Summary
Autophagy allows cells to self-digest portions of their own cytoplasm for a multitude of physiological purposes including innate and adaptive immunity functions. In one of its innate immunity manifestations, autophagy is known to contribute to the killing of intracellular microbes including Mycobacterium tuberculosis, although the molecular mechanisms have been unclear. Here, we delineated sequential steps of the autophagic pathway necessary to control intracellular M. tuberculosis and found that in addition to autophagy initiation and maturation, an accessory autophagy-targeting molecule p62 (A170 or SQSTM1) was required for mycobactericidal activity. The p62 adapter protein delivered specific ribosomal and bulk ubiquitinated cytosolic proteins to autolysosomes where they were proteolytically converted into products capable of killing M. tuberculosis. Thus, p62 brings cytosolic proteins to autolysosomes where they are processed from innocuous precursors into neo-antimicrobial peptides, explaining in part the unique bactericidal properties of autophagic organelles.
Keywords: autophagy, p62, NBR1, ribosome, tuberculosis
Introduction
Autophagy is an evolutionary conserved process whose broad impact on innate and adaptive immunity has only recently been recognized (Deretic and Levine, 2009; Levine and Deretic, 2007; Munz, 2009). In its most generic form, autophagy is a dual purpose homeostatic mechanism that maintains the cytoplasm of eukaryotic cells in good repair and reuses cytosolic components as a nutrient reserve at times of starvation or growth factor withdrawal (Mizushima et al., 2008; Xie and Klionsky, 2007; Yoshimori and Noda, 2008). Autophagy manifests itself morphologically by the formation of double-membrane organelles termed autophagosomes capturing a variety of cytoplasmic components destined for degradation due to damage, excess, or metabolic needs (Mizushima et al., 2008; Xie and Klionsky, 2007; Yoshimori and Noda, 2008). Autophagic targets can be cytoplasmic protein aggregates which are recognized and collected by the adapter molecules p62 (also known as A170 or SQSTM1) and NBR1 (Bjorkoy et al., 2005; Kirkin et al., 2009; Korolchuk et al., 2009; Pankiv et al., 2007), large portions of the cytosol, whole organelles such as ribosomes (Kraft et al., 2008a; Kundu et al., 2008) and mitochondria (Pua et al., 2009), and intracellular microbes whether they are bacteria, protozoa or viruses (Levine and Deretic, 2007). Once autophagosomes surround their cytoplasmic targets, they fully close and mature into acidified, lytic organelles termed autolysosomes that carry out the breakdown of the sequestered contents (Mizushima et al., 2008; Xie and Klionsky, 2007; Yoshimori and Noda, 2008).
Autophagy is controlled by signaling systems; specialized Atg factors that physically execute initiation, elongation and maturation stages of autophagy; and accessory proteins, including p62, which match nascent autophagosomes with targets slated for autophagy (Bjorkoy et al., 2005; Kirkin et al., 2009; Korolchuk et al., 2009; Pankiv et al., 2007). Autophagy induction is governed by two distinct signaling complexes, Tor-Atg13-Atg1 (Chang and Neufeld, 2009; Hosokawa et al., 2009) and Beclin 1 (Atg6)-Atg14-hVPS34 (Itakura et al., 2008; Matsunaga et al., 2009; Wei et al., 2008; Zhong et al., 2009). These systems respond to amino acid and energy starvation cues, absence of growth factors, and receive a variety of stress inputs (hypoxia, genotoxicity, ER stress). They also respond to immune mediators such as cytokines (Harris et al., 2007) and presence of microbial products detected via pattern recognition receptors (Delgado et al., 2009; Xu et al., 2007) or Fcγ receptor (Huang et al., 2009).
The execution stages of autophagosome formation depend on two interconnected protein-protein and protein-lipid conjugation systems (Mizushima et al., 2008; Xie and Klionsky, 2007; Yoshimori and Noda, 2008): first, the Atg5-Atg12 covalent conjugate non-covalently complexed with Atg16 in yeast and in human cells, complexed with its ortholog, the risk factor in inflammatory bowel disease ATG16L1, and secondly, Atg8 (in mammalian systems known as LC3) conjugated at the C-terminus to phosphatidylethanolamine that allows it to associate with and participate in the elongation of the growing autophagosomal membrane. Bridging this basal autophagy apparatus with autophagic targets are proteins that recognize the cytoplasmic objects destined for autophagic degradation, with the best characterized ones being p62 and NBR1. NBR1 and p62 interact with the basal autophagy apparatus through direct association with LC3 and recognize ubiquitin tags on protein aggregates earmarked for autophagic degradation (Bjorkoy et al., 2005; Kirkin et al., 2009; Pankiv et al., 2007). In addition to the above factors, autophagosome acidification, cytoskeletal components (Fass et al., 2006; Kochl et al., 2006), and lysosomal proteases are integral to the proper execution and function of the autophagosomal pathway (Mizushima et al., 2008; Xie and Klionsky, 2007; Yoshimori and Noda, 2008).
Autophagy can eliminate intracellular microbes (Levine and Deretic, 2007). Autophagy kills intracellular Mycobacterium tuberculosis (Alonso et al., 2007; Biswas et al., 2008; Delgado et al., 2008; Gutierrez et al., 2004; Singh et al., 2006; Xu et al., 2007), despite this pathogen's ability to block several antimicrobial mechanisms of the macrophage (Vergne et al., 2004) and despite its capacity to survive in conventional phagolysosomes, a property well known since the classical studies by Hart and colleagues (Armstrong and Hart, 1975). The case of M. tuberculosis brings to the fore the fundamental differences in the microbicidal capacity of autolysosomes vs. phagolysosomes, and highlights the need to understand the specific properties and stages of the complex autophagic pathway that render autophagic organelles particularly microbicidal. In this study, we show that p62, a molecule that targets cytosolic proteins to autophagosomes, is critical for the elimination of the tubercle bacilli. The autophagic adapter protein p62 delivered specific cytosolic components including ribosomal protein S30 (rpS30) and additional ubiquitinated targets to autophagic organelles, where they were processed into mycobactericidal products. Without p62 these neo-antimycobacterial factors were not generated, autophagy was rendered harmless despite its unabated progression through the maturation stages, and intracellular M. tuberculosis was not efficiently eliminated.
Results
Both autophagosome formation and maturation are important for autophagy-mediated killing of the tubercle bacilli
We tested sequential stages and representative factors and processes along the autophagic pathway for their requirement in elimination of mycobacteria. The initial screen included Atg5, autophagosome acidification, cytoskeletal components, and lysosomal proteases, which are needed for autophagosome formation, maturation, and digestion of the captured cargo. We used an established killing assay with short term stimulation of autophagy (Gutierrez et al., 2004; Ponpuak et al., 2009), which has the advantage of avoiding secondary effects potentially associated with longer term incubation (Ponpuak et al., 2009), although the changes in colony forming units (CFU) are lesser in magnitude than in overnight incubations (Alonso et al., 2007). To test whether Atg5 was necessary for autophagic control of mycobacteria, we targeted Atg5 by siRNA in the RAW264.7 macrophage cell line and assayed M. tuberculosis H37Rv survival by plating (Fig. 1A). The Atg5 targeting was verified by immunoblotting (Fig. S1A). Depletion of Atg5 reduced autophagic killing of M. tuberculosis H37Rv in infected cells (Fig. 1A). The same effect was observed in primary, bone-marrow derived macrophages (BMM) isolated from mice with the Atg5 gene conditionally deleted in myeloid cells via the LyzM-Cre Atg5fl/fl system (Fig. 1B). The absence of Atg5 expression in LyzM-Cre Atg5fl/fl macrophages was validated by immunoblots (Fig. S1B). These data indicate that autophagosome formation is important for autophagic restriction of mycobacteria.
Fig 1.
