Abstract
The paraoxonase 1 (PON1) enzyme prevents low density lipoprotein oxidation and also detoxifies the oxon derivatives of certain neurotoxic organophosphate (OP) pesticides. PON1 activity in infants is low compared to adults, rendering them with lower metabolic and antioxidant capacities. We made a longitudinal comparison of the role of genetic variability on control of PON1 phenotypes in Mexican-American mothers and their children at the time of delivery (n=388 and 338, respectively) and again seven years later (n=280 and 281, respectively) using generalized estimating equations models. At age seven, children’s mean PON1 activities were still lower than those of mothers. This difference was larger in children with genotypes associated with low PON1 activities (PON1−108TT, PON1192QQ, and PON1−909CC). In mothers, PON1 activities were elevated at delivery and during pregnancy compared to seven years later when they were not pregnant (p<0.001). In non-pregnant mothers, PON1 polymorphisms and haplotypes accounted for almost 2-fold more variation of arylesterase (AREase) and chlorpyrifos-oxonase (CPOase) activity than in mothers at delivery. In both mothers and children, the five PON1 polymorphisms (192, 55, −108, −909, −162) explained a noticeably larger proportion of variance of paraoxonase activity (62–78%) than AREase activity (12.3–26.6%). Genetic control of PON1 enzymatic activity varies in children compared to adults and is also affected by pregnancy status. In addition to known PON1 polymorphisms, unidentified environmental, genetic, or epigenetic factors may also influence variability of PON1 expression and therefore susceptibility to OPs and oxidative stress.
Keywords: paraoxonase, enzymatic assay, genetic control, organophosphate, pesticides, pregnancy, children
Introduction
Paraoxonase 1 (PON1) is a high-density-lipoprotein (HDL)-associated enzyme, which plays a role in both organophosphate (OP) sensitivity and oxidative stress (Azarsiz et al., 2003). PON1 can metabolize the toxic oxon derivatives of several OP pesticides, which are known to be acutely neurotoxic (Costa et al., 2005a). There is growing evidence that PON1 may play a role in diseases related to oxidative stress including diabetes and heart disease (Li et al., 2003; Li et al., 2005; Bhattacharyya et al., 2008). In vitro and in vivo studies have demonstrated that PON1 has antioxidant properties, preventing LDL and HDL oxidation (Aviram and Rosenblat, 2004) and protecting against atherosclerosis (Tward et al., 2002; Rosenblat et al., 2006). Current studies suggest lipophilic lactones are the primary substrate for PON1 (Draganov et al., 2005; Khersonsky and Tawfik, 2005) and it is through this mechanism that PON1 is involved in lipid peroxidation. Although PON1 was named for its esterase activity towards OPs, the endogenous function of this enzyme is more likely its lipolactonase activity(Draganov et al., 2005). In humans, there is a wide variability of PON1 enzymatic activities among adults (Deakin and James, 2004). Individuals with low PON1 activity may be more susceptible to pesticide exposures and oxidative stress since their metabolic capacity and antioxidant defenses are lower compared to those with average or high PON1 activities. Thus, understanding the determinants of PON1 variability, including genetics and age, and how they confer susceptibility to disease or exposures may have broad public health significance.
Several common polymorphisms in the coding and promoter regions of the PON1 gene influence substrate-specific PON1 enzyme activities (Ferre et al., 2003; Costa et al., 2005b). The single nucleotide polymorphism (SNP) at codon 192 leads to an amino acid substitution from the active-site residue glutamine (Q) to arginine (R) and the catalytic efficiency of the PON1192 R alloform towards the oxon derivatives of OP pesticides parathion and chlorpyrifos is greater than that of the PON1192 Q alloform in in vitro substrate-specific assays. Animal experiments in transgenic mice expressing human PON1192 Q and R alloforms have demonstrated that indeed mice expressing the R alloform are more resistant to chlorpyrifos-oxon (CPO) exposure than mice expression the Q alloform (Cole 2005). For paraoxon however, the catalytic efficiency even in the faster R alloform is too slow to provide any protection from in vivo exposures (Li et al. 2000). Recent studies found that the PON1192 genotype explains a large portion of the variability of in vitro PON1 activity towards paraoxon (POase activity); it accounts for 59% of the variability among Caucasian and African-American adults (Bhattacharyya et al., 2008) and 48% of the variability in a Mexican-American population (Rainwater et al., 2009). Several promoter polymorphisms are also known to influence PON1 expression including PON1− 108, PON1−162, and PON1−909 (positions are relative to the translation start site). The PON1−108 SNP exerts the most noticeable effect on PON1 quantity, as measured by arylesterase activity, accounting for 22.4% of the variability. The PON1−108 CC genotype is associated with two-fold higher PON1 levels compared to the PON1−108 TT genotype (Brophy et al., 2001; Deakin et al., 2003). The association of the SNPs at positions −162 and −909 with AREase activity likely is due in part to their strong linkage disequilibrium (LD) with the PON1−108 SNP (Brophy et al., 2001; Holland et al., 2006). Similarly, the coding SNP PON1L55M is also associated with AREase activity, however most of this effect is attributable to LD with PON1−108 (Brophy et al., 2001). While several studies have described the important genetic contribution of these PON1 SNPs on phenotypic variation in multiple populations, few have characterized how the relative influence of genetic control may change through different stages of childhood and by pregnancy status. Furthermore, although genetic polymorphisms account for a large portion of PON1 variability, it is not sufficient in epidemiological studies to consider PON1 genotypes alone (Richter and Furlong, 1999). PON1 phenotypes range broadly even between individuals with the same PON1 genotypes because enzyme quantity also varies within these groups (Furlong et al., 2006; Holland et al., 2006). Therefore, studies which measure PON1 activities are more informative than studies that rely solely on PON1 genotype data.
Children are particularly vulnerable to environmental exposures because they practice behaviors that can lead to increased exposure and often have lower metabolic capacities than adults (Landrigan et al., 2004; Neri et al., 2006; Wigle et al., 2007). For example, children’s susceptibility to the toxic metabolites of OPs and oxidative stress may be heightened as several studies have demonstrated that PON1 activity is lower in newborns compared to adults (Chen et al., 2003; Holland et al., 2006). Early hypotheses suggested that PON1 developmental expression reaches mature levels at or near age two (Cole et al., 2003), however we recently followed a large cohort of Mexican-American children from birth to age seven and found that their PON1 activities continue to increase past age two until at least age seven (Huen et al., 2009a). This age-dependent increase of PON1 enzymatic activity was modified by genetic polymorphisms. For example, children with PON1192 R alleles and PON1−108 C alleles experienced a steeper rise in activity as they got older compared to children with PON1192 Q alleles and PON1−108 T alleles. These findings suggest that the window of susceptibility to both oxidative stress and OP exposure may be much longer than previously believed and children with certain genotypes may be particularly vulnerable.
Initially, we reported PON1 activity in a subset of 130 mother-child pairs from the Center for Health Assessment in Children and Mothers of Salinas Valley (CHAMACOS) cohort and determined the effects of genotypes and haplotypes on PON1 phenotype and status (Furlong et al., 2006; Holland et al., 2006). In the present study, we performed a longitudinal comparison of the role of genetic control on PON1 enzymatic activities in the entire CHAMACOS birth cohort of mothers and their children at the time of birth and also seven years later. We also compared PON1 activities between mothers and children at both time points and determined differences in PON1 activities in mothers during pregnancy, at delivery, and seven years later when they were not pregnant.
Materials and Methods
Study subjects
CHAMACOS is a longitudinal birth cohort study of the effects of pesticide and other environmental exposures on neurodevelopment, growth, and respiratory disease in children from primarily Mexican-American families (Eskenazi et al., 2003). The Salinas Valley, which is located in Monterey County, CA, is intensively farmed with approximately 200,000 kg of OPs applied annually (DPR, 2007). Six hundred and one pregnant women were enrolled in 1999–2000 and 531 were followed through the birth of a live infant. Mothers were primarily young (M=25.6 ±5.3 years), married, low-income, Mexican-born, and Spanish-speaking. Many were farm workers themselves (44%) and/or lived with farm workers at the time of enrollment (84%). Ethnicity of children and their mothers was based on mothers’ self-report. In this analysis, we included only women and children who were of Hispanic origin, the majority of which were Mexican (>90%), to avoid potential confounding by ethnicity. Study protocols were approved by the University of California, Berkeley Committee for Protection of Human Subjects. Written informed consent was obtained from all mothers and verbal assent was obtained from the children at seven years of age.
