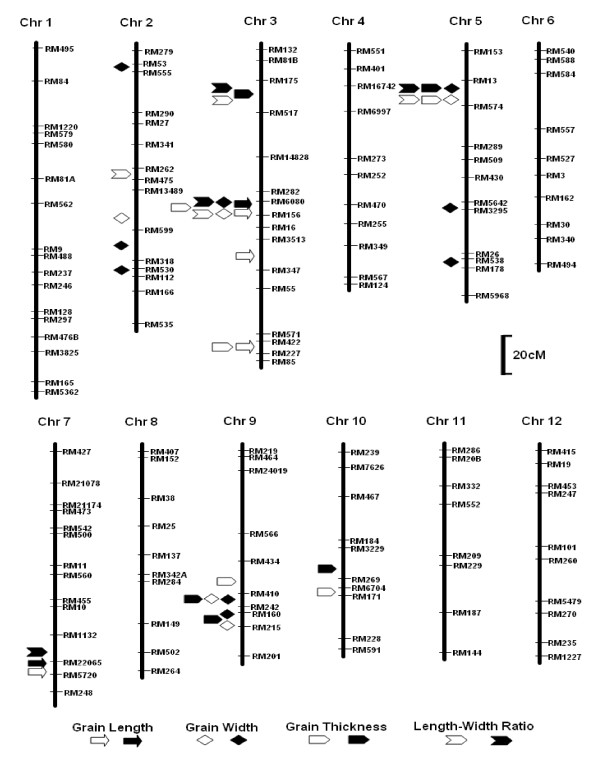
A total of 502 SSR markers were used for screening polymorphic markers between the parents Nanyangzhan and Chuan7. Of these, 262 showed polymorphism between parents. Finally, 164 evenly distributed polymorphic markers were selected for genotyping the RIL population. A genetic linkage map covering a 1635.9-cM genome region was constructed (Figure 3). The average distance between adjacent markers was 9.9 cM.
Figure 3.
Genetic linkage map showing QTL positions detected in the RIL population. Black and white shapes indicate 2006 and 2007, respectively.
QTL for grain shape traits
GL
Four QTLs for GL were detected on chromosomes 3 and 7 in both years (Table 3). qGL3a and qGL7 were commonly detected in both years. qGL3b and qGL3c were detected only in 2007. Nanyangzhan alleles appeared at three QTLs but qGL3b increased GL. The major QTL, qGL3a, in the marker interval between RM6080 and RM156 having a LOD score of more than 46, explained more than 65% of the phenotypic variance in both years. The other QTL individually explained a small GL variation of less than 5%. These three QTLs were located on chromosome 3 and were loosely linked with a distance of more than 20 cM from each other (Figure 3).
Table 3.
QTLs for grain shape detected in the RIL population derived from the cross between Nanyangzhan and Chuan7
| 2006 | 2007 | |||||||
|---|---|---|---|---|---|---|---|---|
| Traits | QTL | Interval | LOD | Aa | Vb % | LOD | Aa | Vb % |
| GL | qGL3a | RM6080-RM156 | 46.2 | 1.20 | 65.8 | 53.4 | 1.30 | 72.2 |
| (mm) | qGL3b | RM3513-RM347 | 4.4 | -0.30 | 4.1 | |||
| qGL3c | RM422-RM227 | 2.3c | 0.22 | 2.4 | 5.7 | 0.30 | 4.9 | |
| qGL7 | RM22065-RM5720 | 4.7 | 0.30 | 5.1 | 5.4 | 0.30 | 4.7 | |
| GW | qGW2a | RM53-RM555 | 3.8 | 0.05 | 4.1 | |||
| (mm) | qGW2b | RM13489-RM599 | 6.0 | 0.10 | 15.6 | |||
| qGW2c | RM599-RM318 | 4.7 | 0.08 | 9.3 | ||||
| qGW2d | RM530-RM112 | 4.3 | 0.06 | 5.9 | ||||
| qGW3 | RM6080-RM156 | 6.2 | -0.08 | 8.9 | 3.4 | -0.07 | 7.0 | |
| qGW5a | RM13-RM574 | 21.0 | 0.17 | 38.1 | 14.0 | 0.17 | 36.3 | |
| qGW5b | RM5642-RM3295 | 5.8 | 0.09 | 11.3 | ||||
| qGW5c | RM538-RM178 | 5.8 | 0.08 | 7.8 | 2.3c | 0.06 | 5.3 | |
| qGW9a | RM410-RM242 | 5.9 | 0.07 | 7.5 | 3.6 | 0.07 | 5.6 | |
| qGW9b | RM160-RM215 | 6.6 | 0.08 | 9.2 | 5.1 | 0.08 | 8.4 | |
| GT | qGT3a | RM175-RM517 | 3.0 | 0.06 | 12.5 | 2.1c | 0.04 | 7.4 |
| (mm) | qGT3b | RM6080-RM156 | 3.3 | 0.04 | 7.3 | |||
| qGT3c | RM422-RM227 | 3.1 | 0.03 | 6.1 | ||||
| qGT5 | RM13-RM574 | 3.7 | 0.05 | 9.1 | 8.9 | 0.06 | 19.1 | |
| qGT9a | RM434-RM410 | 3.1 | 0.03 | 5.3 | ||||
| qGT9b | RM410-RM242 | 4.3 | 0.05 | 9.4 | ||||