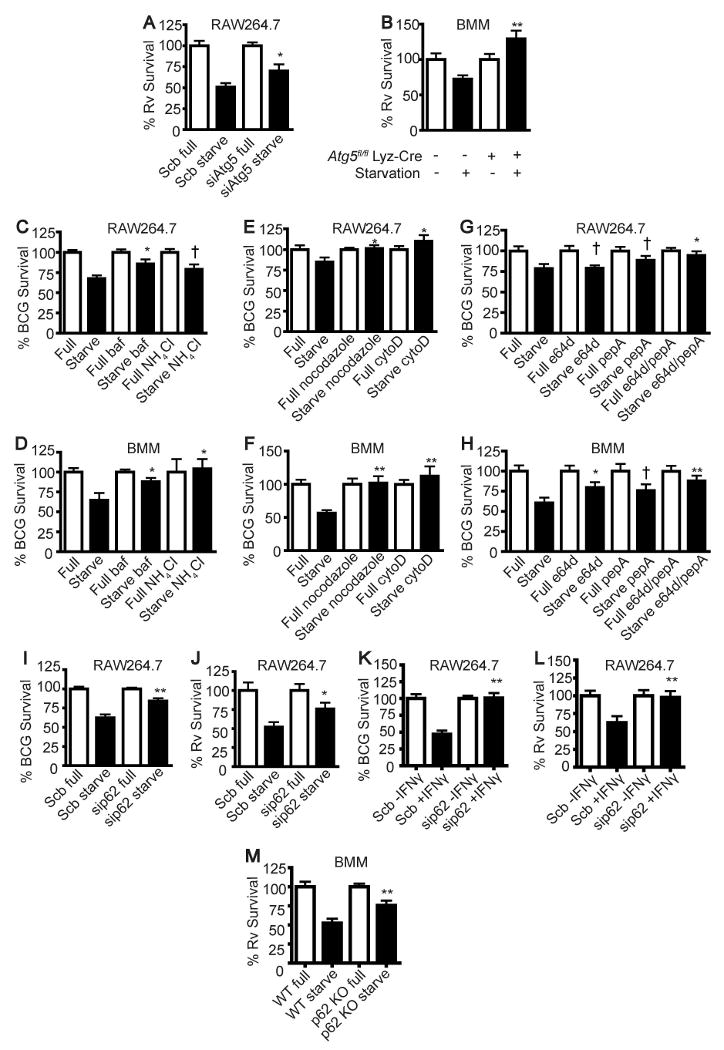
Factors crucial for autophagic control of mycobacteria. (A), RAW264.7 cells were transfected with siRNAs against Atg5 or scramble (Scb) control. At 48 h after transfection, cells were infected with M. tuberculosis H37Rv and subjected to autophagic induction by starvation for 4 h. Cells were then lysed to determine the number of viable bacteria by plating colony forming units (CFU). (B), Atg5-deficient BMM cells were infected with M. tuberculosis H37Rv and subjected to autophagic induction and CFU analysis as in (A). (C-H), RAW264.7 or BMM cells were treated with inhibitors of autophagosome acidification (C and D), or cytoskeleton polymerization (E and F), or lysosomal proteases (G and H) upon autophagic induction for 4 h. Cells were then lysed to determine the number of viable bacteria by plating CFU. (I - K), p62 was depleted from RAW264.7 cells using siRNAs. At 48 h after transfection, cells were infected with M. tuberculosis BCG (I and K) or H37Rv (J and L) and subjected to autophagic induction by starvation for 4 h or IFN-γ for 24 h. Cells were then lysed and the numbers of viable bacteria were determined by plating cfu. (M), p62 deficient (p62 KO) BMM cells were infected with M. tuberculosis H37Rv and subjected to autophagic induction and cfu analysis as in (B). All samples (including those labeled as Full and Starve) in panels C-H contained DMSO as a vehicle used to solubilize compounds. Data, means ± SEM from at least three independent experiments; **p < 0.01, *p < 0.05, †p ≥ 0.05, all relative to starvation control. See also Fig. S1.
We next tested whether autophagosome acidification is required for autophagic killing of the tubercle bacilli. When the infected RAW264.7 or BMM cells were treated with bafilomycin A1, an inhibitor of vacuolar H+ ATPase, or with NH4Cl as a source of the weak base ammonia, autophagic control of the tubercle bacilli was reduced (Fig. 1C and D). Hence, the acidic pH is necessary for autophagy-mediated control of mycobacteria.
The cytoskeletal components, primarily microtubules (Fass et al., 2006; Kochl et al., 2006), and possibly actin (Aplin et al., 1992), have been implicated in the formation and maturation of autophagic vacuoles. When RAW264.7 or BMM cells were infected with mycobacteria and then treated with nocodazole or cytochalasin D, a decrease in autophagic control of the microbe was observed (Fig. 1E and F). Therefore, cytoskeletal elements playing a role in autophagy, including microtubules and F-actin, are needed for autophagic killing of mycobacteria.
Next, we tested whether lysosomal hydrolases are important for autophagic elimination of mycobacteria. When RAW264.7 or BMM were treated with E64d, an inhibitor of cathepsins B and L, and pepstatin A, an inhibitor of cathepsin D, during autophagic induction this protected mycobacteria from autophagic killing (Fig. 1G and H). Thus, lysosomal proteases are important for autophagic restriction of mycobacteria. Collectively, these results show that the elimination of the tubercle bacilli by autophagy requires all typical stages of autophagy, from the formation of autophagosomes to their maturation into functional degradative organelles.
Adaptor protein p62 is required for autophagic control of mycobacteria
The adaptor protein p62 is an autophagy-targeting molecule recognizing ubiquitinated cytoplasmic components and delivering them for degradation (Bjorkoy et al., 2005; Ichimura et al., 2008; Komatsu et al., 2007; Noda et al., 2008; Pankiv et al., 2007; Shvets et al., 2008). Because ubiquitin has been found to be in association with phagosomes (Houde et al., 2003) and bacteria (Perrin et al., 2004) and following autophagic induction there is an accumulation of ubiquitin in phagosomal organelles containing mycobacteria (Alonso et al., 2007), we tested whether p62 might play a role in autophagic killing of the tubercle bacilli. We observed p62 colocalizing with intracellular organelles containing mycobacteria in cells subjected to induction of autophagy by starvation (Fig. 2A and B). Movies S1 and S2 show that p62 puncta dynamically interact with mycobacterial phagosomes. As expected (Komatsu et al., 2007), p62 and endogenous LC3 showed colocalization (Fig. S2A). Moreover, the colocalization of p62 with mycobacterial phagosomes depended on Atg5 (Fig. S2B and C). This accumulation was specific for p62 since we did not find an increase in mycobacterial phagosome colocalization with NBR1, another autophagic receptor protein proposed to act on ubiquitinated targets (Kirkin et al., 2009) (Fig. S2D and E). Survival assays of M. tuberculosis var. bovis BCG (BCG) and virulent M. tuberculosis H37Rv in cells depleted for p62 by siRNA (Fig. S1C) showed decreased autophagic killing (Fig. 1I and J) without affecting bacterial uptake by macrophages (Fig. S1D and E). In addition to the requirement for p62 in starvation-induced autophagic killing, p62 was important for IFN-γ induced killing of BCG (Fig. 1K) and virulent M. tuberculosis H37Rv bacilli (Fig. 1L). BCG showed increased colocalization with p62 in cells treated with IFN-γ (Fig. 2C and D). Furthermore, using BMM from p62-deficient mice (Komatsu et al., 2007), we found that BMM from p62-proficient animals (Sqstm1+/+) killed M. tuberculosis H37Rv more efficiently than BMM from p62-deficient mice (Fig. 1M). BMM from p62-deficient mice did not show any detectable p62 (Fig. S1F) and displayed no change in M. tuberculosis H37Rv uptake relative to wild type mice (Fig. S1G). These observations indicate that p62 is important for autophagy-mediated killing of mycobacteria.
Fig 2.
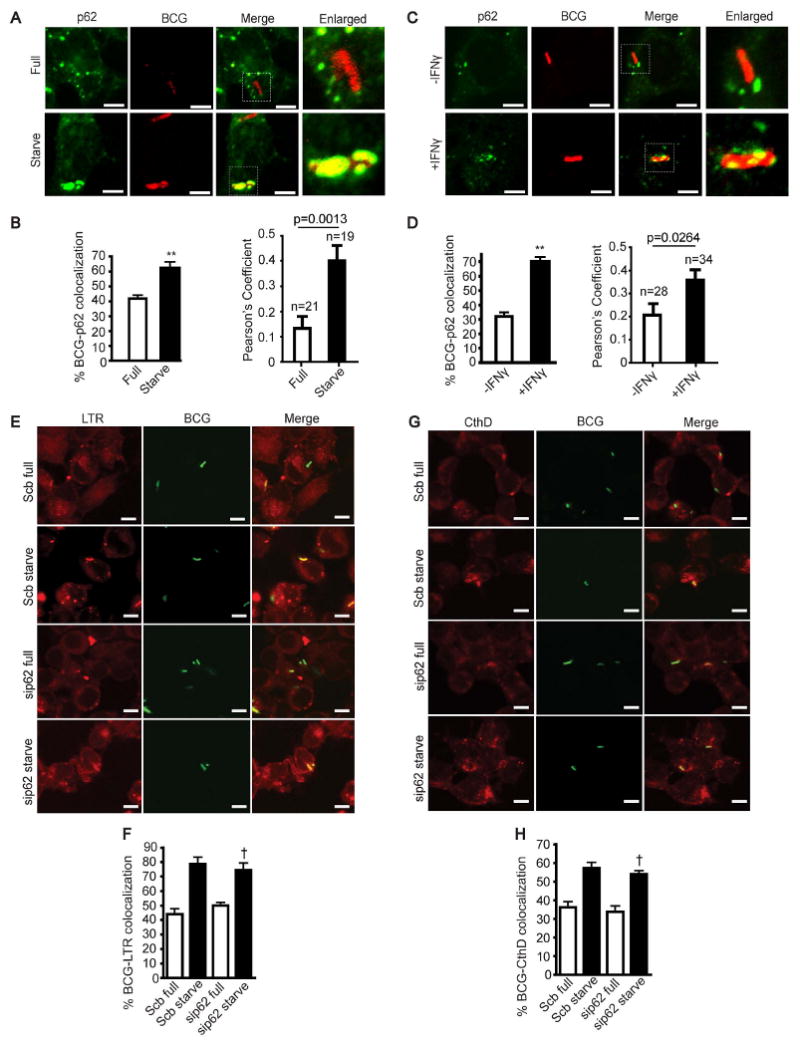
p62 is important for autophagic elimination of tubercle bacilli but does not function in the maturation of mycobacterial phagosomes. (A-D), Autophagy enhances p62 colocalization with mycobacterial phagosomes. RAW264.7 cells were infected with Alexa-568 labeled BCG and chased for 1 h in complete media. Cells were then subjected to autophagic induction for 4 h by starvation (A and B) or 24 h by IFN-γ treatment (C and D). Cells were fixed and stained for p62. (E - H) p62 does not play a role in mycobacterial phagosome maturation. RAW264.7 cells were transfected with siRNAs against p62 or scramble control. At 48 h after transfection, cells were infected with Alexa-488 labeled mycobacteria and chased for 1 h in complete media. Cells were subjected to autophagic induction by starvation for 4 h and stained with LysoTracker Red (E and F) or cathepsin D (G and H) to determine the acidification and acquisition of a lysosomal protease by mycobacterial phagosomes. Data, means ± SEM from at least three independent experiments. At least 50 phagosomes per category per independent experiment, were quantified using % BCG-marker colocalization; **, p < 0.01, *, p < 0.05, †, p ≥ 0.05, all relative to starvation control. For Pearson's colocalization coefficient (defined in Experimental Procedures) exact p values are shown and number of phagosomes analysed in each category indicated. See also Figs. S2 and S3, and Movies S1 and S2.