Blood collection and processing
Blood specimens were collected from mothers during pregnancy at the time of their glucose tolerance test (approximately 26 weeks gestation) and also at the hospital shortly before or after delivery. They were collected from children at the time of delivery (umbilical cord blood) and when the children were approximately seven years old (mean ± SD:7.05 ± 0.15 yr). Heparinized whole blood was collected in BD vacutainers® (Becton, Dickinson and Company, Franklin Lakes, NJ), centrifuged, divided into plasma, buffy coats and red blood cells, and stored at −80°C. Serum and blood clots were collected in vacutainers containing no anticoagulant. DNA was isolated from clots as described previously (Holland et al., 2006).
Determination of PON1 genotypes
DNA isolated from clots was available for genotyping for 431 mothers and 434 children. The coding polymorphisms, PON1192 and PON155, and the promoter polymorphism, PON1−162, were genotyped using the Taqman real-time PCR method. Primers for the nucleotide sequence flanking the SNP, and probes specific for the SNPs were custom-designed by Applied Biosystems, Inc. (Foster City, CA). The promoter SNPs, PON1−909 and PON1−108, were genotyped using a fluorogenic allele-specific genotyping assay (Amplifluor). The PON1−108 assay required a two-part nested PCR strategy, where the region surrounding the SNP was pre-amplified using non-allelic flanking primers. The amplicon was then diluted and used as the template for the Amplifluor assay. Quality assurance procedures for genotyping all five PON1 SNPs included assessment of randomly distributed blank samples in each plate and duplicates of randomly selected samples with independently isolated DNA from the same subjects. Repeated analysis (4% of samples) in several runs showed a high degree (>99%) of concordance. All discrepancies were resolved with additional genotyping.
Determination of PON1 enzymatic activities
PON1 enzyme activity was measured in heparinized plasma samples from 275 pregnant mothers (26 weeks gestation), 388 mothers and 338 newborns at delivery (312 complete mother-child pairs), and from 300 mothers and 281 children at age seven (246 complete mother-child pairs). In this paper, the 26 weeks gestation time point is referred to as pregnancy, measurements made in mothers at the time of delivery are referred to as delivery, and measurements made in umbilical cord blood are referred to as birth or newborns. At the seven-year collection, 21 mothers were pregnant and were excluded from the analysis. PON1 enzyme measurements were obtained from 481 mothers in total; of those, 228 had measurements at two time points, and 127 had measurements at all 3 time points. Enzyme measurements were available from 428 children of whom 191 had measurements at both time points. All samples which were previously assayed for PON1 activity in 130 mother-child pairs (Holland et al., 2006) were completely re-assayed simultaneously with all remaining samples in the CHAMACOS cohort. We found high correlations (r ~0.51–0.79, p<0.0005) with previous measures as reported in Huen et al. (2009b) although re-assayed samples had lower activities likely due to storage duration.
We measured PON1 enzyme activities against three different substrates (paraoxon (PO), phenyl acetate (ARE), and chlorpyrifos-oxon (CPO)) in plasma samples using spectrophotometric methods as described previously (Huen et al., 2009b). In this paper, we use these three measurements as markers of PON1 molecular phenotype. The arylesterase (AREase) assay serves as an indirect measure of PON1 enzyme quantity and AREase rates do not vary between PON1192 Q and R alloforms as they do for paraoxon hydrolysis (POase). PON1 quantity measured using ELISA and Western blot based methods are highly correlated with AREase activity (r > 0.85)(Kujiraoka et al., 2000; Connelly et al., 2008). In contrast, the paraoxonase (POase) and chlorpyrifos-oxonase (CPOase) substrate specific activities reflect both quantity and catalytic efficiency of the enzyme. All assays were performed in triplicate. Quality assurance included assessment of repeat samples (separate aliquots of the same sample run on different days) and internal controls (aliquots of the same sample run on all assay plates). We found a high degree of concordance between repeated samples (3% of samples were repeated). The average coefficient of variation (CV) for repeated samples was 6–9% and the correlation coefficients between repeated runs were between 0.91–0.98 for all three assays. The same internal controls samples were used on every plate for every assay and the inter-assay variability (average CV) for these samples ranged from 7 and 9%.
Storage duration of specimens can affect PON1 activity, particularly in specimens that had been stored for extended periods of time (Brackley et al., 1983; Stenzel et al., 2007; Huen et al., 2009b). For instance, PON1 activity declined on average by 17.1%, 39.4%, and 37.6% for AREase, CPOase, and POase, respectively after five years of storage (p<0.001 in all 3 assays). In our study, specimens collected at delivery were generally stored for longer periods of time than specimens collected during the seven year assessment. In order to compare PON1 activity differences between time points (delivery versus seven years later and pregnancy versus seven years later) while taking into account the potential effects of storage duration, we developed a storage duration correction factor using pilot data from a previous study (Huen et al., 2009b). Parallel aliquots of specimens from the same subjects (n=95) were assayed after two and seven years of storage at −80°C. We used these data to construct linear models that predicted the change in PON1 activity (%) as a function of storage duration (years) and storage duration*assay temperature. The models, which included an interaction between storage duration and assay temperature had the lowest Akaike Information Criterion (AIC) and were selected to be used to determine the correction factors. The intercept was constrained to equal zero. Applying the coefficients from these models as correction factors, PON1 activity (AREase, CPOase, POase) was then predicted for a storage time of zero years and these storage duration corrected PON1 activity measures were then used in our generalized estimating equation (GEE) models comparing (1) pregnant mothers versus seven years later and (2) mothers at delivery versus seven years later. The comparison between pregnancy at 26 weeks gestation and at delivery did not require correction for storage duration as the time span was relatively short (~10 weeks).
Statistical analysis
To determine whether there were systematic differences between participants and non-participants in this longitudinal study, we used logistic regression to assess whether missingness was associated with numerous variables related to sociodemographics, exposures, and genotype frequency including: mother’s country of birth, length of time in the United States, sex of the child, poverty levels, alcohol and tobacco use, and OP exposure (dialkylphosphate urinary metabolites). These factors were not associated with missingness with the exception of the number of years the mother spent in the United States. At the 7-year collection, the odds that mothers and their children remained in the study were 0.22 and 0.18-fold higher for mothers who had lived in the United States longer (p=0.01 and p=0.03 for mothers and children respectively).
Previous studies have reported associations of PON1 activity with factors such as nutrition, and alcohol and tobacco consumption (Deakin and James, 2004; Costa et al., 2005b). Therefore, we also explored whether variables such as OP exposure (urinary dialkylphosphate metabolites), alcohol, and tobacco consumption, and some sociodemographic factors were also associated PON1 activity in our CHAMACOS subjects using univariate regression models. The number of years that the mother lived in the United States, which was previously shown to be correlated with differences in nutrition in the CHAMACOS cohort (Harley et al., 2005), was negatively associated with AREase and POase activity in newborns (p=0.04 and p=0.02, respectively) and maternal smoking during pregnancy was associated with lower POase activity in seven-year olds (p=0.026). However, in this paper we focused on the effects of genotypes, age, and pregnancy on PON1 activity; inclusion of maternal smoking during pregnancy and length of time lived in the United States in statistical models did not significantly change the relationship between the main variables of interest and PON1 enzymatic activities (data not shown).
