Abstract
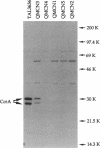
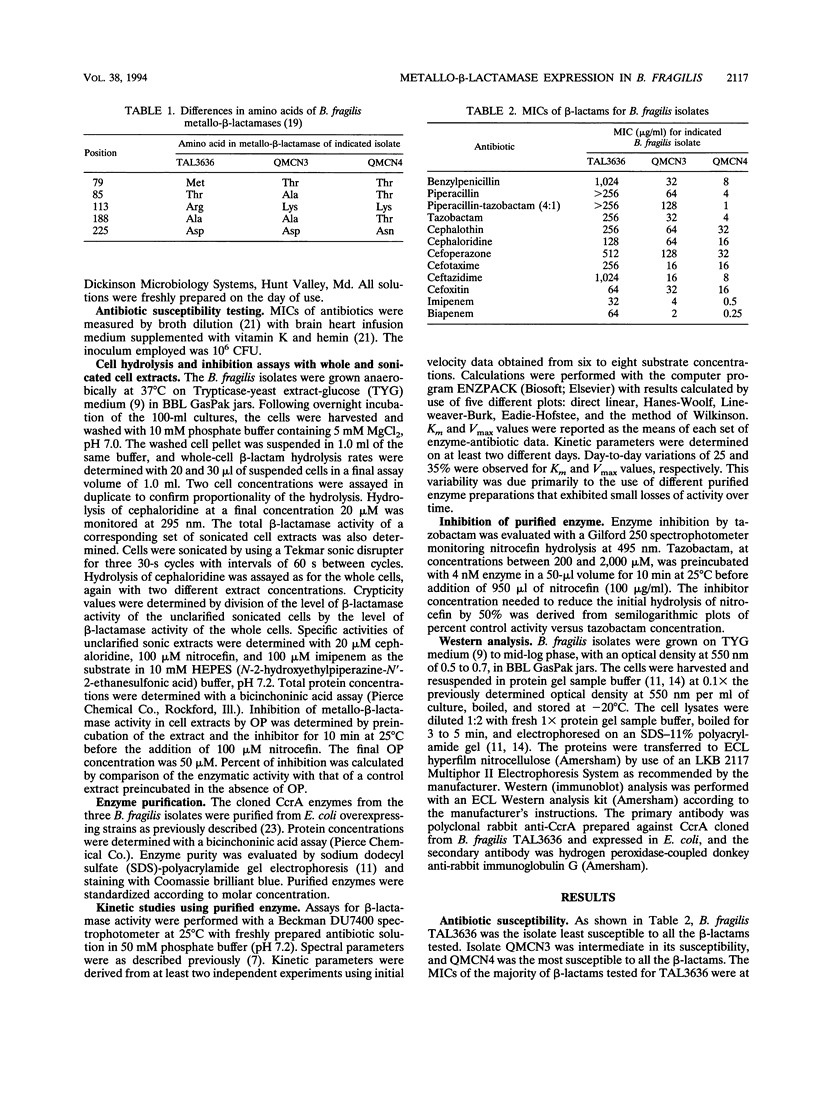
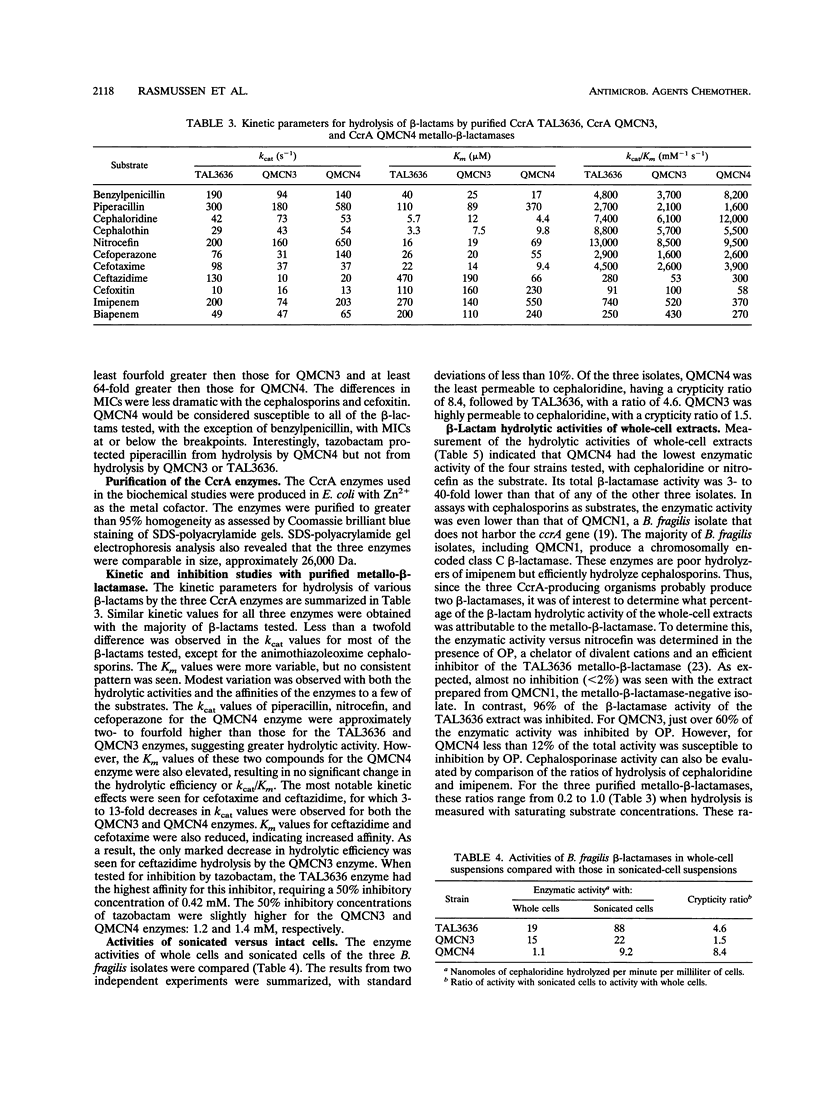
The metallo-beta-lactamase gene, ccrA, has been cloned from three clinical isolates of Bacteroides fragilis, TAL3636, QMCN3, and QMCN4. Although all three isolates harbored a gene encoding a potent beta-lactamase, the MICs of benzylpenicillin, piperacillin, cefotaxime, ceftazidime, imipenem, and biapenem for the three isolates varied from 4- to > 128-fold. QMCN4 was the most susceptible of the three isolates, followed by QMCN3. TAL3636 was resistant to all of the beta-lactams. Previous DNA sequence analysis of the three ccrA genes revealed that the enzymes differed at 5 amino acid residues (B. A. Rasmussen, Y. Gluzman, and F. P. Tally, Mol. Microbiol. 5:1211-1219, 1991). Biochemical characterization of the three enzymes revealed only small differences in kcat and Km values for the majority of beta-lactams tested. Thus, the 5 amino acid substitutions affected the hydrolyzing activity of the enzymes only modestly. Crypticity differences between the three isolates showed that QMCN4 was the least permeable of the isolates to cephaloridine, followed by TAL3636, and that QMCN3 was highly permeable to cephaloridine. Therefore, neither catalytic activity nor permeability was a major contributor to the dramatic differences in the MICs. Instead, microbiological susceptibility was closely related to the level of metallo-beta-lactamase present in each isolate. Both biochemical and physical studies indicated that TAL3636 produced 5- to 10-fold and 50- to 100-fold more metallo-beta-lactamase than QMCN3 and QMCN4, respectively. Therefore, the level of CcrA enzyme production is the dominant contributing factor to high-level resistance among strains harboring a ccrA gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Daniel