Autophagic adaptor p62 does not influence lysosomal maturation of compartments harboring mycobacteria
We tested whether gene targeting of p62 exerted its mycobacteria-sparing effects by affecting maturation of intracellular compartments containing mycobacteria. First, we examined whether p62 gene targeting altered autophagy in general, as assessed by starvation-induced proteolysis of stable proteins. Autophagic proteolysis did not change with p62 targeting (Fig. S3A) in keeping with the previous reports (Komatsu et al., 2007). When p62 was targeted in RAW264.7 cells, mycobacterial phagosome acidification and acquisition of a lysosomal protease, cathepsin D, were not altered (Fig. 2 E-H). Autophagy induction still increased acidification of mycobacterial phagosomes in cells treated with p62 siRNA, as determined by LysoTracker Red staining (Fig. 2E and F). Similarly, immunofluorescence analyses indicated that p62 depletion by siRNA did not inhibit enhanced delivery of cathepsin D to mycobacterial phagosomes associated by autophagy (Fig. 2G and H). Thus, p62 requirement in autophagic elimination of the tubercle bacilli is independent of organelle maturation, ruling out the simplest explanation that p62 depletion exerted its mycobacteria-protective effect by inhibiting delivery of BCG to autophagic organelles such as autolysosomes. It has also been reported that absence of p62 can affect NF-κB expression (Mathew et al., 2009), and because this cytokine plays a role in several immune processes we tested whether NF-κB expression was altered in our experiments. Targeting of p62 did change the NF-κB activation as determined by pNF-κB:Luc probe, in cells incubated in starvation or full medium or treated with IFN-γ (Fig. S3B and C). However, NF-κB was not a factor in starvation induced autophagic killing of mycobacteria because when p65 was depleted by siRNA (Fig. S3D) in cells stimulated for autophagy by starvation or IFN-γ treatment, there was no alteration in M. tuberculosis H37Rv survival (Fig. S3E and F). These observations ruled out indirect effects of p62 on mycobacterial survival, and, taken together with the results indicating that p62 was not required for mycobacterial transfer to autolysosomes, suggested a less predictable role for p62 function in autophagic elimination of tubercle bacilli.
Adaptor p62 functions in autophagic delivery of microbicidal factors to the mycobacterial phagosome
We next tested an alternative hypothesis that p62 brings specialized autophagic microbicidal components to the mycobacterial phagosome. One distinguishable feature of autophagosomes is that they collect cytosolic components including ribosomes (Kraft et al., 2008b). Importantly, a subset of ribosomal proteins have been previously reported as possessing microbicidal activity against Gram-positive and Gram-negative bacteria (Howell et al., 2003; Tollin et al., 2003). The ribosomal proteins in question include L30, S19, and S30. S30 is also known as rpS30 or ubiquicidin, and is referred herein as rpS30. How these ribosomal proteins meet their targets has not been examined in the past studies, although rpS30 has been identified as an antimicrobial polypeptide within the cytosolic fraction of RAW264.7 macrophages activated with interferon-γ (IFN-γ) (Hiemstra et al., 1999), a cytokine now known to be an autophagic inducer (Deretic and Levine, 2009; Levine and Deretic, 2007).
We examined if any of the above ribosomal proteins were recruited to mycobacterial phagosomes upon induction of autophagy. Interestingly, rpS30 is synthesized as a precursor protein (Fig. S4A) called Fau (a fusion of rpS30 with a ubiquitin-like domain at its N-terminus) (Olvera and Wool, 1993). A substantial increase was observed in Fau localization with mycobacterial phagosomes upon autophagic induction (Fig. 3A and B, and Supplementary movie M3). Fau colocalization with BCG phagosomes showed two morphological types: (i) punctate (Fig. 3A, middle panel of images and Supplementary movie M3); and (ii) homogenious distribution (Fig. 3A, bottom panel of images). Immunofluorescence analysis of Fau and endogenous LC3 indicated that Fau was delivered to LC3-positive compartments (Fig. S4B). The colocalization of Fau with mycobacterial phagosomes in cells induced for autophagy by starvation was dependent on Atg5 (Fig. S4C, D) as shown by comparing BMM from wild type and LyzM-Cre ATG5fl/fl mice that have lost Atg5 specifically in myeloid cells (Zhao et al., 2008). An increase in Fau-BCG phagosome colocalization was also detected in cells stimulated with IFN-γ (Fig. S4E and F). In contrast, no increase in colocalization of ribosomal proteins S19 or L30 with the tubercle bacilli was detected in cells undergoing autophagy (Fig. 3C–F). The absence of colocalization with rpL30 and rpS19 indicated specificity for Fau in this process. When p62 was depleted, a decrease in Fau delivery to mycobacterial phagosomes was noted (Fig. 3G and H). These data indicate that p62 is required for the trafficking of Fau to the mycobacterial phagosome.
Fig 3.