We used a chi-squared goodness of fit test to assess whether allele frequencies for each polymorphism deviate from Hardy-Weinberg equilibrium. Chi-squared tests were also used to determine whether allele and genotype frequencies in mothers and children were significantly different. The Cuzick’s non-parametric test for trend was used to assess trends in PON1 activity (AREase, CPOase, and POase) by PON1 genotypes. Since we performed numerous tests over many time points and three measurements (35 in children, 45 in mothers), we used Bonferonni correction in which a p-value < 0.001 was considered to be significant after adjusting for multiple testing. To infer haplotypes containing all five PON1 polymorphisms (PON1−909, PON1−162, PON1−108, PON155, PON1192) from genotype data, we used PHASE 2.1 (Stephens et al., 2001; Stephens and Scheet, 2005) software, which utilizes a Bayesian-based approach. To determine the proportion of variance (R2) explained, separate regression models were constructed for dependent variables AREase, CPOase, and POase. Independent variables included in the model were either the five PON1 genotypes or the imputed haplotypes. For each of the five polymorphisms, genotype variables were coded 0, 1, or 2 for the number of alleles present. Similarly, for the haplotype regression model, each haplotype with > 1% frequency was coded as a variable in the linear regression model, where the values 0, 1, or 2 denoted the presence of zero, one, or two copies of the haplotype for each subject. Haplotypes with < 1% frequency were pooled into one group for this analysis. Spearman correlation coefficients were calculated to assess correlations of PON1 activities between children at both time points and also between mothers and children at each time point for AREase, CPOase, and POase.
We performed GEE augmentation of linear models to compare AREase, CPOase, and POase activity (1) in mothers during pregnancy and delivery, (2) mothers during pregnancy and seven years later, (3) mothers at delivery and seven years later (4) in mothers at delivery versus children at birth, and (5) in both mothers and children at the seven-year collection. We used an exchangeable correlation structure to account for repeated measures on the same individuals or within the same family. Separate models for each of the three substrate-specific outcomes were generated. To compare PON1 activity in mothers at delivery versus pregnancy, we created an indicator variable where the reference group, mothers at pregnancy, was coded as 0 and mothers at delivery were coded as 1, which served as the independent variable. The other two comparisons, pregnancy versus seven years later and delivery versus seven years later, were treated similarly. To compare measures in mothers versus children at the same time point, we also used an indicator variable (child coded as 1, mother coded as 0) as the independent variable. Since we previously found that assay temperature can affect the PON1 activity measurement (Huen et al., 2009b), assay temperature was included as a covariate in all models. Finally, to determine whether the differences between mothers and children or between mothers at different time points are modified by PON1 genotypes, we also generated models for each of the five PON1 SNPs, that included the number of variant alleles (0, 1, or 2) and an interaction term for the indicator variable of the main comparison multiplied by the number of alleles. All analyses were performed in STATA 10.0 (College Station, TX). P-values less than 0.05 were considered significant and p-values less than 0.10 were reported as marginally significant.
Results
PON1 polymorphisms and haplotypes
Genotype distributions did not deviate significantly from Hardy-Weinberg equilibrium. Allelic frequencies in mothers and children for all five SNPs are presented in Table 1. For SNPs PON1−909, PON1 −108, and PON1192 allele frequencies were approximately equal in this population. The frequencies of the major allele for PON1−162 (G) and PON1 55 (L) were 81% and 82%, respectively. Allele and genotype frequencies did not differ significantly between mothers and children (χ2 test: p-value > 0.05 for all five SNPs) (Table 1, Table 2, and Table 3 and Supplemental Tables 1 and 2). Inferred haplotypes containing all five SNPs were reported in the following order: PON1−909 C/G, PON1−162 A/G, PON1 −108 C/T, PON1 55 L/M, PON1192 Q/R (Table 4). Haplotype analysis revealed 20 different combinations of alleles. Of these, 11 haplotype combinations accounted for more than 98% of the haplotypes present in the CHAMACOS population. The most common haplotype (GGCTR) was observed in over 25% of mothers and children.
Table 1.
PON1 allelic frequencies (%) in Mexican-American mothers and their children
| SNP | Allele | Mothers (n=431) |
Children (n=434) |
Total Populationa (n=865) |
|---|---|---|---|---|
| 192 | Q | 51.1 | 50.3 | 50.6 |
| R | 48.9 | 49.7 | 49.4 | |
| 55 | L | 81.3 | 82.2 | 82.0 |
| M | 18.7 | 17.8 | 17.0 | |
| −108 | C | 53.1 | 54.3 | 53.9 |
| T | 46.9 | 45.7 | 46.1 | |
| −162 | A | 19.1 | 19.2 | 19.0 |
| G | 80.9 | 80.8 | 81.0 | |
| −909 | C | 45.5 | 45.9 | 46.5 |
| G | 54.5 | 54.1 | 53.5 |
There were no significant differences observed between PON1 allele frequencies in mothers and children. All distributions did not deviate from Hardy-Weinberg equilibrium.
Table 2.
Summary of PON1 Enzyme Activity by PON1 Polymorphisms in Children at Birth (n=335) and Age 7 (n=268)
| PON1 | AREasec (U/mL) | CPOased (U/L) | POasee (U/L) | ||||
|---|---|---|---|---|---|---|---|
| Frequency (%) |
Birth Mean(Range) |
7 Years Mean(Range) |
Birth Mean(Range) |
7 Years Mean(Range) |
Birth Mean(Range) |
7 Years Mean(Range) |
|
| 192 | |||||||
| 24.4 | 31.7(3.9–70.9)a | 129.5(52.9–246.3) | 1811.3(175.3–4397.6)ab | 6885.8(2074.8–11181.3) | 110.0(13.1–519.7)ab | 316.4(119.5–954.7)ab | |
| QR | 51.6 | 32.2(3.8–98.0) | 119.9(14.5–184.6) | 1974.7(153.9–5255.2) | 6985.0(904.2–11195.7) | 253.7(7.5–682.1) | 881.3(133.6–1444.2) |
| RR | 24.0 | 36.7(9.7–73.6) | 117.6(61.1–190.4) | 2334.1(569.3–5159.5) | 7321.5(3858.3–11576.7) | 408.3(62.1–1017.5) | 1461.8(782.0–2504.5) |
| −108 | |||||||
| CC | 27.8 | 42.2(13.9–90.3)ab | 138.5(73.3–246.3)ab | 2607.6(823.4–4397.6)ab | 8286.1(3737.3–11576.7)ab | 348.8(38.3–742.8)ab | 1218.2(166.0–2504.5)ab |
| CT | 53.2 | 32.0(3.8–98.0) | 119.6(14.5–198.9) | 1939.1(153.9–5255.2) | 6863.8(904.2–11195.7) | 241.9(7.5–1017.5) | 844.5(119.5–2005.8) |
| TT | 19.0 | 23.2(3.9–54.7) | 101.7(33.6–159.9) | 1389.1(175.3–3409.0) | 5734.6(1896.8–7967.2) | 157.9(13.1–485.6) | 568.2(155.5–1468.2) |
| All | 33.1(3.84–98.0) | 121.5(14.5–246.3) | 2020.2(4153.9–5255.2) | 7035.8(904.2–11576.7) | 257.1(7.5–1017.5) | 894.2(119.5–2504.5) | |
In children, the PON1192 genotype was significantly associated with POase activity at both time points. Children with the RR genotype had the highest mean POase activities compared to QR and QQ children. The promoter polymorphism, PON1−108, was associated with all three substrate-specific activities at birth and age 7. On average, children with the TT genotype had lower activities than those with the CT and CC genotypes.
ptrend for genotype <0.05, determined by Cuzick’s non-parametric test
ptrend for genotype significant after Bonferonni correction for multiple testing (n=35 tests, p≤0.001)
AREase = arylesterase activity
CPOase = chlorpyrifos-oxonase activity
POase = paraoxonase activity
Table 3.