M., Fleming J., Hermoso J. M., Pang C., Waley S. G. The amino acid sequence of the zinc-requiring beta-lactamase II from the bacterium Bacillus cereus 569. FEBS Lett. 1985 Sep 23;189(2):207–211. doi: 10.1016/0014-5793(85)81024-3. [DOI] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Baldwin G. S., Edwards G. F., Kiener P. A., Tully M. J., Waley S. G., Abraham E. P. Production of a variant of beta-lactamase II with selectively decreased cephalosporinase activity by a mutant of Bacillus cereus 569/H/9. Biochem J. 1980 Oct 1;191(1):111–116. doi: 10.1042/bj1910111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandoh K., Watanabe K., Muto Y., Tanaka Y., Kato N., Ueno K. Conjugal transfer of imipenem resistance in Bacteroides fragilis. J Antibiot (Tokyo) 1992 Apr;45(4):542–547. doi: 10.7164/antibiotics.45.542. [DOI] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 2c, 2d, 2e, 3, and 4. Antimicrob Agents Chemother. 1989 Mar;33(3):271–276. doi: 10.1128/aac.33.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Singer S. B. Biochemical characteristics of extended broad spectrum beta-lactamases. Infection. 1989 Nov-Dec;17(6):429–433. doi: 10.1007/BF01645566. [DOI] [PubMed] [Google Scholar]
- Cuchural G. J., Jr, Malamy M. H., Tally F. P. Beta-lactamase-mediated imipenem resistance in Bacteroides fragilis. Antimicrob Agents Chemother. 1986 Nov;30(5):645–648. doi: 10.1128/aac.30.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Carlino A., Madonna M. J., Lampen J. O. Cloning and sequencing of the metallothioprotein beta-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J Bacteriol. 1985 Oct;164(1):223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lim H. M., Pène J. J. Mutations affecting the catalytic activity of Bacillus cereus 5/B/6 beta-lactamase II. J Biol Chem. 1989 Jul 15;264(20):11682–11687. [PubMed] [Google Scholar]