| qGT9c | RM160-RM215 | 8.9 | 0.07 | 17.1 | 2.6c | 0.03 | 5.2 | |
| qGT10a | RM3229-RM6704 | 3.4 | 0.04 | 6.1 | ||||
| qGT10b | RM6704-RM171 | 4.4 | 0.04 | 10.4 | ||||
| LWR | qLWR2 | RM262-RM475 | 3.8 | -0.14 | 3.7 | |||
| qLWR3a | RM175-RM517 | 4.2 | 0.19 | 6.0 | 3.4 | 0.22 | 8.9 | |
| qLWR3b | RM6080-RM156 | 48.0 | 0.50 | 57.0 | 51.3 | 0.53 | 64.2 | |
| qLWR5 | RM13-RM574 | 13.4 | -0.25 | 13.5 | 5.5 | -0.16 | 5.3 | |
| qLWR7 | RM1132-RM22065 | 3.2 | 0.13 | 3.5 | ||||
aA, additive effect, positive additive effect means Nanyangzhan allele increasing trait values.
bV, variance explained by the QTL.
cQTL with LOD ≥ 2.8 in one year but 2.0 ≤ LOD ≤ 2.8 in the other year are also listed.
GW
Ten QTLs were detected for GW (Table 3). One, two, three, and four QTLs were detected on chromosomes 3, 9, 5, and 2, respectively (Figure 3). Only four QTLs (qGW3, qGW5a, qGW9a and qGW9b) could be commonly detected in both years. A major QTL, qGW5a, was located in the marker interval between RM13 and RM574 with a high LOD score of more than 14 and explained more than one third of the GW variance in both years. qGW9a and qGW9b were located in the linked region about 15 cM away. qGW5a, qGW5b, and qGW5c were sparsely located on chromosome 5. qGW2a was far away from qGW2b, qGW2c, and qGW2d, which were located in a neighboring interval. Of the 10 QTLs, Nanyangzhan alleles at the other nine QTLs except for qGW3a had a positive effect on GW.
GT
Nine QTLs were identified for GT (Table 3 and Figure 3). Nanyangzhan alleles of all the nine QTLs increased GT. Only the major QTL, qGT5, was commonly detected in both years and explained 9.1% and 19.1% of phenotypic variation, respectively. The other eight QTLs were detected only in one year. Three linked QTLs and three sparsely distributed QTLs were identified on chromosomes 9 and 3, respectively. qGT10a and qGT10b were mapped to the neighboring regions in 2006 and 2007, respectively. Although qGT9c with a large LOD score of 8.9 could explain 17.1% of phenotypic variation in 2006, it was not detected in 2007.
LWR
Five QTLs were located on four chromosomes (Table 3 and Figure 3). Two QTLs were detected on chromosome 3 in both years. Nanyangzhan alleles increased LWR. qLWR5 was observed in both years, but Nanyangzhan alleles decreased LWR. qLWR2 and qLWR7 were detected only in one year. The major QTLs, qLWR3b and qLWR5, shared the same intervals with major QTLs, qGL3a and qGW5a, respectively.
In summary, 28 QTLs were found for the four grain shape traits. Three QTL hotspot regions were found on chromosomes 3, 5, and 9. No QTL for grain shape was identified on chromosomes 1, 4, 6, 8, 11, and 12.