Summary of PON1 Enzyme Activity by PON1 Polymorphisms in Mothers During Pregnancy (270), Delivery (n=382), and 7 Years Later (n=263)
| PON1 | AREasec (U/mL) | CPOased (U/L) | POasee (U/L) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Frequency (%) |
Pregnancy Mean(Range) |
Delivery Mean(Range) |
7 Years Later Mean(Range) |
Pregnancy Mean(Range) |
Delivery Mean(Range) |
7 Years Later Mean(Range) |
Pregnancy Mean(Range) |
Delivery Mean(Range) |
7 Years Later Mean(Range) |
|
| 192 | ||||||||||
| 27.5 | 141.1(44.9–263.2)ab | 143.8(29.9–346.6)a | 129.3(56.6–247.0) | 6635.5(2197.1–10623.3) | 7198.0(1766.1–13948.6)a | 7025.6(2946.1–12302.8) | 340.6(93.0–1167.3)ab | 360.7(75.2–2694.9)ab | 336.1(144.5–919.9)ab | |
| QR | 45.5 | 127.7(31.3–272.4) | 134.8(40.1–255.9) | 130.2(60.4–348.5) | 6832.9(1856.0–14810.3) | 7576.0(2376.9–13807.1) | 7546.8(2937.9–12759.4) | 938.7(145.6–1953.7) | 989.2(174.8–2856.4) | 989.4(243.9–2299.2) |
| RR | 27.0 | 118.7(57.2–263.3) | 129.3(27.1–285.9) | 120.4(54.5–181.6) | 6666.1(3811.5–11735.5) | 8067.8(1927.7–14675.8) | 7420.9(3368.5–11429.9) | 1503.2(699.0–3757.1) | 1648.1(294.3–3537.7) | 1555.7(666.1–2390.8) |
| −108 | ||||||||||
| CC | 29.6 | 145.8(44.9–272.4)ab | 151.6(40.1–289.1)ab | 143.8(54.5–348.5)ab | 7661.3(2197.1–14810.3)ab | 8628.8(2376.9–14675.8)ab | 8497.1(3442.2–12759.4) | 1228.8(93.0–3757.1)ab | 1347.7(148.2–3537.7)ab | 1324.5(144.5–2390.8)ab |
| CT | 48.0 | 127.4(31.3–263.2) | 134.5(27.1–265.1) | 127.0(63.6–182.9) | 6596.2(1856.0–10623.3) | 7617.6(1766.1–12931.6) | 7411.5(2937.9–12302.8) | 861.6(145.6–3256.7) | 928.1(75.2–2856.4) | 915.6(204.2–2299.2) |
| TT | 22.4 | 114.9(52.6–269.3) | 118.3(34.5–346.6) | 106.0(56.6–206.4) | 6032.9(3191.7–11735.5) | 6310.0(2067.7–13948.6) | 5891.1(2946.1–8981.8) | 730.2(138.2–2387.3) | 680.5(134.3–2564.2) | 639.3(146.5–1491.3) |
| All | 129.1(31.3–272.4) | 136.3(27.1–346.6) | 127.3(54.5–348.5) | 6725.4(1856–14810.3) | 7613.2(1766.1–14675.8) | 7376.0(2937.9–12759.4) | 912.1(93.0–3757.1) | 987.2(75.2–3537.7) | 965.6(144.5–2390.8) | |
In mothers, the PON1192 SNP was associated with POase activity at all 3 time points with the highest mean activities in RR mothers and the lowest mean activities in QQ mothers. The coding SNP PON1−108, was associated with all three substrate-specific activities at all 3 time point with the highest mean acitivities in CC mothers and the lowest mean activities in TT mothers.
ptrend for genotype <0.05, determined by Cuzick’s non-parametric test
ptrend for genotype significant after Bonferonni correction for multiple testing (n=45 tests, p≤0.001)
AREase = arylesterase activity
CPOase = chlorpyrifos-oxonase activity
POase = paraoxonase activity
Table 4.
PON1 Haplotype frequencies (%) in Mexican-American Mothers (n=431) and Children (n=434)
| Haplotypea,b | Mothers | Children |
|---|---|---|
| GGCLR | 25.1 | 26.4 |
| CGTMQ | 15.2 | 14.9 |
| CGTLR | 15.2 | 14.7 |
| CGTLQ | 14.3 | 14.3 |
| GACLQ | 11.2 | 10.8 |
| GGCLQ | 6.6 | 6.7 |
| GACLR | 5.7 | 6.4 |
| CGCLR | 1.7 | 1.3 |
| GGCMQ | 0.9 | 1.3 |
| GATLQ | 1.4 | 1.1 |
| GGCMR | 1.0 | 0.6 |
Polymorphisms in five loci of PON1 gene are listed in the haplotypes using the following order: −909(C/G), −162(A/G), −108(C/T), 55(L/M),192(Q/R)
The eleven haplotypes listed represent 98.4% and 98.5% of the haplotypes present in CHAMACOS mothers and children, respectively. The remaining nine haplotypes (not shown) had frequencies lower than 1%.
PON1 haplotypes were inferred using PHASE 2.1 software.
Effects of genotypes and haplotypes on PON1 activity
The means and ranges of AREase, CPOase, and POase activity by genotype are shown in Table 2 (children) and Table 3 (mothers) for PON1192 and PON1−108. Supplemental Tables 1 and 2 describe similar data for PON1−909, PON1−162, and PON155. All three promoter SNPs (−108, −162, and −909) were associated with AREase and CPOase activity for mothers and children at all time points (ptrend <0.05). The highest mean AREase and CPOase activities were observed in individuals with the PON1−108 CC, PON1−162 AA, and PON155 LL genotypes. PON−108 and PON1−909 were also significantly associated with POase activity at all time points (ptrend <0.05).
The coding SNP, PON1192, was associated with POase activity at all time points and CPOase activity at birth in both mothers and children, with QQ individuals showing the lowest activity levels and RR the highest. Patterns for AREase activity were less consistent; at birth, QQ newborns had the lowest mean PON1 activity levels (p<0.005) but QQ mothers had the highest activity levels (p=0.015 and p<0.0005 for mothers at delivery and during pregnancy, respectively). No significant trends in AREase activity were observed for PON1192 at 7 years.
All three substrate-specific measures in both mothers and children were highest in those with the PON155 LL genotype and lowest in those with the PON155 MM genotype (Supplemental Tables 1 and 2). For AREase activity, this trend did not reach statistical significance (p<0.05) in mothers at the time of delivery and in seven-year old children, but was significant at all other time points.
The relative genetic contribution of the five PON1 SNPs (genotypes and haplotypes) on the variation of PON1 phenotypes in mothers and children are shown in Table 5. In some instances, haplotypes can be better predictors of phenotype than genotypes because they incorporate multiple polymorphisms on a single chromosome (Stephens et al., 2001). However, in our study, PON1 haplotypes did not provide improved assessment of the effects of PON1 genetic variability on enzyme activities in comparison to PON1 genotypes. The five PON1 genotypes explained 26% of the variance of AREase in newborns and seven-year olds. The same genotypes explained less variability in mothers, particularly at delivery (12%). In fact, for all three PON1 measures (POase, AREase, and CPOase), the contribution of these genetic determinants was much lower – almost half for AREase and CPOase – at delivery versus seven years later. At birth, the five PON1 SNPs explained noticeably less variation of POase both in mothers and children (63% and 49%, respectively) in comparison to the seven-year measures, where POase variability was primarily due to these genetic polymorphisms (almost 80% variation explained), especially PON1192.
Table 5.
Proportion of variance explained for PON1 enzyme activity by genotype and haplotype regression models
| Children | Mothers | |||||||
|---|---|---|---|---|---|---|---|---|
| AREase | CPOase | POase | AREase | CPOase | POase | |||
| n | R2 | R2 | R2 | n | R2 | R2 | R2 | |
| Birth | 330 | 395 | ||||||
| 5 PON1 Genotypes | 0.266 | 0.323 | 0.490 | 0.123 | 0.157 | 0.626 | ||
| Haplotypes | 0.295 | 0.355 | 0.510 | 0.126 | 0.170 | 0.619 | ||
| 7 yr | 266 | 253 | ||||||
| 5 PON1 Genotypes | 0.260 | 0.297 | 0.782 | 0.201 | 0.303 | 0.778 | ||
| Haplotypes | 0.272 | 0.319 | 0.775 | 0.222 | 0.355 | 0.780 | ||
The proportion of variance (R2) was determined by running separate regression models for each substrate-specific assay. AREase refers to the arylesterase assay which utilizes a phenyl acetate substrate. CPOase is the chlorpyrifos-oxonase assay, which uses the substrate chlorpyrifos-oxon. POase is the paraoxonase assay using the paraoxon substrate. Models either included a variable for each of 5 PON1 polymorphisms: −909(C/G), −162(A/G), −108(C/T), 55(L/M),192(Q/R) or ordinal variables for haplotypes containing all 5 polymorphisms. These models were performed both in samples collected in mothers and children at time of the child’s birth and in samples collected from both mothers and children when children were 7 years old.