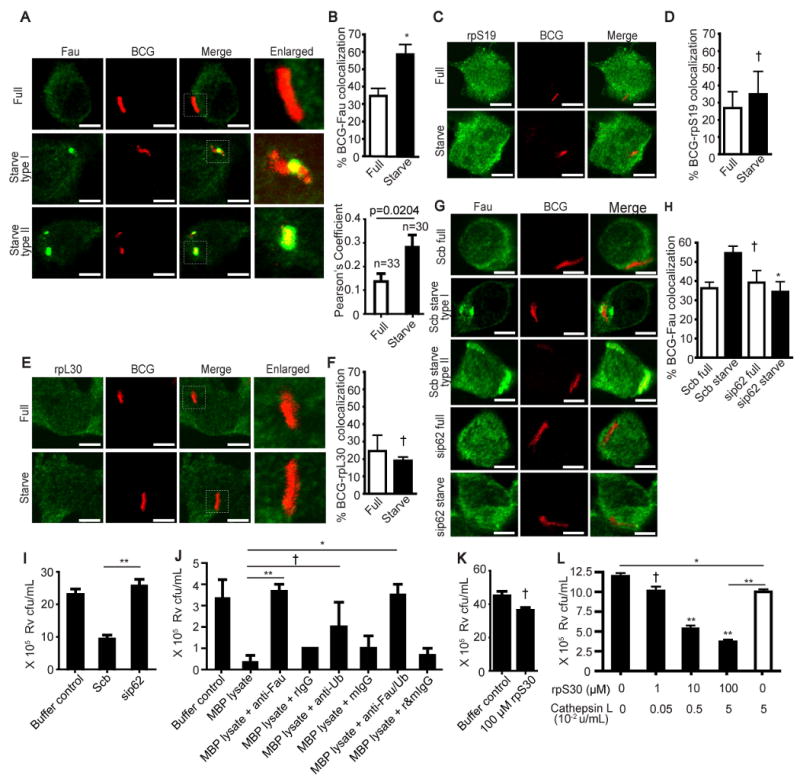
p62 functions by delivering microbicidal ribosomal protein S30 (rpS30) to the mycobacterial phagosomes. (A - B) Autophagy increases Fau (a fusion precursor protein consisting of rpS30 and a ubiquitin-like domain) colocalization with mycobacterial phagosomes. RAW264.7 cells were infected with Alexa-568 labeled BCG and chased for 1 h in complete media. Cells were then subjected to autophagic induction for 4 h by starvation. Cells were fixed and stained for rpS30. (C – F) Autophagy does not enhance the delivery of ribosomal proteins S19 (rpS19) or L30 (rpL30) to mycobacterial phagosomes. Cells were treated as in (A and B) but stained for rpS19 (C and D) or rpL30 (E and F). (G - H) p62 is required for autophagic translocation of Fau to mycobacterial phagosomes. p62 was depleted from RAW264.7 cells with siRNAs. At 48 h after transfection, cells were infected with Alexa-568 labeled BCG, subjected to autophagic induction, and stained for Fau. Type I (punctate distribution) and type II (homogeneous distribution) refer to two different morphotype patterns observed in colocalization between Fau and BCG. At least 50 phagosomes per category per independent experiment (n=3) were quantified using % BCG-marker colocalization; **, p < 0.01, *, p < 0.05, †, p ≥ 0.05; in panel H, relative to corresponding scrambled siRNA-treated controls. For Pearson's colocalization coefficient (defined in Experimental Procedures) exact p values are shown and number of phagosomes analyzed in each category indicated. (I) p62 is important for cidal activity of phagosomal compartments. p62 was depleted from RAW264.7 cells with siRNAs. At 48 h after transfection, cells were infected with mixture of magnetic beads and mycobacteria for 1 h and subjected for autophagic induction by starvation for 4 h. Phagosomal contents were purified as described in methods and used in in vitro killing assays. The numbers of viable bacteria were determined by plating CFU. (J) Fau contributes to the mycobactericidal capacity of phagosomal compartments. Phagosomal contents were purified and subjected for immunodepletion of Fau or Ubiquitin. The resulting lysates were then used in in vitro killing assays and bacteria survival was determined by plating cfu. (I-J) rpS30 fragments contain antimycobactericidal activity. rpS30 was subjected to proteolysis overnight with cathepsin L. rpS30 peptides at the indicated concentration were then incubated with M. tuberculosis H37Rv for 2d. Serial dilutions were then plated for cfu quantification. Data, means ± SEM from at least three independent experiments. **p < 0.01, *p < 0.05, †p ≥ 0.05, all relative to control. See also Fig. S4, Movie S3 and Table S1.
In the next set of experiments, we developed an in vitro killing assay with extracts from phagosomes purifed form cells induced for autophagy (KEPPA). For KEPPA, we prepared phagosomal compartments (purified by manetic beads as described in Experimental Procedures) from cells induced for autophagy by starvation. These organelles were then subjected to release of the lumenal content by mild detergent treatment (as described in Experimental Procedures) and the obtained material was used for in vitro killing assay with M. tuberculosis H37Rv. When these extracts were prepared from cells knocked down for p62, the killing capacity was abrogated (Fig. 3I) indicating that p62 was important for delivery of mycobactericidal ingredients to the lytic organelles.
Applying the in vitro killing KEPPA assay described above, we next immunodepleted Fau (Fig. 3J) from phagosomal lumenal extracts before subjecting M. tuberculosis H37Rv to killing. Mycobactericidal capacity of the extract from the lytic organelles was abrogated by immunodepletion of Fau (Fig, 3J, bars 1-4). Thus, Fau supplies at least a portion of bactericidal activity in autophagolysosmal organelles.
Based on our results that Fau is accumulated in mycobacterial phagosomes upon autophagic induction, that its delivery depends on p62, and that Fau supplies important killing activity, we examined if its portion corresponding to rpS30, previously shown to posses antibacterial activity (Hiemstra et al., 1999; Howell et al., 2003; Tollin et al., 2003) may be improtant. Although previous studies have not investigated the mechanism for delivery of rpS30 to bacteria and have not tested it against mycobacteria, having now a plausable delivery process and data showing that its precursor Fau enhances kiling activity, we tested whether rpS30 possessed activity against mycobacteria. A 59-residue polypeptide corresponding to rpS30 was synthesized and tested for its ability to kill mycobacteria. Intact rpS30 could not kill virulent M. tuberculosis H37Rv (Fig. 3K). However, incubation of rpS30 with lysosomal proteinase cathepsin L resulted in a liberation of mycobactericidal activity from the protein (Fig. 3L). In titration experiments, where both components (Cathepsin L and rpS30) were simultaneously diluted in 10-fold increments, the killing activity was abrogated in samples with 1/100 dilution. Cathepsin L alone, when used at the highest concentration (0.05 u/ml) showed only 17% decrease in mycobacterial viability (Fig. 3L last bar). At the same concentration of Cathepsin L, samples containing 100 or 10 μM rpS30 showed a 3-fold and a 2-fold drop in viability relative to control (Fig. 3L; bars 3 and 4). We also examined the proteolytic products by mass spectrometry and identifed 3 fragments (rpS30-1, AKQEKKKKK; rpS30-2, NRRFNVVPTFGK; and rpS30-3, GKKKGPNANS) specifically liberated from rpS30 by cathepsin L (Table S1). Of these three fragments, rpS30-2 efficiently killed virulent M. tuberculosis H37Rv (Table S1). Altogether, these data indicate that rpS30 peptide fragments kill mycobacteria and may contribute to the autophagic elimination of the tubercle bacilli due to delivery of rpS30 to autophagic organelles.
We also observed increased colocalization of ubiquitinated proteins with mycobacterial phagosomes upon induction of autophagy by starvation (Fig. 4A and B) in keeping with the observation by Alonso et al. (Alonso et al., 2007). This colocalization was dependent on Atg5 (Fig. S5A and B). As with starvation, colocalization between ubiquitinated proteins and BCG was detected in cells treated with IFN-γ (Fig. S5C and D). As previously reported (Kim et al., 2008), endogenous LC3 and ubiquitin showed colocalization (Fig. S5E). More importantly, we detected a decrease in the delivery of ubiquitinated proteins to mycobacterial phagosomes (Fig. 4C and D) upon p62 depletion. In the KEPPA assay, we observed a trend with ubiquitin immunodepletion, albeit the statistical significance cutoff was not reached (p = 0.23) (Fig. 3J, bar 5). A combination of Fau and ubiquitin immunodepletion showed, as expected based on immunodepletion of Fau alone (Fig. 3J bar 3), a strong reduction in mycobactericidal potential (Fig. 3J, bar 7). Collectively, these data indicate that p62 contributes to autophagic targeting of several cytosolic proteins including the ribosomal precursor protein Fau and ubiquitin-positive material to the mycobacterial phagosome.
Fig 4.
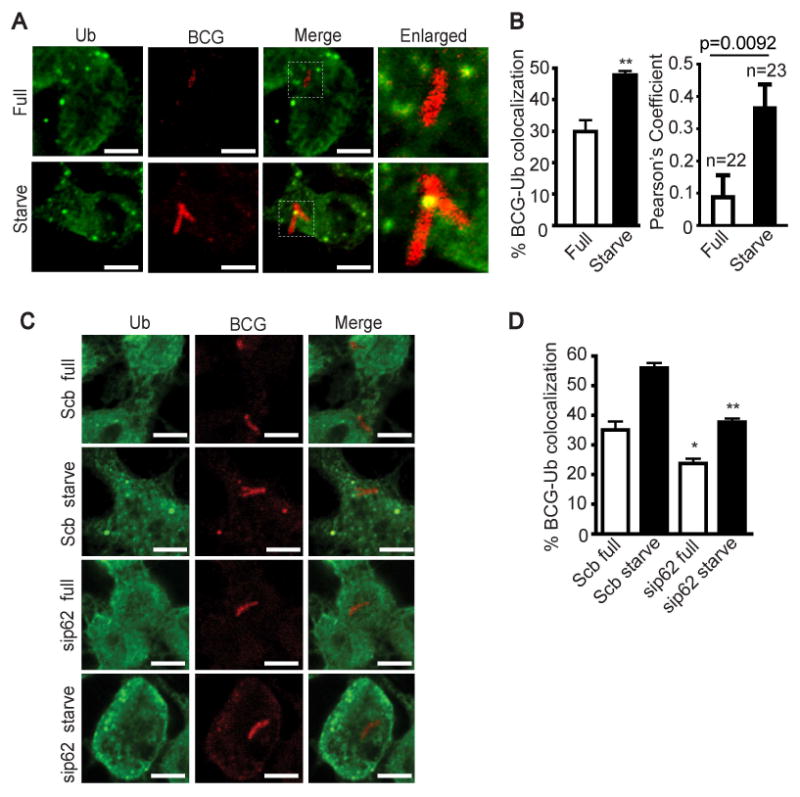
p62 delivers ubiquitin and microbicidal ubiquitin-derived peptides to the mycobacterial phagosomes. (A - B), Autophagy promotes ubiquitin colocalization with mycobacterial phagosomes. RAW264.7 cells were infected with Alexa-568 labeled BCG and chased for 1 h in complete media. Cells were then subjected to autophagic induction for 4 h by starvation. Cells were fixed and stained for ubiquitin conjugates. (C - D) p62 is necessary for the autophagic targeting of ubiquitin conjugates to the mycobacterial phagosomes. p62 was depleted from RAW264.7 cells with siRNAs. At 48 h after transfection, cells were infected with Alexa-568 labeled BCG, subjected to autophagic induction, and stained for ubiquitin conjugates as in (A and B). Data, means ± SEM from at least three independent experiments. At least 50 phagosomes per category per independent experiment were quantified using % BCG-marker colocalization; **, p < 0.01, *, p < 0.05, all relative to control. For Pearson's colocalization coefficient (defined in Experimental Procedures) exact p values are shown and number of phagosomes analyzed in each category indicated. See also Fig. S5.