Digenic interaction
QTL genotypes were replaced with close-linked marker genotypes when QTL-by-QTL digenic interaction was analyzed. All possible digenic interactions among all 28 QTLs were analyzed by two-way ANOVA. However, no significant digenic interaction was observed at the level of P = 0.01%.
Validation of qGL7
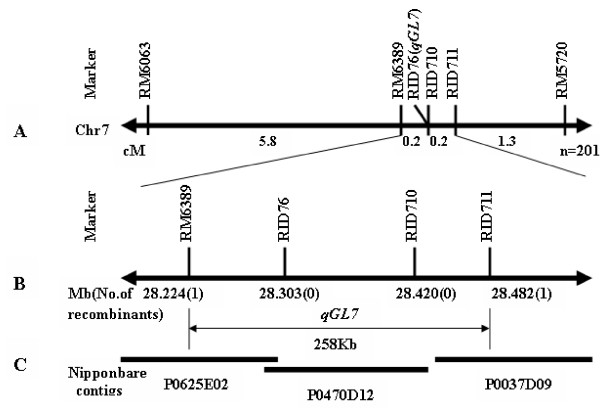
A total of 201 individuals of the NIL-F2 population were genotyped by three SSR markers and three InDel markers. A local genetic linkage map of qGL7 region was constructed (Figure 4A). GL showed a bimodal distribution of 7.1 mm as the boundary in the F2 population. In contrast, GW and GT exhibited normal distributions. Significant correlations were observed between the five traits in the F2 population (data not shown). A positive correlation (r = 0.43, P = 0.01) between GL and GW was also detected, which is opposite to the case in the RIL population. Again, the qGL7 with a LOD score of 42.2 was mapped to the interval between markers RID76 and RID711, whose 1-LOD confidence interval spanned a 0.4-cM region from RM6389 to RID711. The Nanyangzhan allele increased GL and could explain 61.5% of the phenotypic variance in GL (Table 4).
Figure 4.
Fine mapping of qGL7 and its candidate region. (A) Local genetic map of qGL7 constructed with qGL7-NILs. (B) The physical positions of markers close linked to qGL7. The physical distance was determined from the sequence of Nipponbare and the known positions of the markers on the sequence. Number in parentheses indicates the number of recombinants between the marker and qGL7. (C) A contig consisted of the PAC, P0625E02, P0470D12 and P0037D09, contained qGL7.
Table 4.
qGL7 effects on grain shape and grain yield traits in the NIL-F2 population
| Traits | Intervals | LOD | LOD peak | Confidence interval (cM)a | Addb | Domc | Var (%)d |
|---|---|---|---|---|---|---|---|
| GL (mm) | RID76-RID711 | 42.2 | RID76 | 5.8-6.2 | 0.24 | 0.14 | 61.5 |
| GW (mm) | RID76-RID711 | 10.6 | RID76 | 5.0-6.3 | 0.04 | 0.04 | 21.5 |
| GT (mm) | RID76-RID711 | 32.1 | RID76 | 5.8-6.3 | 0.05 | 0.03 | 51.7 |
| SPP(n) | RID76-RID711 | 15.5 | RID76 | 4.0-6.2 | -16.6 | -13.4 | 29.3 |
| TGW(g) | RID76-RID711 | 66.8 | RID76 | 5.8-6.2 | 1.71 | 1.14 | 77.9 |
aGenetic position in the local linkage map (Figure 4A).
bAdd additive effect, positive additive effect means Nanyangzhan allele increasing trait values.
cDom dominance effect.
dVar variation explained by QTL
On the basis of progeny tests, all 201 families could be divided into three subclasses: 62 families showed identical long grain within family, 55 families showed identical short grain within family, and 84 families showed varied GL. The ratio of the number of plants in short grain and long grain groups agreed with the expected segregation of a single mendelian factor (χ2 = 5.91, P > 0.05), which is indicative of one gene controlling the GL variation. These three subclasses corresponded to the three genotypes at qGL7, Nanyangzhan homozygotes (NN), Chuan7 homozygotes (CC), and heterozygotes (NC), respectively (Table 5). The averaged GL of NN and NC classes were 7.4 and 7.3 mm, respectively, significantly longer than that of CC class with an average GL of 6.9 mm (Figure 1). Thus, CC genotypes were easily distinguished from NN and NC. Genotypes NN and NC had similar ranges, means, and standard deviations in GL, but they were still distinguishable because the progenies from a single NC F2 plant segregated in GL, which was clearly evident since individual F3 families of this class had higher standard deviations around the mean GL than individual families of the NN class, which did not segregate. Then qGL7, regarded as a co-dominant marker, was mapped to the same interval of RM6389-RID711 and co-segregated with RID76 and RID710. The interval of RM6389-RID711 covered a 258-kb region in Nipponbare (Figure 4B). The Nanyangzhan allele of qGL7 controlling long grains was dominant against Chuan7 allele.