Correlations
Correlations of enzyme activity between mothers and their children are presented in Supplemental Table 3. In general, children’s PON1 activities were weakly correlated with those of their mothers with the exception of newborns and mothers at the time of delivery. As expected since their PON1 activities were closer to those of adults, seven year old children’s activities (r = 0.16–0.51) were more highly correlated to their mothers than when they were newborns (r=0.04–0.39). Correlations of enzyme activity between time points (in the same individuals) are shown in Supplemental Table 4. Correlations of enzyme activities in mothers at birth and seven years later were statistically significant and much higher for POase (r=0.86) than for AREase (r=0.38) and CPOase and (r=0.44). For AREase and CPOase activities in children at the two time points, correlations were higher than those in mothers (r= 0.47 and 0.58, respectively) but were slightly lower for POase (r=0.80).
PON1 activity in children
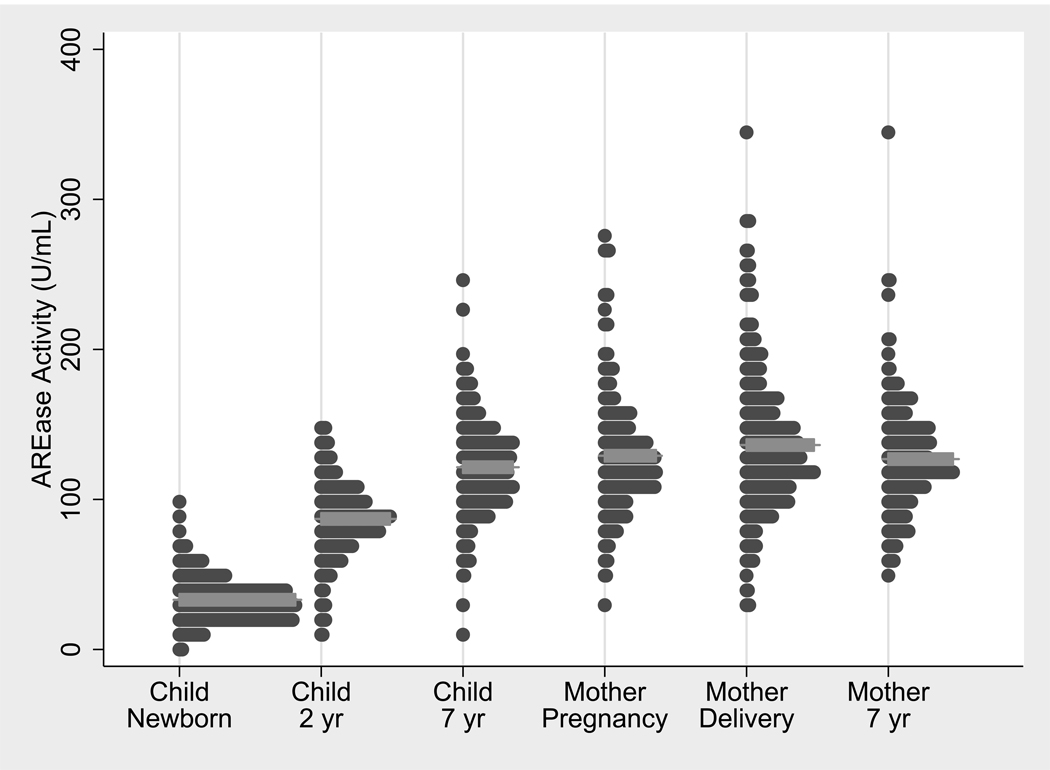
AREase, CPOase, and POase activities in children at birth and age seven are described in Table 2. Overall, mean AREase activity increased 3.7-fold and mean CPOase and POase activities both increased 3.5-fold between birth and year seven. We previously reported an age-dependent increase in children’s PON1 activities from birth through ages one, two, five and seven (Huen et al., 2009a) and here we focus on the two time points which coincide with maternal sampling (birth and age seven). A graph presenting these data and also PON1 activities in mothers is shown in Figure 1. The graph clearly demonstrates a wide inter-individual variability between study participants at all time points. Among newborns, there was a 38-fold difference between the highest and lowest AREase levels. When we compared the lowest AREase levels in newborns to the highest levels in seven year olds, there was a 64-fold difference. Since cord blood measurements reflect both fetal and placental expression, we explored whether the mother’s genotype or phenotype influenced PON1 activity in newborns. Among newborns sharing the same PON1−108 genotype, neither maternal AREase activity nor maternal PON1−108 genotype affected the AREase activity of their newborns. For instance, among PON1−108 CC newborns, their mothers’ PON1−108 genotype was not significantly associated with the newborns’ AREase activity (p>0.05). Similar results for POase activity were found such that among newborns sharing the same PON1192 genotype, neither PON1192 genotype nor POase activity of their mothers affected the newborns’ POase activities.
Figure 1.
The distribution of AREase in the CHAMACOS children at birth, age 2, and age seven and in their mothers during pregnancy (26 weeks gestation), at the time of delivery, and when their children were seven years old. Mean AREase activities as indicated by the grey horizontal bars, were 33.1, 89.2, 121.5 U/mL in children at birth, 2, and 7 years of age, respectively. In mothers, mean AREase activities were 129.1, 136.3, and 127.3 U/mL at pregnancy, delivery and seven years later, respectively. PON1 enzymatic activity was very low in newborns and continued to increase over time. At age seven, activities were still significantly lower than that of mothers at the same time point (p= 0.05). PON1 activities were elevated in mothers at the time of delivery (p<0.0005). This figure contains some of the children’s PON1 activity data (children at age two) modified from Huen et al. (2009a).
PON1 activity in mothers
AREase, CPOase, and POase activities in mothers during pregnancy, at delivery, and seven years later are described in Table 3. In comparison to pregnancy, PON1 activity at delivery was slightly higher for all three measures and these differences were statistically significant in the GEE models (p≤0.001) (Supplemental Table 5). Before adjusting for storage duration, PON1 activity during pregnancy (26 weeks gestation) appeared slightly lower than seven years later. However after adjustment for storage duration, we found higher AREase, CPOase, and POase activities during pregnancy (p≤0.005, GEE models). Even before applying the storage duration correction factors, means for AREase, CPOase, and POase were elevated in mothers at the time of delivery in comparison to seven years later (7.4, 3.8, and 2.2%, higher respectively). After applying the storage duration correction factors, the difference between delivery and seven years later became even more apparent. Mean AREase and CPOase and median POase activities were 29.0, 49.1, and 20.8% higher at the delivery time point compared to seven years later and this difference was statistically significant in the GEE models (p<0.001 for all three measures). Results were similar when we adjusted for each of the five PON1 SNPs.
PON1 activity in children compared to mothers
Previously we reported that newborns have significantly lower PON1 activities than their mothers and that as children age, their activities increase over time (Holland et al., 2006; Huen et al., 2009a). In this expanded dataset, mean AREase and CPOase and median POase activities at birth were about four-fold lower in newborns than mothers (p<0.005 in all three GEE models) (Supplemental Table 5). At age seven, children’s PON1 activities were much closer to those in mothers (mean AREase: 4.3%; mean CPOase: 4.1%; median POase: 1.8% lower in children than in mothers). Although children’s enzymatic activities at this age were more comparable to mothers, the GEE models indicated the difference in PON1 activity between mothers and children remained statistically significant for AREase (p=0.05) and CPOase (p=0.02) and marginally significant for POase (p=0.08).