The PB1, LIR, and UBA domains of p62 are necessary for autophagic delivery of Fau and ubiquitin to the mycobacterial phagosome
Next, specific domains (Fig. S6A) of p62 of importance for the above processes were studied. The N-terminal PB1 domain enables p62 to interact with other PB1 domain-containing proteins and p62 itself to form heterodimers and oligomers (Lamark et al., 2003). At its C-terminus, p62 has a UBA domain which binds to ubiquitin (Kirkin et al., 2009). Recently, the middle region of p62, designated LIR, has been shown to bind directly to autophagosomal proteins, LC3 and GABARAP (Ichimura et al., 2008; Pankiv et al., 2007). Both PB1 and UBA domains of p62 are required for ubiquitinated-protein aggregate formation and LIR is needed for p62-LC3 interaction, allowing recognition of ubiquitinated protein aggregate and its targeting for degradation by autophagy (Bjorkoy et al., 2005; Ichimura et al., 2008; Pankiv et al., 2007). We transfected GFP-tagged p62 constructs with a mutated PB1, ΔLIR, or ΔUBA domain into RAW264.7 macrophages and examined effect on ubiquitin delivery to mycobacterial phagosomes. The expression of p62 mutants was assessed by immunfluorescence (Fig. 5A and B) and by immunoblots (Fig. S6B), and their relationship to endogenous LC3 was determined by fluorescence microscopy (Fig. S6C). The mutated PB1, ΔLIR, and ΔUBA p62 were previously shown to act as dominant negative mutants in autophagic assays (Bjorkoy et al., 2005; Pankiv et al., 2007). Overexpression of all p62 mutants interfered with the traffic of ubiquitin to mycobacterial phagosomes following autophagy induction by starvation (Fig. 5A and C and Fig. S6D). The PB1 domain p62 mutant does not form aggregates while mutant p62ΔLIR forms aggregates, but both interfered with delivery of ubiquitin to BCG phagosomes, and thus the results of the interference assay cannot be attributed to aggregate formation. Similar domain requirement was observed for Fau delivery to BCG phagosomes (Fig. 5B and D and Fig. S6E). These results indicate that p62 oligomerization, interaction with LC3, and recognition of ubiquitin are all needed for proper delivery of Fau and ubiquitin complexes to mycobacterial phagosomes, confirming the importance of p62 in this autophagic trafficking process.
Fig 5.
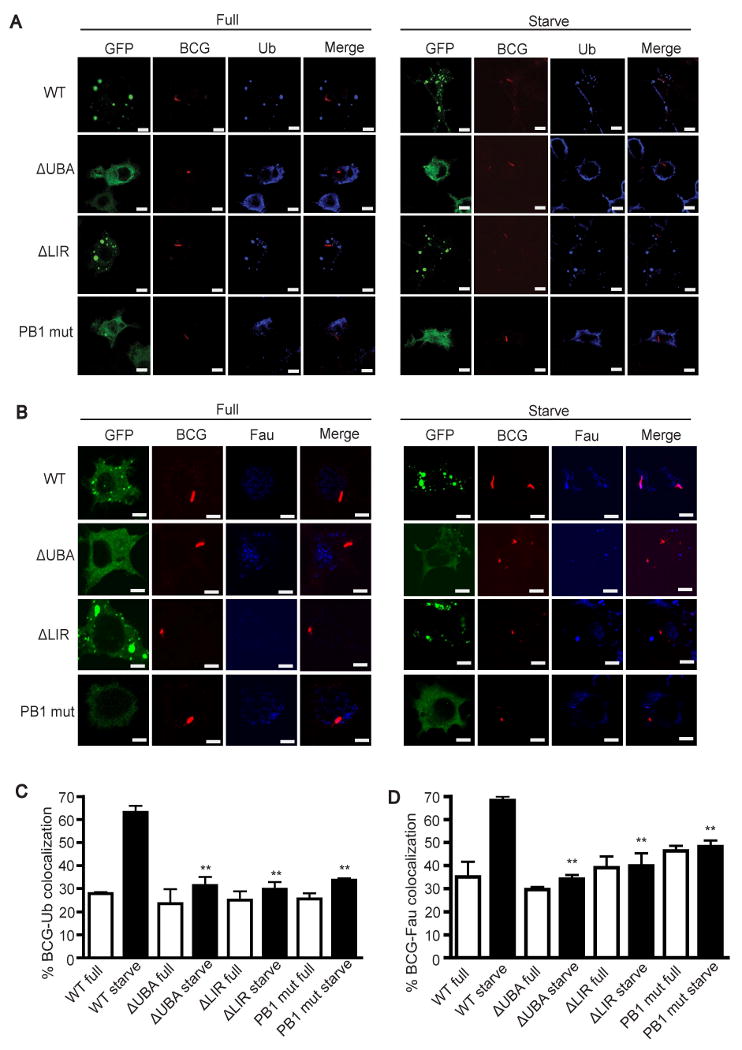
PB1, LIR, and UBA domains of p62 are vital for ubiquitin and Fau delivery to the mycobacterial phagosomes. (A-D), RAW264.7 cells were transfected with GFP tagged p62 constructs with mutation in PB1, or ΔLIR, or ΔUBA domains. At 24 h after transfection, cells were infected with Alexa-568 labeled BCG and chased for 1 h in complete media. Cells were then subjected to autophagic induction by starvation for 4 h and stained for ubiquitin conjugates (A and C) or Fau (Band D). Data, means ± SEM from at least three independent experiments. For each experiment, at least 50 phagosomes were quantified per condition. **p < 0.01, all relative to starvation control. See also Fig. S6.
Biochemical analysis of p62 in delivery of Fau and ubiquitin conjugates to phagosomes
We isolated magnetic bead phagosomes (MBP) from RAW264.7 cells undergoing autophagy and probed for associated proteins by immunoblotting (Fig. 6A). Both Fau and ubiquitin conjugates were found to be associated with MBP (Fig. 6A, lane 1 and 2). Upon induction of autophagy, there was an increase in amounts of both Fau and ubiquitin conjugates associated with MBP (Fig. 6A, lane 3 and 4, top and bottom sets). As expected, polyubiquitinated material appeared as multiple bands or a smear on immunoblots (Fig. 6A, bottom set) These biochemical data reinforce our immunofluorescence microscopy findings showing accumulation of Fau and ubiquitinated proteins in phagosomes upon autophagic induction. The Fau delivered to these organelles was undergoing degradation, albeit with slower kinetics than ubiquitin conjugates and p62, as seen when amounts of all three substrates were compared to samples subjected to bafilomycin-conferred protection from degradation (Fig. S7).
Fig 6.
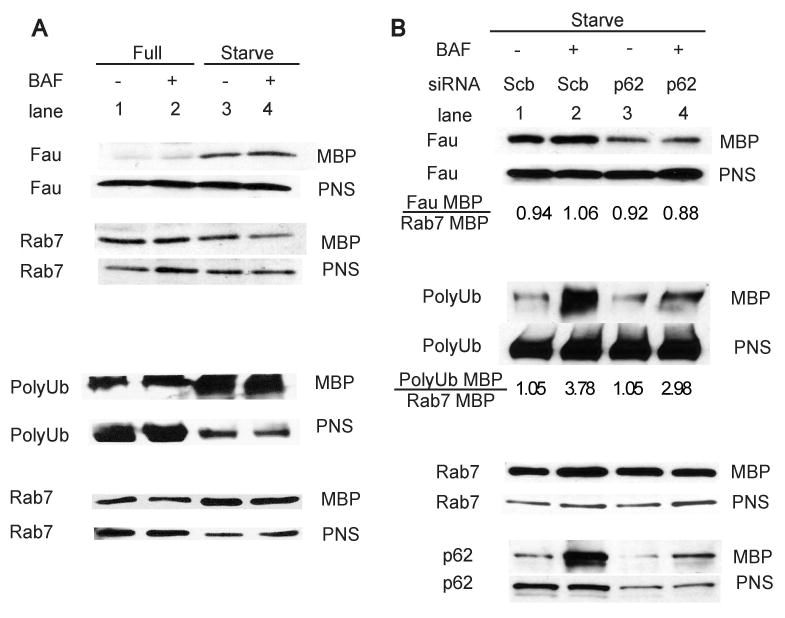
p62 is important for the trafficking of Fau and ubiquitin conjugates to the phagosomes. (A), Autophagy increases Fau and ubiquitin conjugates translocation to magnetic bead phagosomes. RAW264.7 cells were infected with mycobacteria in a mixture with magnetic beads and subjected to autophagic induction by 2 h starvation in the presence or absence of bafilomycin A1 treatment to inhibit autophagic degradation. Magnetic bead phagosomes (MBP) and post-nuclear-supernatant less of MBP (PNS) were then isolated and subjected to immunoblot analysis for indicated proteins. (B), p62 is vital for Fau and ubiquitin conjugate delivery to the phagosomes. Cells were depleted of p62 using siRNAs and subjected to the same treatments as in (A) under starvation condition. MBP and PNS were then isolated and analyzed by immunoblotting. See also Fig. S7.