Table 5.
Trait performance of the three genotypes at qGL7 on the basis of progeny tests
| NN (62 families) | NC (84 families) | CC (55 families) | ||||
|---|---|---|---|---|---|---|
| Traits | Mean ± SD | Range | Mean ± SD | Range | Mean ± SD | Range |
| GL (mm) | 7.4 ± 0.12 | 7.1-7.7 | 7.3 ± 0.11 | 7.1-7.6 | 6.8 ± 0.15 | 6.5-7.0 |
| GW (mm) | 3.0 ± 0.07 | 2.8-3.2 | 3.0 ± 0.07 | 2.7-3.1 | 2.9 ± 0.07 | 2.8-3.1 |
| GT (mm) | 2.0 ± 0.03 | 1.8-2.0 | 2.0 ± 0.04 | 1.9-2.0 | 1.9 ± 0.04 | 1.8-2.0 |
| SPP (n) | 117 ± 18 | 90-138 | 115 ± 17 | 91-140 | 152 ± 22 | 116-201 |
| TGW(g) | 19.1 ± 0.74 | 17.0-21.6 | 18.5 ± 0.79 | 16.9-20.1 | 15.6 ± 0.82 | 14.3-16.4 |
NN homozygous for Nanyangzhan allele, NC heterozygous, CC homozygous for Chuan7 allele, GL grain length, GW grain width, GT grain thickness, SPP spikeletes per panicle, TGW 1000-grain weight
Pleiotropic effects of qGL7
QTLs for the other four traits were also detected in the same interval in the NIL-F2 population. Their corresponding LOD peaks were exactly mapped to the maker locus of RID76/RID710 (Table 4). Their 1-LOD confidence intervals largely overlapped. A negative correlation was detected between SPP and TGW (r = -0.42, P = 0.01). That is, the panicle with long grains always had fewer secondary branches than that of the panicle with short grains (data not shown), and vice versa. The average TGWs of Nanyangzhan and Chuan7 homozygous genotypes at qGL7 in the F2 population were 19.1 g and 15.6 g, respectively. Their corresponding SPP averages were 117 and 152. The QTLs could explain 77.9% and 29.3% of the phenotypic variance in TGW and SPP. The Nanyangzhan allele increased GL and TGW but decreased SPP (Table 4). Accordingly, the progenies could be classified into three subclasses based on SPP and TGW performance: 62 families had identical small panicles with larger TGW but fewer SPP, 55 families showed identical large panicles with smaller TGW but more SPP, and 84 families showed segregation in TGW and SPP within each family. SPP was co-segregated with GL and TGW in the NIL-F2 population. Moreover, these three subclasses were identical to the three genotypes at qGL7, NN, CC, and NC, respectively (Table 5). Therefore, QTLs for SPP and TGW were co-segregated with qGL7.