Since we previously found that the age-dependent increase of PON1 activity is modified by PON1 genotype in young children (Huen et al., 2009a), we sought to determine whether the differences in activity between mothers and children are also modified by PON1 genotype. When we adjusted for PON1 genotypes and included an interaction term (PON1 genotype × indicator variable for child) in the GEE models, we found that children’s PON1 activities were only significantly lower than those of mothers for specific child genotypes. For instance, among children with the PON1−108 CC genotype (the genotype associated with increased AREase activity compared to the TT genotype), children’s mean AREase activity was not significantly different than that of mothers (includes mothers of all genotypes). However, among children with the PON1−108 CT genotype, children’s AREase activities were significantly lower than those of all mothers and this difference was even larger among children with the TT genotype (p-value for interaction term=0.07). Similarly, for CPOase and POase, activities in children with the CC genotype were not significantly different than mothers, however the significant interaction terms (p=0.009 and 0.03, respectively) indicated that activities for CT and TT children were significantly lower than those of mothers (β for interaction term= −458.6 and −100.3 for CPOase and POase, respectively). When we limited the comparison to mothers and children with the same genotype, children’s AREase and CPOase activities were lower for all three genotypes however this difference was only statistically significant for the PON1−108 TT genotype on AREase activity (p=0.01). For POase activity, children’s activities were lower than mothers for CC and CT genotypes but this did not reach statistical significance.
We also identified a significant interaction for the promoter SNP at position −909 when comparing CPOase and POase activity between mothers and children (β for interaction term = − 473.8 and −106.2; p=0.008 and 0.03, respectively). Mean activity for PON1−909 CG and CC children was significantly lower than mean activity in all mothers but this difference was not significant in PON1−909 GG children (p=0.38 and 0.58 for CPOase and POase, respectively). For the coding SNP at position 192, mean POase activity in QQ children was 289.4 U/L lower than that in mothers (p<0.0005), yet the significant interaction term (β=216.3; p<0.0005) indicated that this difference was much smaller in QR children and the mean POase activity in RR children was actually higher than the overall mean in mothers. However, when we limited comparisons to mothers and children with the same PON1192 genotype, children’s POase activities were lower in all three genotype groups however, this difference was only statistically significant for the RR genotype (β indicator variable for children =−130.8, p=0.05).
DISCUSSION
In this study, three substrate-specific measures of PON1 quantity and activity were measured in over 400 mothers and children at the time of delivery, when the children were seven years old, and also in mothers during pregnancy. Although children’s PON1 activities increased about 3.5-fold between birth and age seven, they still remained 1.8–4.3% lower than levels measured in their non-pregnant mothers. Mothers had significantly higher levels of PON activity during pregnancy than seven years later when they were not pregnant. PON1 activity was particularly high at the time of delivery compared both to 26 weeks gestation (means were 1.9– 8.5% higher at delivery) and seven years later (means were 20.8–49.1% higher at delivery). Using regression analysis, we found that 5 PON1 genotypes explained a similar proportion of variance compared to inferred haplotypes. The genetic contribution of the 5 PON1 SNPs on phenotypic variation differed in children between birth and age seven and also between pregnant and non-pregnant mothers suggesting additional factors may affect PON1 expression depending on age and pregnancy status.
In an extended analysis of mothers and newborns, we confirmed our previous data in 130 mother-child pairs that PON1 activities are four-fold lower at birth in comparison to adults (Holland et al., 2006). Therefore, newborns and young infants may have increased vulnerabilities to OP exposure and oxidative stress. At age seven, children’s PON1 activities on average remained significantly lower than in their adult, non-pregnant mothers suggesting that even at school-age, children may remain more susceptible to pesticide exposure and oxidative stress than adults. When adjusting for PON1 genotypes, we found that PON1 activities in children with PON1192 RR and PON1−108 CC genotypes (genotypes related to increased catalytic efficiency and enzyme quantity, respectively) were much more similar to adults by age seven, and children with other genotypes (those associated with lower quantity and efficiency of the enzyme) experienced a pronounced lag in developmental expression with significantly lower PON1 activities than mothers at age seven. It is not yet clear if PON1 activities in children with susceptible genotypes will eventually reach average mothers’ levels as they continue to age or if their levels will remain lower throughout their lives.
Similar patterns of differential developmental expression have been observed in other metabolic and antioxidant enzymes. For instance, pharmacokinetic studies and in vitro data demonstrate that many cytochrome P-450 enzymes (CYPs) at birth are quite low (about 30% that of adults) and then increase quickly until they reach adult levels within six months to a year (Ginsberg et al., 2004). In rat livers, several antioxidant enzymes involved in detoxification of xenobiotics (GPx, GST, and GR) were very low at birth and did not begin to reach adult levels until puberty (Elbarbry 2009). Previously, we reported that whole blood cholinesterase levels in CHAMACOS newborns were 25% lower than in their mothers at delivery, and the difference was even larger (33%) when comparing with maternal levels at 26 weeks of pregnancy (Eskenazi et al., 2004). Data on the ontogeny of different enzymes related to xenobiotic metabolism, included that reported here in PON1, can be used by toxicokinetic models and risk assessment to account for potential susceptibilities during development.
In addition to OPs, variability of PON1 activity may also affect susceptibility to other environmental exposures. As mentioned earlier, it is likely that the endogenous function of PON1 is related to its antioxidant activities involved in lipid peroxidation. Exposure to a wide variety of toxicants including arsenic, polybrominated diphenyl ethers (PBDEs), and bisphenol A (BPA) has been shown to induce oxidative stress in humans and/or human cell lines (Barchowsky et al., 1999; Franco et al., 2009; Gao et al., 2009; Hong et al., 2009; Tagliaferri et al., 2009; Yang et al., 2009). Through this mechanism, variability of PON1 activity and levels may affect vulnerabilities to multiple environmental exposures. For example, two recent studies of arsenic exposure suggest PON1 may modify the effect of arsenic on cardiovascular outcomes (Li et al., 2009; Liao et al., 2009). Therefore young children with low PON1 activity may experience increased susceptibility not only to OP exposures, but to a multitude of environmental hazards due to reduced antioxidant capacity.
In addition to age-related changes observed in children, we also found PON1 activity varied by pregnancy status in mothers; all three substrate-specific activities were elevated during pregnancy, particularly at the time of delivery, in comparison to the seven year time point. Two other studies in pregnant and non-pregnant women corroborate our findings that PON1 activity (AREase and POase) is elevated during pregnancy (Roy et al., 1994; Carpintero et al., 1996). Others studies have reported increased levels of lipid hydroperoxides (markers of oxidative stress), triglycerides, and lipoproteins in late pregnancy accompanied by higher total antioxidant capacity reflected by greater levels of uric acid, and vitamins C and E (Toescu et al., 2002). Increased levels of PON1 during late pregnancy and at delivery suggest yet another mechanism by which the body may maintain oxidant balance during late pregnancy when free radical generation may be accelerated.
We previously determined that 5 PON1 SNPs account for 23.1% and 8.1% of the variability of AREase activity in newborns and their mothers, respectively (Holland et al., 2006). In this study, we included all Mexican-American mothers and children from the CHAMACOS cohort (for whom specimens were available) in the analysis and determined the genetic contribution of phenotypic variation both at birth and age seven; results at birth were very similar to those we previously reported in the subset of 130 mother-child pairs. At birth, fifty percent of POase activity was explained by these five genotypes and at age seven, the genetic contribution was even higher (78%). Furthermore, this was primarily due to the PON1192 polymorphism (partial correlation coefficient r = 0.58 and 0.82 for newborns and seven-year olds, respectively) which has previously been shown to account for a large proportion of variance in other recent studies (48% and 58.5%, respectively) of POase activity (Bhattacharyya et al., 2008; Rainwater et al., 2009). This finding suggests that the strong genetic control exerted by the PON1192 SNP on POase activity is weaker at birth, and other factors may play a significant role at this time.