To assess p62 role in trafficking of Fau and ubiquitin conjugates into phagosomes, we depleted p62 from cells undergoing autophagy (Fig. 6B). The amount of Fau and polyubiquitin conjugates were diminished in cells subjected to p62 knockdown and treated with bafilomycin A1 (Fig. 6B, lane 4) compared to control cells treated only with bafilomycin A1 (Fig. 6B, lane 2). These results show that p62 is required in the autophagic delivery of Fau and ubiquitin conjugates to the phagosomes.
Fau Forms a Complex with p62
The interaction between p62 and ubiquitin has been extensively characterized (Kirkin et al., 2009; Vadlamudi et al., 1996). In contrast, it is not known whether or how p62 and Fau interact. Since Fau contains a ubiquitin like domain at its N-terminus, we hypothesize that this domain may mediate Fau interaction with p62. Alternatively, Fau itself could be modified by ubiquitin conjugation thus facilitating interaction with p62. We performed immunocolocalization between endogeneous Fau, p62, and ubiquitin (Fig. 7A). The Fau staining pattern was diffuse in the cytosol but additionally showed puctate distribution. The Fau puncta colocalized with a subset of p62 and ubiquitin pucta in cells (with or without autophagic stimulation). To determine if endogeneous Fau could associate with p62, we carried out coimmunoprecipitation experiments. Using anti-p62 for immunoprecipitation, we observed that endogeneous Fau coprecipitated with p62 (Fig. 7B). Another Fau antibody-reactive band corresponding to the size of monoubiquitinated Fau was seen. When we reprobed the blot with mono- and poly-ubiquitin antibodies, this band was identified as monoubiquitinated Fau. Hence, both ubiquitin-free Fau and monoubiquitinated Fau are found in complexes with p62.
Fig 7.
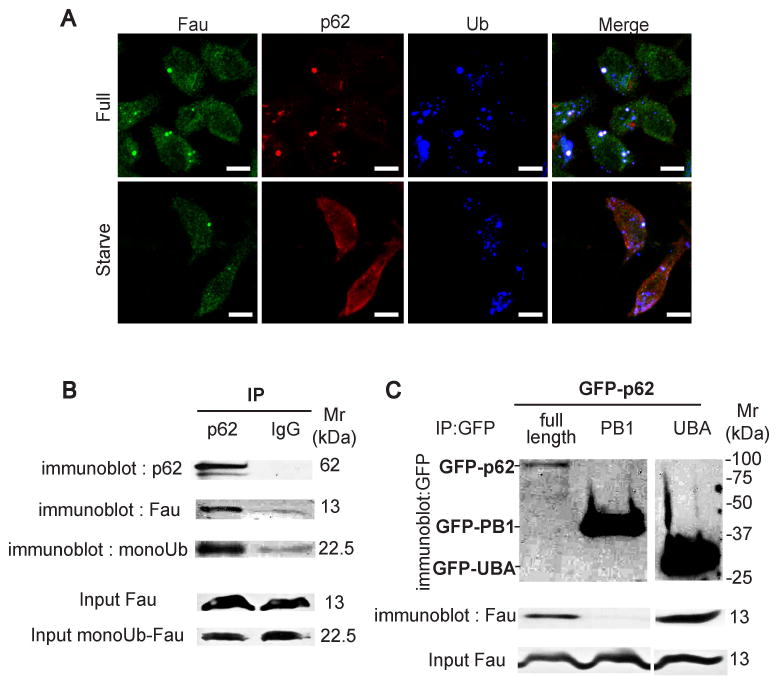
Fau interacts with p62 in vivo. (A), Fau, p62, and ubiquitin conjugates are colocalizing in cells with or without undergoing autophagy. RAW264.7 cells were subjected to autophagic induction by starvation for 2 h and immunostained for endogeneous Fau, p62, and ubiquitin conjugates. (B), Fau is found in the same immune complexes with p62. RAW264.7 cells were infected with mycobacteria for 1 h, washed, and harvested. Cell lysates were prepared and subjected for immunoprecipitation experiments and the immune complexes were analyzed by Western blotting with indicated antibodies. 100 μg of input also analyzed in parallel. (C) The ubiquitin associate domain (UBA) of p62 interacts with Fau in vivo. P62 full-length or PB1 domain containing R21A/D69A mutation (to prevent oligomerization) or UBA domain tagged with GFP plasmids were tranfected into RAW264.7 cells. 48 h after transfection, cells were subjected for immunoprecipitation and the immune complexes and cell lysates were analyzed by immunoblotting.
Additional immunoprecipitation experiments using individual p62 domains fused to GFP showed that the UBA domain of p62 interacted with endogenous Fau (Fig. 7C). This interaction was specific to the UBA domain as we did not observe coimmunoprecipitation of Fau with the PB1 domain of p62 (Fig. 7C, lane 2). These data indicated that p62 binds to Fau via its ubiquitin associated (UBA) portion.
Based on these experiments, antimycobacterial properties of cathepsin fragments of Fau, and enhanced colocalization of Fau with mycobacteria, we conclude that Fau and additional ubiquitinated protein complexes are delivered by p62 to compartments where their processing upon induction of autophagy allows them to exert their antimycobacterial action. Thus, p62 is a hitherto unappreciated mediator of innate immunity.
Discussion
In this work we have uncovered two previously unappreciated key mechanisms explaining how autophagy exerts its direct antibacterial function that sets it apart from conventional bactericidal compartments such as phagolysosomes: First, p62, a protein previously known to assists autophagic clean-up of protein aggregates toxic to the cell of importance in neurodegenerative disease (Bjorkoy et al., 2005; Komatsu et al., 2007), is used for immunological purposes in macrophages (and likely in other cells) to deliver cytosolic proteins to autolysosomes where they are converted proteolytically into neo-antimicrobial peptides; and secondly, this p62-dependent process unleashes a cryptic capacity of cytosolic proteins to give rise to antibacterial peptides, upon autophagic conversion from their innocuous forms supporting routine cellular functions. This is an example of a resourceful utilization of cellular components, normally engaged in anabolic processes (e.g. protein synthesis on ribosomes) or destined for degradation (ubiquitinated complexes), into innate immunity effectors available in principle to every cell within its own cytoplasm in protection against microbial invasion.
The first principal finding of our studies is that p62, a protein previously implicated in autophagic removal of cytoplasmic protein aggregates in non-immunological contexts (Bjorkoy et al., 2005; Ichimura et al., 2008; Noda et al., 2008; Pankiv et al., 2007; Shvets et al., 2008), has an innate immunity function. It appears that p62 shows specialization for this role, as its paralog NBR1, which has similar properties (albeit with regulatory and distribution differences) in terms of interaction with LC3 and ubiquitinated protein aggregates (Bjorkoy et al., 2005; Kirkin et al., 2009; Pankiv et al., 2007), showed no recruitment to mycobacterial phagosomes upon induction of autophagy while p62 did. This differential activity is mirrored in observations reported by Kirkin et al. (Kirkin et al., 2009), indicating partial complementarity rather than full redundance of p62 and NBR1 distribution and functions. A further finding regarding p62 role is that acquisition of lysosomal hydrolases and acidification of compartments harboring intracellular mycobacetria in macrophages induced for authoagy are independent of p62. This separation of functions underscores the special role that p62 plays in conferring microbicidal properties on autophagic organelles.