- Lim H. M., Pène J. J., Shaw R. W. Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 beta-lactamase II structural gene. J Bacteriol. 1988 Jun;170(6):2873–2878. doi: 10.1128/jb.170.6.2873-2878.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podglajen I., Breuil J., Bordon F., Gutmann L., Collatz E. A silent carbapenemase gene in strains of Bacteroides fragilis can be expressed after a one-step mutation. FEMS Microbiol Lett. 1992 Feb 1;70(1):21–29. doi: 10.1016/0378-1097(92)90557-5. [DOI] [PubMed] [Google Scholar]
- Rasmussen B. A., Bush K., Tally F. P. Antimicrobial resistance in Bacteroides. Clin Infect Dis. 1993 Jun;16 (Suppl 4):S390–S400. doi: 10.1093/clinids/16.supplement_4.s390. [DOI] [PubMed] [Google Scholar]
- Rasmussen B. A., Gluzman Y., Tally F. P. Cloning and sequencing of the class B beta-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1990 Aug;34(8):1590–1592. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen B. A., Gluzman Y., Tally F. P. Escherichia coli chromosomal mutations that permit direct cloning of the Bacteroides fragilis metallo-beta-lactamase gene, ccrA. Mol Microbiol. 1991 May;5(5):1211–1219. doi: 10.1111/j.1365-2958.1991.tb01895.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Tally F. P., Jacobus N. V., Bartlett J. G., Gorbach S. L. Susceptibility of anaerobes to cefoxitin and other cephalosporins. Antimicrob Agents Chemother. 1975 Feb;7(2):128–132. doi: 10.1128/aac.7.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Malamy M. H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J Bacteriol. 1990 May;172(5):2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Rasmussen B. A., Bush K. Biochemical characterization of the metallo-beta-lactamase CcrA from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1992 May;36(5):1155–1157. doi: 10.1128/aac.36.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]