At the time of delivery, we found that a much lower proportion of PON1 variability (particularly for AREase and CPOase activity) was explained by genotype in mothers. Thus, child birth may represent a time in which environmental and physiological factors or other unidentified genetic factors have a stronger influence on PON1 phenotype. In both mothers and children, PON1192 genotype accounted for the majority of POase activity (up to 80%) yet the five PON1 polymorphisms, including three promoter SNPs only accounted for about 12–26% of AREase activity (PON1 quantity). Therefore, other genetic polymorphisms in the PON1 gene and elsewhere in metabolic networks or other types of unidentified factors like epigenetic modifications and environmental exposures may serve as additional determinants of PON1 expression. Other factors that have been shown to contribute to variability PON1 enzyme activity in adults include environmental and behavioral factors such as alcohol consumption, cigarette smoke, and diet (Li et al., 2003). Physiological conditions such as diabetes, Alzheimer’s disease, and cardiovascular disease have also been linked to reduced PON1 activity (Deakin and James, 2004), but it is not yet clear whether depressed PON1 activity leads to these conditions or conversely if these conditions trigger lower activity.
In summary, we examined PON1 genotypes and phenotypes in Mexican-American mothers and their children living in an agricultural community when the children were born and also when the children reached seven years of age. For many children, especially those with genotypes associated with decreased PON1 activities, their PON1 levels remain lower than those of their mothers even at age seven. This finding suggests that young children through school-age, particularly those with the most vulnerable genotypes, PON1−108 TT, PON1192 QQ, and PON1−909 CC, do not have mature levels of the protective PON1 enzyme and may continue to experience increased susceptibility to OP exposure and oxidative stress. Furthermore, this increased vulnerability due to low PON1 activity may also be relevant to additional environmental toxicants like BPA and PBDEs via the oxidative stress pathway.
Supplementary Material
Acknowledgements
We are grateful to the laboratory and clinical staff and participants of the CHAMACOS study for their contributions. We would also like to thank Dr. Kenneth Beckman for his assistance with PON1 genotyping and Dr. Clement Furlong and Rebecca Richter for advice on PON1 enzymatic assays. This publication was made possible by grant numbers R826886 and R82670901 from the U.S. Environmental Protection Agency (EPA) and R01ESO12503-03 and PO1 ES009605 from the National Institute of Environmental Health Science (NIEHS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS and the EPA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aviram M, Rosenblat M. Paraoxonases 1, 2, and 3, oxidative stress, and macrophage foam cell formation during atherosclerosis development. Free Radic Biol Med. 2004;37:1304–1316. doi: 10.1016/j.freeradbiomed.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Azarsiz E, Kayikcioglu M, Payzin S, Yildirim Sozmen E. PON1 activities and oxidative markers of LDL in patients with angiographically proven coronary artery disease. Int J Cardiol. 2003;91:43–51. doi: 10.1016/s0167-5273(02)00595-8. [DOI] [PubMed] [Google Scholar]
- Barchowsky A, Klei LR, Dudek EJ, Swartz HM, James PE. Stimulation of reactive oxygen, but not reactive nitrogen species, in vascular endothelial cells exposed to low levels of arsenite. Free Radic Biol Med. 1999;27:1405–1412. doi: 10.1016/s0891-5849(99)00186-0. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya T, Nicholls SJ, Topol EJ, Zhang R, Yang X, Schmitt D, Fu X, Shao M, Brennan DM, Ellis SG, Brennan ML, Allayee H, Lusis AJ, Hazen SL. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. Jama. 2008;299:1265–1276. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackley M, Carro-Ciampi G, Stewart DJ, Lowden JA, Ray AK, Kalow W. Stability of the paraoxonase phenotyping ratio in collections of human sera with differing storage times. Res Commun Chem Pathol Pharmacol. 1983;41:65–78. [PubMed] [Google Scholar]
- Brophy VH, Jampsa RL, Clendenning JB, McKinstry LA, Jarvik GP, Furlong CE. Effects of 5' regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. Am J Hum Genet. 2001;68:1428–1436. doi: 10.1086/320600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpintero A, Sanchez-Martin MM, Cabezas-Delamare MJ, Cabezas JA. Variation in serum arylesterase, beta-glucuronidase, cathepsin L and plasminogen activators during pregnancy. Clin Chim Acta. 1996;255:153–164. doi: 10.1016/0009-8981(96)06403-0. [DOI] [PubMed] [Google Scholar]
- Chen J, Kumar M, Chan W, Berkowitz G, Wetmur JG. Increased influence of genetic variation on PON1 activity in neonates. Environ Health Perspect. 2003;111:1403–1409. doi: 10.1289/ehp.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TB, Jampsa RL, Walter BJ, Arndt TL, Richter RJ, Shih DM, Tward A, Lusis AJ, Jack RM, Costa LG, Furlong CE. Expression of human paraoxonase (PON1) during development. Pharmacogenetics. 2003;13:357–364. doi: 10.1097/00008571-200306000-00007. [DOI] [PubMed] [Google Scholar]
- Connelly PW, Maguire GF, Picardo CM, Teiber JF, Draganov D. Development of an immunoblot assay with infrared fluorescence to quantify paraoxonase 1 in serum and plasma. J Lipid Res. 2008;49:245–250. doi: 10.1194/jlr.D700022-JLR200. [DOI] [PubMed] [Google Scholar]
- Costa LG, Cole TB, Vitalone A, Furlong CE. Measurement of paraoxonase (PON1) status as a potential biomarker of susceptibility to organophosphate toxicity. Clin Chim Acta. 2005a;352:37–47. doi: 10.1016/j.cccn.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Costa LG, Vitalone A, Cole TB, Furlong CE. Modulation of paraoxonase (PON1) activity. Biochem Pharmacol. 2005b;69:541–550. doi: 10.1016/j.bcp.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Deakin S, Leviev I, Brulhart-Meynet MC, James RW. Paraoxonase-1 promoter haplotypes and serum paraoxonase: a predominant role for polymorphic position - 107, implicating the Sp1 transcription factor. Biochem J. 2003;372:643–649. doi: 10.1042/BJ20021670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin SP, James RW. Genetic and environmental factors modulating serum concentrations and activities of the antioxidant enzyme paraoxonase-1. Clin Sci (Lond) 2004;107:435–447. doi: 10.1042/CS20040187. [DOI] [PubMed] [Google Scholar]
- DPR. Pesticide Use Report, Annual 2007. Sacramento, CA: Department of Pesticide Regulation, California Environmental Protection Agency; 2007. [Google Scholar]
- Draganov DI, Teiber JF, Speelman A, Osawa Y, Sunahara R, La Du BN. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J Lipid Res. 2005;46:1239–1247. doi: 10.1194/jlr.M400511-JLR200. [DOI] [PubMed] [Google Scholar]
- Eskenazi B, Bradman A, Gladstone E, Jaramillo S, Birch K, Holland N. CHAMACOS, a longitudinal birth cohort study: lessons from the fields. J Childrens Healt. 2003;1:3–27. [Google Scholar]
- Eskenazi B, Harley K, Bradman A, Weltzien E, Jewell NP, Barr DB, Furlong CE, Holland NT. Association of in utero organophosphate pesticide exposure and fetal growth and length of gestation in an agricultural population. Environ Health Perspect. 2004;112:1116–1124. doi: 10.1289/ehp.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre N, Camps J, Fernandez-Ballart J, Arija V, Murphy MM, Ceruelo S, Biarnes E, Vilella E, Tous M, Joven J. Regulation of serum paraoxonase activity by genetic, nutritional, and lifestyle factors in the general population. Clin Chem. 2003;49:1491–1497. doi: 10.1373/49.9.1491. [DOI] [PubMed] [Google Scholar]