The second principal finding of our work is that cytoplasmic proteins with no overt immunological function have an “afterlife” following their autophagic degradation. At least one aspect of this has already become well appreciated - the MHC-II restricted endogeonous antigen presentation with important functions in central tolerance (Nedjic et al., 2008) and potential role in adaptive immunity against viruses (Schmid and Munz, 2007). In these processes, autophagy captures and processes cytosolic proteins for loading onto MHC II molecules (Schmid and Munz, 2007). A previous study by Alonso et al, has suggested that ubiquitin fragments can be enriched in compartments harboring M. tuberculosis upon induction of autophagy (Alonso et al., 2007), but how this occurs remained unclear. The same puzzle applies to the earlier reports that a subset of ribosomal proteins L30, S19, and S30 can yield cryptic microbicidal activities against Escherichia coli, Bacillus megaterium, Candida albicans, Listeria monocytogenes, Salmonella typhimurium, Staphylococcus aureus, and Yersinia enterocolitica (Hiemstra et al., 1999; Howell et al., 2003; Tollin et al., 2003). The data in our studies now show that p62 ferries ribosomal proteins and ubiquitinated complexes as inactive precursors to autophagosomes where they give rise to neo-antimicrobial peptides upon digestion. Whether this occurs in a single step of delivery to mycobacteria-containing organelles, or is delivered first to a separate pool of autolysosomal organelles or vesicles that subsequently fuse with mycobacterial phagosomes remains to be determined. Regardless of the precise routing order, a subset of Fau, p62, and ubiquitin profiles were colocalizing with the autophagosomal marker LC3, suggesting involvement of bona fide autophagic organelles. We also noted that not all p62 cargo delivered to degradative compartments showed equal degradation rates. In these experiments, Fau was degraded slower than ubiquitin conjugates and p62, although it was a substrate for degradation as seen by appreciable bafilomycin-conferred protection from degradation. This is in keeping with the observations that Fau appears both punctate on and uniformly distributed within a substantial fraction of mycobacterial phagosomes, unlike p62 and ubiquitin which are detectable only in punctate profiles associating with mycobacterial compartments.
The generic cytoplasmic sources of bactericidal peptides such as ubiquitin (Kieffer et al., 2003; Wang et al., 2002) recently implicated in M. tuberculosis killing (Alonso et al., 2007) and ribosomal proteins (Hiemstra et al., 1999; Howell et al., 2003; Tollin et al., 2003) can now be added to the list of antimycobacterial peptides (Liu et al., 2007; Miyakawa et al., 1996; Sharma et al., 2000). Interestingly, although rpS30 is conserved in eukaryotic cells form yeast to man, the synthesis of rpS30 as a fusion protein is evolutionally restricted to metazoa and the equivalent ribosomal protein is produced free of the ubiquitin-like domain FUb in yeast, plants, and protozoa (Baker et al., 1996). This evolutionary restriction of the ubiquitin-like domain of Fau in metazoa coincides with the fact that p62 is only found in multicellular animals. The coevolution of the two proteins, therefore, strongly suggests their linked functions in support of their newly found role in innate immunity against M. tuberculosis uncovered in this work.
Because the cytoplasm is a rich source of a variety of proteins, the combinatorial wealth of generating bioactive peptides, such as those with antimicrobial action shown here, is possibly an important cellular resource that may not be limited to direct bactericidal action and immune control. Thus, in addition to autophagic killing factors identified here, we predict that there are further components and activities with immunological and non-immunological functions produced by autophagic organelles from cytosolic proteins and other cytoplasmic constituents that may be of interest to examine for cryptic biological functions.
Experimental Procedures
Cell and bacterial culture
RAW264.7 macrophages (ATCC, No. TIB-71) were maintained in DMEM (Gibco), 10% FBS (Hyclone), and 4 mM L-glutamine (Gibco) (Full media). Murine BMM cells were isolated from femurs of C57/BL6 or Atg5fl/fl and Atg5fl/fl Lyz-Cre mice or p62-defficient and p62-proficient mice as previously described (Komatsu et al., 2007; Zhao et al., 2008) and maintained in DMEM, 20% FBS, 4 mM L-glutamine, and m-CSF. M. tuberculosis H37Rv and BCG were grown in Middlebrook 7H9 broth or on 7H11 plates with 0.05% Tween 80, 0.2% glycerol, and albumin dextrose catalase (ADC) enrichment (+ oleic acid for H37Rv) (BD Biosciences) and homogenized to generate single cell suspension.
Inhibitors, fluorescent dyes, antibodies, and DNA constructs
All inhibitors used in this study are from Sigma unless otherwise indicated. Bafilomycin A1 (LC laboratories) was used at 100 nM. NH4Cl was used at 20 mM. Nocodazole and cytochalasin D were used at 10 μM. E64d and pepstatin A were used at 10 μg/mL. LysoTracker Red (Invitrogen) was used at 1:4000. For immunofluorescence assays, polyclonal antibodies against cathepsin D (R&D Systems) were used at 1:10, monoclonal antibody against ubiquitin conjugates (FK2, Biomol) was used at 1:100, polyclonal antibodies against p62 (Progen) were used at 1:500, polyclonal antibodies against LC3 (from Z. Elazar, The Weizmann Institute of Science, Israel) and T. Ueno (Juntendo University School of Medicine, Japan) were used at 1:50, and polyclonal antibodies against rpS19, rpL30, and Fau (Abcam) were used at 1:50. For immunoblotting, polyclonal antibodies against p62 (Progen) were used at 1:5000, polyclonal antibodies against Atg5 (Novus) were used at 1:1000, monoclonal antibody against Rab7 (Sigma) was used at 1:1000, monoclonal antibody against mono and polyubiquitin conjugates (FK2, Biomol) was used at 1:1000, monoclonal antibody against polyubiquitin conjugates (FK1, Biomol) was used at 1:1000, polyclonal antibodies agaist Fau were used at 1:100, and monoclonal antibody against actin (Abcam) was used at 1:2000, monoclonal antibody against p65 (Santa Cruz) was used at 1:500, monoclonal antibody against GFP (Abcam) was used at 1:1000. Plasmid constructs used in this study were previously described (Bjorkoy et al., 2005; Lamark et al., 2003; Pankiv et al., 2007).
Macrophage transfection
RAW264.7 cells were transfected based on manufacturer's protocol (Amaxa). Briefly, cells were passaged at 1:3 dilution into new flasks 24 h before transfection. Cells were harvested and 10 -15 × 106 cells were resuspended in electroporation buffer solution V. Plasmid DNAs (5 μg) or siRNAs (1.5 μg, Dharmacon) were mixed with 0.1 mL of cell suspension, transferred to a cuvet, and nucleoporated with Amaxa Nucleofector apparatus. Cells were then transferred into 12 mL complete media and cultured at 37 °C for either 24 h (transfected with plasmid DNAs) or 48 h (transfected with siRNAs) before being used in assays.
Infection, microbial survival and immunofluorescence microscopy methods
Determination of autophagic killing of intarcellular mycobacetria was carried out as described (Ponpuak et al., 2009). For confocal microscopy, RAW264.7 cells were plated onto coverslips in 12-well plates (3 × 105 cells per well) 24 h before infections. Cells were infected with 3 × 106 Alexa 488- or Alexa 568-labeled mycobacteria per well for 15 min at 37°C, washed three times with complete media, and chased in complete media for 1 h. Autophagy was induced by washing cells three times with 2 mL of either complete or starvation media or complete media supplemented with 500 u/mL murine IFN-γ (Sigma) and cells incubated for 4 h (starvation) or 24 h (IFN-γ) at 37°C. Cells were then fixed with 4% paraformaldehyde/PBS and permeabilized with 0.1% Triton X-100/PBS or 100 μg/mL digitonin/PBS, or fixed with methanol and permeabilized with acetone. Cells were blocked for 1 h in PBS containing 3% BSA and incubated with primary antibodies according to the manufacturer's recommendation. Cells were then washed three times with PBS and incubated in appropriate secondary antibodies (Invitrogen). The coverslips were mounted in mounting medium and analyzed by confocal microscopy using the Zeiss 510 Laser Scanning Microscope. For LysoTracker Red staining, cells were pre-stained in complete media containing LysoTracker Red for 2 h at 37°C before infection. Subsequent steps were carried out as described above but in the presence of LysoTracker Red until the cells were fixed. For quantification, % BCG-marker colocalization was fraction of total BCG phagosomes examined scored as positive when one or more puncta or homogeneously distributed marker were observed overlaping, on, or in contact with mycobacterial phagosome. At least 50 phagosomes per experimental condition in three independent experiments were quantified. Where Pearsons' colocalization coefficient (Rr=Σ[(S1i-S1avg)×(S2i-S2avg)]/[Σ(S1i-S1avg)2×Σ(S2i-S2avg)2]1/2; with Rr values ranges of ≥-1 Rr≤+1) was displayed, it was determined using SLIDEBOOK 5.0 (Intelligent Imaging Innovations) applying the SLIDEBOOK 5 default algorithm command “OR”. The OR command allows the pixels in an object to be binned and counting as positive green and red profiles that do not absolutely overlap but are tethered and only partially overlap (see Movies S1-S3, showing how these objects are tethered and associated dynamically with the phagosomes).