Discussion
More QTLs identified in the RIL population than in previous single populations
One important finding in this study was to identify 28 QTLs for grain shape using a population with wide variations in grain shape. Although grain shape has received increased attention since the 1990s, most mapping populations used in grain shape-targeted QTL mapping studies were derived from parents whose cultivars usually produce common sized grains (TGW from 20 to 32 g) [2,7,8,11,31-34]. Populations derived from wild rice and cultivar were also used to determine the genetic basis of grain shape. Yoon et al. identified two grain size QTL hotspots located on chromosomes 6 and 11 using an advanced backcross population that was derived from wild rice, Oryza grandiglumis and japonica cultivar Hwaseongbyeo [35]. QTL analyses for grain shape showed that it was mainly controlled by GS3, GW2, GW5, and GT2 [18,19,35]. In general, a mapping population derived from two contrasting parents in grain shape is expected to detect more QTLs. In this study, the mapping population was derived from two indica parents with contrasting grain shape, long slender and short round. Their genetic backgrounds were also diverse (52% of investigated markers showed polymorphism) and provided us the potential to detect more QTLs for grain shape. The 28 QTLs identified in the population is a higher total than found in any other single population and proved this potentiality. Among them, two major QTLs, qGL3a and qGW5a, were repeatedly identified [11,14,15]. Also, a minor QTL, qGW3, for GW on chromosome 3 was also detected by Yoon et al. [35]. Interestingly, a minor QTL, qGW2a, identified in this study is located in the similar region of GW2 that was cloned by Song et al. as a major QTL [19]. This indicated that NIL populations derived from two different parental alleles at a given QTL will result increased or decreased QTL effects because different parents carry different functional alleles. Seven novel QTLs, including qGL3b, qGW5b, qGW9b, qGT3a, qGT3c, qGT10b, and qLWR7, were also detected in this study. Therefore, the RIL population in this study could identify more QTLs than a previous single population. In addition, besides the major QTLs and the QTL hotspots located on the long arm of chromosome 9, a minor QTL, qGL7, was repeatedly identified in two different environments. Other QTLs were detected only in one year, but some of them could also be detected if the LOD threshold decreased to 2.0, an empirical threshold in rice [36]. This indicated that a minor QTL could interact with the environment and was reliable only in certain conditions.
Pleiotropic effects of QTLs
In the NIL-F2 population, QTLs for TGW, SPP, GW, and GT were detected in the same target region of qGL7. Their 1-LOD confidence intervals largely overlapped. This suggested the possibility that qGL7 had pleiotropic effects on all five traits. Furthermore, progeny tests showed that qGL7 co-segregated with the QTLs for TGW and SPP, which indicated qGL7 may simultaneously control GL, TGW, and SPP. Although qGL7 showed pleiotropic effects in the NIL-F2 population, a limited resolution of mapping due to the small size of F2 population still cannot clearly discern two closely linked genes or one pleiotropic gene in a small region. However, QTLs with pleiotropic effects on grain shape were frequently found in rice. Fan et al. reported that the GS3 gene had a major effect on GL and a minor effect on GW and GT in NILs [17]. Similarly, Song et al., using transgenic plants of GW2, a QTL that controls GW, confirmed that it also had an opposite effect on grain number per panicle [19]. Thus, qGL7 was a grain shape gene. Based on our current results, we tend to believe that qGL7 has pleiotropic effects on TGW and SPP, though in opposite directions. Moreover, due to its counteracting effects on TGW and SPP, qGL7 has no effect on grain yield. This implies that qGL7 may have an important role in balancing the distribution between SPP and TGW. That is, more distribution to SPP results in a decrease in TGW, and vice versa.
Fine mapping and cloning of minor QTLs
Several QTLs for grain shape (grain weight) such as gw3.1/GS3, gw8.1, gw9.1, GW2, and gw-5 [16,17,19,22,23,37] were fine mapped in the last 5 years. So far, all cloned grain shape QTLs had major effects. Of great interest, two major QTLs, qGL3a and qGW5a in this study, shared similar positions with cloned genes GS3 and qSW5/GW5 [17,20,37]. It is therefore tempting to speculate that they are exactly the cloned genes and the minor QTLs of qGL7, commonly detected in both years, used for fine mapping. Obviously, cloning of minor QTLs detected in a primary population is more difficult. Development of QTL-based NILs revealed that most QTLs have major effects in homogeneous genetic backgrounds [38-40]. Thus, the construction of high-quality NILs became a vital step for fine mapping and cloning minor QTLs.
Rice grains are covered by hulls. The GL and GW are fixed as long as the panicle is normally differentiated. Thus, rice GL and GW are mainly controlled by genotype and have higher heritability. However, GT is greatly affected by filling degree, which is considerably affected by environment. Hence, for pleiotropic QTLs for grain shape, GL and GW provided more reliable trait values for gene cloning. Although the minor QTL, qGL7, explained only about 5% of GL variation in RILs, it clearly expressed the character of a single mendelian factor and explained most parts of the trait variation in the NIL-F2 population. It is promising to narrow down qGL7 to a small region given a large NIL-F2 population. Characterization of qGL7 clearly indicated that a minor QTL can be treated as a single mendelian factor for further functional research on quantitative traits especially when major QTLs have been cloned.