- Franco R, Sanchez-Olea R, Reyes-Reyes EM, Panayiotidis MI. Environmental toxicity, oxidative stress and apoptosis: menage a trois. Mutat Res. 2009;674:3–22. doi: 10.1016/j.mrgentox.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Furlong CE, Holland N, Richter RJ, Bradman A, Ho A, Eskenazi B. PON1 status of farmworker mothers and children as a predictor of organophosphate sensitivity. Pharmacogenet Genomics. 2006;16:183–190. doi: 10.1097/01.fpc.0000189796.21770.d3. [DOI] [PubMed] [Google Scholar]
- Gao P, He P, Wang A, Xia T, Xu Z, Niu Q, Guo L, Chen X. [Effects of PCB153 on oxidative stress and 8-OHdG content induced by PBDE-47 in human neuroblastoma cells in vitro] Wei Sheng Yan Jiu. 2009;38:513–515. [PubMed] [Google Scholar]
- Ginsberg G, Hattis D, Sonawane B. Incorporating pharmacokinetic differences between children and adults in assessing children's risks to environmental toxicants. Toxicol Appl Pharmacol. 2004;198:164–183. doi: 10.1016/j.taap.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Harley K, Eskenazi B, Block G. The association of time in the US and diet during pregnancy in low-income women of Mexican descent. Paediatr Perinat Epidemiol. 2005;19:125–134. doi: 10.1111/j.1365-3016.2005.00640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland N, Furlong C, Bastaki M, Richter R, Bradman A, Huen K, Beckman K, Eskenazi B. Paraoxonase polymorphisms, haplotypes, and enzyme activity in Latino mothers and newborns. Environ Health Perspect. 2006;114:985–991. doi: 10.1289/ehp.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YC, Park EY, Park MS, Ko JA, Oh SY, Kim H, Lee KH, Leem JH, Ha EH. Community level exposure to chemicals and oxidative stress in adult population. Toxicol Lett. 2009;184:139–144. doi: 10.1016/j.toxlet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Huen K, Harley K, Brooks J, Hubbard A, Bradman A, Eskenazi B, Holland N. Developmental Changes in PON1 Enzyme Activity in Young Children and Effects of PON1 Polymorphisms. Environmental Health Perspectives. 2009a;117:1632–1638. doi: 10.1289/ehp.0900870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen K, Richter R, Furlong C, Eskenazi B, Holland N. Validation of PON1 enzyme activity assays for longitudinal studies. Clin Chim Acta. 2009b;402:67–74. doi: 10.1016/j.cca.2008.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS. Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry. 2005;44:6371–6382. doi: 10.1021/bi047440d. [DOI] [PubMed] [Google Scholar]
- Kujiraoka T, Oka T, Ishihara M, Egashira T, Fujioka T, Saito E, Saito S, Miller NE, Hattori H. A sandwich enzyme-linked immunosorbent assay for human serum paraoxonase concentration. J Lipid Res. 2000;41:1358–1363. [PubMed] [Google Scholar]
- Landrigan PJ, Kimmel CA, Correa A, Eskenazi B. Children's health and the environment: public health issues and challenges for risk assessment. Environ Health Perspect. 2004;112:257–265. doi: 10.1289/ehp.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Liu DP, Liang CC. Paraoxonase gene polymorphisms, oxidative stress, and diseases. J Mol Med. 2003;81:766–779. doi: 10.1007/s00109-003-0481-4. [DOI] [PubMed] [Google Scholar]
- Li J, Wang X, Huo Y, Niu T, Chen C, Zhu G, Huang Y, Chen D, Xu X. PON1 polymorphism, diabetes mellitus, obesity, and risk of myocardial infarction: Modifying effect of diabetes mellitus and obesity on the association between PON1 polymorphism and myocardial infarction. Genet Med. 2005;7:58–63. doi: 10.1097/01.gim.0000151152.78092.ca. [DOI] [PubMed] [Google Scholar]
- Li WF, Sun CW, Cheng TJ, Chang KH, Chen CJ, Wang SL. Risk of carotid atherosclerosis is associated with low serum paraoxonase (PON1) activity among arsenic exposed residents in Southwestern Taiwan. Toxicol Appl Pharmacol. 2009;236:246–253. doi: 10.1016/j.taap.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Liao YT, Li WF, Chen CJ, Prineas RJ, Chen WJ, Zhang ZM, Sun CW, Wang SL. Synergistic effect of polymorphisms of paraoxonase gene cluster and arsenic exposure on electrocardiogram abnormality. Toxicol Appl Pharmacol. 2009;239:178–183. doi: 10.1016/j.taap.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Neri M, Bonassi S, Knudsen LE, Sram RJ, Holland N, Ugolini D, Merlo DF. Children's exposure to environmental pollutants and biomarkers of genetic damage. I. Overview and critical issues. Mutat Res. 2006;612:1–13. doi: 10.1016/j.mrrev.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Rainwater DL, Rutherford S, Dyer TD, Rainwater ED, Cole SA, Vandeberg JL, Almasy L, Blangero J, Maccluer JW, Mahaney MC. Determinants of variation in human serum paraoxonase activity. Heredity. 2009;102:147–154. doi: 10.1038/hdy.2008.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter RJ, Furlong CE. Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics. 1999;9:745–753. [PubMed] [Google Scholar]
- Rosenblat M, Gaidukov L, Khersonsky O, Vaya J, Oren R, Tawfik DS, Aviram M. The catalytic histidine dyad of high density lipoprotein-associated serum paraoxonase-1 (PON1) is essential for PON1-mediated inhibition of low density lipoprotein oxidation and stimulation of macrophage cholesterol efflux. J Biol Chem. 2006;281:7657–7665. doi: 10.1074/jbc.M512595200. [DOI] [PubMed] [Google Scholar]
- Roy AC, Loke DF, Saha N, Viegas OA, Tay JS, Ratnam SS. Interrelationships of serum paraoxonase, serum lipids and apolipoproteins in normal pregnancy. A longitudinal study. Gynecol Obstet Invest. 1994;38:10–13. doi: 10.1159/000292435. [DOI] [PubMed] [Google Scholar]
- Stenzel J, Worek F, Eyer P. Preparation and characterization of dialkylphosphoryl-obidoxime conjugates, potent anticholinesterase derivatives that are quickly hydrolyzed by human paraoxonase (PON1192Q) Biochem Pharmacol. 2007;74:1390–1400. doi: 10.1016/j.bcp.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, Duan J, Carr JL, Lee MS, Koshy B, Kumar AM, Zhang G, Newell WR, Windemuth A, Xu C, Kalbfleisch TS, Shaner SL, Arnold K, Schulz V, Drysdale CM, Nandabalan K, Judson RS, Ruano G, Vovis GF. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliaferri S, Caglieri A, Goldoni M, Pinelli S, Alinovi R, Poli D, Pellacani C, Giordano G, Mutti A, Costa LG. Low concentrations of the brominated flame retardants BDE-47 and BDE-99 induce synergistic oxidative stress-mediated neurotoxicity in human neuroblastoma cells. Toxicol in vitro. 2009 doi: 10.1016/j.tiv.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Toescu V, Nuttall SL, Martin U, Kendall MJ, Dunne F. Oxidative stress and normal pregnancy. Clin Endocrinol (Oxf) 2002;57:609–613. doi: 10.1046/j.1365-2265.2002.01638.x. [DOI] [PubMed] [Google Scholar]
- Tward A, Xia YR, Wang XP, Shi YS, Park C, Castellani LW, Lusis AJ, Shih DM. Decreased atherosclerotic lesion formation in human serum paraoxonase transgenic mice. Circulation. 2002;106:484–490. doi: 10.1161/01.cir.0000023623.87083.4f. [DOI] [PubMed] [Google Scholar]
- Wigle DT, Arbuckle TE, Walker M, Wade MG, Liu S, Krewski D. Environmental hazards: evidence for effects on child health. J Toxicol Environ Health B Crit Rev. 2007;10:3–39. doi: 10.1080/10937400601034563. [DOI] [PubMed] [Google Scholar]
- Yang YJ, Hong YC, Oh SY, Park MS, Kim H, Leem JH, Ha EH. Bisphenol A exposure is associated with oxidative stress and inflammation in postmenopausal women. Environ Res. 2009;109:797–801. doi: 10.1016/j.envres.2009.04.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.