Purification of magnetic bead phagosomes and in vitro killing (KEPPA) assay
Magnetic bead phagosomes were prepared as follows: RAW264.7 macrophages were infected with mycobacteria at MOI=10 mixed with magnetic particles (Polysciences) (1/45, v/v) in complete media for 30 min at 37°C. After three washes with cold PBS, cells were incubated for 2 h at 37°C in either complete or starvation media in the absence or presence of 100 nM bafilomycin A1, washed with cold PBS, and lysed in homogenization buffer (HB) (250 mM sucrose, 0.5 mM EGTA, 3 mM imidazole pH 7.2 supplemented with protease inhibitors) by passing through 22-gauge needles connected to a two-syringe apparatus. Post-nuclear-supernatant (PNS) was generated and magnetic bead phagosomes (MBP) were isolated using a magnetic separator (Polysciences) and washed four times with HB. PNS (25 μg) and MBP (10 μg) were analyzed by immunoblotting. For p62 gene targeting experiments, RAW264.7 macrophages were transfected for 48 h with scramble or p62 siRNAs and phagosomes were prepared as described above. 5 μg of MBP associated proteins were analyzed by immunoblotting. For in vitro killing assay (KEPPA; in vitro killing assay with extracts from phagosomes purifed from cells induced for autophagy), phagosomes were prepared as described above from cells subjected to 4 h starvation medium. MBP contents were released using 20 mM sodium acetate buffer (pH 5.5) containing 1% Tween 20. This lysate was then centrifuged at 100,000 × g, 4°C, for 1 h. Protein concentration was then meas ured using BCA methods. Lysates (150 μg/mL) were then incubated with H37Rv (1 × 105 cfu/mL) for 2-4 d at 37°C before plating. For immunodepleti on, MBP lysates were prepared from cells at time points (2-4 h) when full size Fau was detectable and was not degraded (compare Fig. 6A lanes 3 and 4) as described above and then subjected to 1.5 h incubation with antibodies against Fau (Abcam) or Ubiquitin (FK2, Biomol) or respective IgG controls at 4°C. Protein G agarose (Santa Cruz) was added to the sample for 0.5 h. Lysates were centrifuged and the supernatants were then used in in vitro killing assays with H37Rv as described above.
Immunoprecipitations
RAW264.7 macrophages were infected with mycobacteria at MOI=10 in complete media for 1 h at 37°C. After three washes with cold PBS, cells were harvested and resuspended in immunoprecipitation buffer (IP buffer) containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% TritonX-100, and protease inhibitors. Cell lysate was generated by sonication at 4°C 3× with a small probe, 5 sec pulse and 1 min rest between each pulse. Protein concentration was then determined by standard BCA assays (Pierce) according to the manufracturer's protocol and 7 mg of total protein was incubated either with anti-p62 (Progen) or anti-GFP (Abcam) antibodies or their respective IgG control at 1:1000, 4 °C for 1 h. 30 μL of ProteinG conjugated magnetic bead was then added and incubated further at 4 °C for 1 h. Immune complexes were isolated using a magnetic separator (Polysciences) and washed three times with IP buffer. Immune complexes were solubilized in SDS loading buffer and were analyzed by immunoblotting. 100 μg of protein input from each condition was analyzed.
Statistics
All data were analyzed with Prism software (Graphpad) using two-tailed unpaired Student's t tests. All experiments were performed at least three times and the data pooled for presentation using +/- S.E.M.
Highlights.
Autophagy in its innate immunity role converts cytoplasmic proteins into antimicrobial peptides
Autophagic adapter p62 delivers ribosomal protein precursor Fau and ubiquitinated complexes to autolysosomes
Ribosomal proteins and ubiquitin digested by lysosomal hydrolases give rise to neo-antibacterial peptides
Autophagic conversion of innocuous cytoplasmic proteins into bactericidal peptides kills intracellular Mycobacterium tuberculosis
Supplementary Material
Acknowledgments
We thank M. Komatsu, Fukushima Medical University School of Medicine, Japan for p62-deficient and -sufficient mice and Z. Elazar, The Weizmann Institute of Science, Israel and Takashi Ueno, Juntendo University School of Medicine, Japan for LC3 antibodies. This work was supported by NIH grants A1069345, A145148, and A142999 to VD and AI057160 Project 5 to HV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA., Jr Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol. 1992;152:458–466. doi: 10.1002/jcp.1041520304. [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Hart PD. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J Exp Med. 1975;142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RT, Williamson NA, Wettenhall RE. The yeast homolog of mammalian ribosomal protein S30 is expressed from a duplicated gene without a ubiquitin-like protein fusion sequence. Evolutionary implications. J Biol Chem. 1996;271:13549–13555. doi: 10.1074/jbc.271.23.13549. [DOI] [PubMed] [Google Scholar]
- Biswas D, Qureshi OS, Lee WY, Croudace JE, Mura M, Lammas DA. ATP-induced autophagy is associated with rapid killing of intracellular mycobacteria within human monocytes/macrophages. BMC Immunol. 2008;9:35. doi: 10.1186/1471-2172-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado M, Singh S, De Haro S, Master S, Ponpuak M, Dinkins C, Ornatowski W, Vergne I, Deretic V. Autophagy and pattern recognition receptors in innate immunity. Immunol Rev. 2009;227:189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. Embo J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006;281:36303–36316. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Hiemstra PS, van den Barselaar MT, Roest M, Nibbering PH, van Furth R. Ubiquicidin, a novel murine microbicidal protein present in the cytosolic fraction of macrophages. J Leukoc Biol. 1999;66:423–428. doi: 10.1002/jlb.66.3.423. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M, Bertholet S, Gagnon E, Brunet S, Goyette G, Laplante A, Princiotta MF, Thibault P, Sacks D, Desjardins M. Phagosomes are competent organelles for antigen cross-presentation. Nature. 2003;425:402–406. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- Howell SJ, Wilk D, Yadav SP, Bevins CL. Antimicrobial polypeptides of the human colonic epithelium. Peptides. 2003;24:1763–1770. doi: 10.1016/j.peptides.2003.07.028. [DOI] [PubMed] [Google Scholar]
- Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural Basis for Sorting Mechanism of p62 in Selective Autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer AE, Goumon Y, Ruh O, Chasserot-Golaz S, Nullans G, Gasnier C, Aunis D, Metz-Boutigue MH. The N- and C-terminal fragments of ubiquitin are important for the antimicrobial activities. Faseb J. 2003;17:776–778. doi: 10.1096/fj.02-0699fje. [DOI] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008a;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Kraft M, Adler KB, Ingram JL, Crews AL, Atkinson TP, Cairns CB, Krause DC, Chu HW. Mycoplasma pneumoniae induces airway epithelial cell expression of MUC5AC in asthma. Eur Respir J. 2008b;31:43–46. doi: 10.1183/09031936.00103307. [DOI] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PT, Stenger S, Tang DH, Modlin RL. Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J Immunol. 2007;179:2060–2063. doi: 10.4049/jimmunol.179.4.2060. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Miyakawa Y, Ratnakar P, Rao AG, Costello ML, Mathieu-Costello O, Lehrer RI, Catanzaro A. In vitro activity of the antimicrobial peptides human and rabbit defensins and porcine leukocyte protegrin against Mycobacterium tuberculosis. Infect Immun. 1996;64:926–932. doi: 10.1128/iai.64.3.926-932.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- Olvera J, Wool IG. The carboxyl extension of a ubiquitin-like protein is rat ribosomal protein S30. J Biol Chem. 1993;268:17967–17974. [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Ponpuak M, Delgado MA, Elmaoued RA, Deretic V. Monitoring autophagy during Mycobacterium tuberculosis infection. Methods Enzymol. 2009;452:345–361. doi: 10.1016/S0076-6879(08)03621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Verma I, Khuller GK. Antibacterial activity of human neutrophil peptide-1 against Mycobacterium tuberculosis H37Rv: in vitro and ex vivo study. Eur Respir J. 2000;16:112–117. doi: 10.1034/j.1399-3003.2000.16a20.x. [DOI] [PubMed] [Google Scholar]
- Shvets E, Fass E, Scherz-Shouval R, Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Tollin M, Bergman P, Svenberg T, Jornvall H, Gudmundsson GH, Agerberth B. Antimicrobial peptides in the first line defence of human colon mucosa. Peptides. 2003;24:523–530. doi: 10.1016/s0196-9781(03)00114-1. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- Vergne I, Chua J, Singh SB, Deretic V. Cell biology of mycobacterium tuberculosis phagosome. Annu Rev Cell Dev Biol. 2004;20:367–394. doi: 10.1146/annurev.cellbio.20.010403.114015. [DOI] [PubMed] [Google Scholar]
- Wang Y, Griffiths WJ, Jornvall H, Agerberth B, Johansson J. Antibacterial peptides in stimulated human granulocytes: characterization of ubiquitinated histone H1A. Eur J Biochem. 2002;269:512–518. doi: 10.1046/j.0014-2956.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T, Noda T. Toward unraveling membrane biogenesis in mammalian autophagy. Curr Opin Cell Biol. 2008;20:401–407. doi: 10.1016/j.ceb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