Molecular breeding design for rice grain appearance improvement
Grain appearance, which is mainly evaluated by the parameter of LWR, is an essential component of rice quality. Comprehensive understanding of the genetic basis of grain shape will be particularly helpful in improving its appearance quality. No transgressive segregation was observed for GL, GW, and GT in RILs, which indicated that positive alleles were almost all from Nanyangzhan. Actually, among the 28 QTLs for GL, GW, and GT, only at two minor QTLs of parental Chuan7 were trait values increased. On the other hand, positive alleles were not always favored in rice improvement. For example, consumers in East Asia prefer short and round rice. Especially long and slender grains (large GL and large LWR) always decreased head rice percentage when rice was milled. In our study, 4 and 10 QTLs were identified for GL and GW, respectively. The major QTLs were, of course, first considered for improving grain shape. For example, if we focused on the four QTLs of qGL3a, qGL7, qGW5a, and qGW9b, their combinations can produce 16 homozygous genotypes. If the plant fixed with Nanyangzhan alleles at qGL3a and qGL7, and Chuan7 alleles at qGW5a and qGW9b, its ratio of GL/GW would produce the largest LWR of 3.86; however, in the reciprocal case, the ratio would change to the smallest LWR of 2.26 (Table 6). Among the 16 genotypes, a diverse LWR variation, ranging from 2.26 to 3.86, provides breeders flexibility in designing new varieties for people's preference in different regions. The flanking markers of the four QTLs can be directly used for marker-aided selection.
Table 6.
Ratios of GL to GW in the gene combinations among four grain shape QTLs
| Genotypes | qGW5aqGW9b | |||
|---|---|---|---|---|
| qGL3aqGL7 | NNNN | NNCC | CCNN | CCCC |
| NNNN | 3.36 | 3.43 | 3.64 | 3.86 |
| NNCC | 2.83 | 2.89 | 3.06 | 3.25 |
| CCNN | 2.48 | 2.53 | 2.68 | 2.84 |
| CCCC | 2.26 | 2.30 | 2.44 | 2.59 |
NN homozygous for Nanyangzhan allele, CC homozygous for Chuan7 allele.
Conclusion
In this study, we performed QTL analysis for grain shape using an RIL population derived from two varieties with contrasts in grain shape. Twenty-eight QTLs were identified. Seven of them were detected for the first time. These results demonstrated that the mapping population derived from parents with contrasting phenotypes can be used for detecting more QTLs. Diverse materials are recommended to develop mapping populations for powerful QTL detection. In the present study, a minor QTL of qGL7 was validated with pleiotropic effects on GL, GW, TGW, SPP, and GT in an NIL-F2 population. It is suggested that minor QTL of a highly heritable trait could be isolated following the strategy of map-based cloning in a large NIL-F2 population. Combinations of different QTLs were expected to produce diverse grain shapes from long slender to short and round rice, which provides more flexibility to breed rice varieties for different preferences.
Authors' contributions
BXF conducted the experiment, developed the genetic linkage map, and analyzed the data. LLJ developed the RIL population. YWH, KMR, and ZW partially contributed in phenotypic data collection and population genotyping. XYZ designed the work and wrote the manuscript together with BXF.
All authors have read and approved the final manuscript.
Contributor Information
Xufeng Bai, Email: a020809@webmail.hzau.edu.cn.
Lijun Luo, Email: lijun@sagc.org.cn.
Wenhao Yan, Email: yanwenhao@webmail.hzau.edu.cn.
Mallikarjuna Rao Kovi, Email: mallik@mail.hzau.edu.cn.
Wei Zhan, Email: kbm00850713@hotmail.com.
Yongzhong Xing, Email: yzxing@mail.hzau.edu.cn.
Acknowledgements
The authors kindly thank farm technician Mr. JB Wang for his excellent field work. We thank Dr. Jianhong Chen for improving the language. This work was supported by a grant from the National Natural Science Foundation, a grant from the National Key Program of Basic and Development, and a grant from the National Program on the Development of Hi-tech Research (2007AA10Z103).
References
- Unnevehr LJ, Duff B, Juliano BO. Consumer demand for rice grain quality. International Rice Research Institute, Manila, The Philippines/International Development Research Centre, Ottawa, Canada; 1992. [Google Scholar]
- Wan XY, Wan JM, Jiang L, Wang JK, Zhai HQ, Weng JF, Wang HL, Lei CL, Wang JL, Zhang X, Cheng ZJ, Guo XP. QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor Appl Genet. 2006;112:1258–1270. doi: 10.1007/s00122-006-0227-0. [DOI] [PubMed] [Google Scholar]
- Juliano BO, Villareal. Grain quality evaluation of world rices. International Rice Research Institute. Manila, The Philippines; 1993. [Google Scholar]
- Yang LS, Bai YS, Zhang PJ, Xu CW, Hu XM, Wang WM, She DH, Chen GZ. Studies on the correlation between grain shape and grain quality in rice. Hybrid Rice. 2001;16:48–50. [in Chinese] [Google Scholar]
- Luo YK, Zhu ZW, Chen N, Duan BW, Zhang LP. Grain types and related quality characteristics of rice in China. Chinese J Rice Sci. 2004;18:135–139. [Google Scholar]
- Evans LT. Storage capacity as a limitation on grain yield in rice breeding. International Rice Research Institute, Manila, The Philippines; 1972. [Google Scholar]
- Xu JL, Xue QZ, Luo LJ, Li ZK. Genetic dissection of grain weight and its related traits in rice (Oryza sativa L.) Chinese J Rice Sci. 2002;16:6–10. [Google Scholar]
- Lin LH, Wu WR. Mapping of QTL underlying grain shape and grain weight in rice. Mol Plant Breed. 2003;1:337–342. [Google Scholar]
- Takeda K, Saito K. Major genes responsible for grain shape in rice. Jpn J Breed. 1980;30:280–282. [Google Scholar]
- Takeda K, Saito K. The inheritance and character expression of the minute gene derived from a rice genetic tester minute. Bull Fac Agric Hirosaki Univ. 1977;27:1–29. [Google Scholar]
- Tan YF, Xing YZ, Li JX, Yu SB, Xu CG, Zhang QF. Genetic bases of appearance quality of rice grain in Shanyou63, an elite rice hybrid. Theor Appl Genet. 2000;101:823–829. doi: 10.1007/s001220051549. [DOI] [PubMed] [Google Scholar]
- Kubo T, Takano-kai N, Yoshimura A. RFLP mapping of genes for long kernel and awn on chromosome 3 in rice. Rice Genet Newsl. 2001;18:26–28. [Google Scholar]
- Li J, Xiao J, Grandillo S, Jiang L, Wan Y, Deng Q, Yuan L, McCouch SR. QTL detection for rice grain quality traits using an interspecific back-cross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S.) rice. Genome. 2004;47:697–704. doi: 10.1139/g04-029. [DOI] [PubMed] [Google Scholar]
- Wan XY, Wan JM, Weng JF, Jiang L, Bi JC, Wang CM, Zhai HQ. Stability of QTL for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor Appl Genet. 2005;110:1334–1346. doi: 10.1007/s00122-005-1976-x. [DOI] [PubMed] [Google Scholar]
- Xing YZ, Tan YF, Xu CG, Hua JP, Sun XL. Mapping quantitative trait loci for grain appearance traits of rice using a recombinant inbred line population. Acta Bot Sin. 2001;43:840–845. [Google Scholar]
- Li JM, Thomson M, McCouch SR. Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics. 2004;168:2187–2195. doi: 10.1534/genetics.104.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- Zhang QF. Strategies for developing Green Super Rice. Proc Natl Acad Sci USA. 2007;104:16402–16409. doi: 10.1073/pnas.0708013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- Weng JF, Gu SH, Wan XY, Gao H, Guo T, Su N, Lei CL, Zhang X, Cheng ZJ, Guo XP, Wang JL, Jiang L, Zhai HQ, Wan JM. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- Xie XB, Jin FX, Song MH, Suh JP, Hwang HG, Kim YG, McCouch SR, Ahn SN. Fine mapping of a yield-enhancing QTL cluster associated with transgressive variation in an Oryza sativa × O. rufipogon cross. Theor Appl Genet. 2008;116:613–622. doi: 10.1007/s00122-007-0695-x. [DOI] [PubMed] [Google Scholar]
- Xie XB, Song MH, Jin FX, Ahn SN, Suh JP, Hwang HG, McCouch SR. Fine mapping of a grain weight quantitative trait locus on rice chromosome 8 using near-isogenic lines derived from a cross between Oryza sativa and Oryza rufipogon. Theor Appl Genet. 2006;113:885–894. doi: 10.1007/s00122-006-0348-5. [DOI] [PubMed] [Google Scholar]
- Tuinstra MR, Ejeta G, Goldsbrough PB. Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet. 1997;95:1005–1011. doi: 10.1007/s001220050654. [DOI] [Google Scholar]
- Cho YG, Blair MW, Panaud O, McCouch SR. Cloning and mapping of variety-specific rice genomic DNA sequences: amplified fragment length polymorphisms (AFLP) from silver stained polyacrylamide gels. Genome. 1996;39:373–378. doi: 10.1139/g96-048. [DOI] [PubMed] [Google Scholar]
- Wu KS, Tanksley SD. Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol Gen Genet. 1993;241(1-2):225–235. doi: 10.1007/BF00280220. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlaw A, Daly M, Lincoln SE, Newburg L. MAPMAKER: An interactive computer package for maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. The combination of linkage values, and the calculation of distances between the loci of linked factors. J Genet. 1919;8:299–309. doi: 10.1007/BF02983270. [DOI] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC; 2007. [Google Scholar]
- StatSoft Inc. Statistica, Tulsa, OK. 2004.
- Yan CJ, Liang GH, Chen F, Li X, Tang SZ, Yi CD, Tian S, Lu JF, Gu MH. Mapping quantitative trait loci associated with rice grain shape based on an indica/japonica backcross population. Acta Genet Sin. 2003;30:711–716. [PubMed] [Google Scholar]
- Jiang GH, Hong XY, Xu CG, Li XH, He YQ. Identification of quantitative trait loci for grain appearance and milling quality using a doubled-haploid rice population. J Integr Plant Biol. 2005;47:1391–1403. doi: 10.1111/j.1744-7909.2005.00089.x. [DOI] [Google Scholar]
- Amarawathi Y, Singh R, Singh AK, Singh VP, Mohapatra T, Sharma TR, Singh NK. Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.) Mol Breeding. 2008;21:49–65. doi: 10.1007/s11032-007-9108-8. [DOI] [Google Scholar]
- Lei DY, Xie FM, Xu JL, Chen LY. QTL mapping and epistasis analysis for grain shape and chalkiness degree of rice. Chinese J Rice Sci. 2008;22:255–260. [Google Scholar]
- Yoon DB, Kang KH, Kim HJ, Ju HG, Kwon SJ, Suh JP, Jeong OY, Ahn SN. Mapping quantitative trait loci for yield components and morphological traits in an advanced backcross population between Oryza grandiglumis and the O. sativa japonica cultivar Hwaseongbyeo. Theor Appl Genet. 2006;112:1052–1062. doi: 10.1007/s00122-006-0207-4. [DOI] [PubMed] [Google Scholar]
- Vanooijen JW. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity. 1999;83:613–624. doi: 10.1038/sj.hdy.6886230. [DOI] [PubMed] [Google Scholar]
- Wan XY, Weng JF, Zhai HQ, Wang JK, Lei CL, Liu XL, Guo T, Jiang L, Su N, Wan JM. Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on the chromosome 5. Genetics. 2008;179:2239–2252. doi: 10.1534/genetics.108.089862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S, Tuberosa R. To clone or not to clone plant QTL: present and future challenges. Trends Plants Sci. 2005;10:297–304. doi: 10.1016/j.tplants.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Zhang YS, Luo LJ, Xu CG, Zhang Q, Xing YZ. Quantitative trait loci for panicle size, heading date and plant height co-segregating in trait-performance derived near-isogenic lines of rice (Oryza sativa) Theor Appl Genet. 2006;113:361–368. doi: 10.1007/s00122-006-0305-3. [DOI] [PubMed] [Google Scholar]
- Zhang YS, Luo LJ, Liu TM, Xu CG, Xing YZ. Four rice QTL controlling number of spikelets per panicle expressed the characteristics of single Mendelian gene in near isogenic backgrounds. Theor Appl Genet. 2009;118:1035–1044. doi: 10.1007/s00122-008-0960-7. [DOI] [PubMed] [Google Scholar]