Abstract
The mammalian sirtuin SIRT6 is a site-specific histone deacetylase that regulates chromatin structure. SIRT6 is implicated in fundamental biological processes in aging, including maintaining telomere integrity, fine-tuning aging-associated gene expression programs, preventing genomic instability, and maintaining metabolic homeostasis. Despite these important functions, the basic molecular determinants of SIRT6 enzymatic function—including the mechanistic and regulatory roles of specific domains of SIRT6—are not well understood. Sirtuin proteins consist of a conserved central ‘sirtuin domain’—thought to comprise an enzymatic core—flanked by variable N- and C-terminal extensions. Here, we report the identification of novel functions for the N- and C-terminal domains of the human SIRT6 protein. We show that the C-terminal extension (CTE) of SIRT6 contributes to proper nuclear localization but is dispensable for enzymatic activity. In contrast, the N-terminal extension (NTE) of SIRT6 is critical for chromatin association and intrinsic catalytic activity. Surprisingly, mutation of a conserved catalytic histidine residue in the core sirtuin domain not only abrogates SIRT6 enzymatic activity but also leads to impaired chromatin association in cells. Together, our observations define important biochemical and cellular roles of specific SIRT6 domains, and provide mechanistic insight into the potential role of these domains as targets for physiologic and pharmacologic modulation.
Keywords: sirtuin, SIRT6, chromatin, histone deacetylation, aging
1. Introduction
Saccharomyces cerevisiae Sir2 is the founding member of an evolutionarily conserved family of sirtuin proteins present in organisms ranging from bacteria to humans. As an NAD-dependent histone deacetylase, Sir2 deacetylates lysines in the amino terminal ‘tails’ of histones H3 and H4, as well as on the globular core of histone H3 (Imai et al., 2000; Landry et al., 2000; Smith et al., 2000; Xu et al., 2007). In this context, Sir2 modulates the assembly and spreading of heterochromatin at telomeres, silent mating type loci, and ribosomal DNA repeats. In turn, these activities of Sir2 impact on genomic stability, gene silencing, and yeast lifespan (Denu, 2003).
In mammalian genomes, there are seven SIR2 family members, dubbed SIRT1-SIRT7 (Frye, 1999; Frye, 2000). SIRT6 has recently emerged as a critical regulator of transcription, genome stability, telomere integrity, DNA repair, and metabolic homeostasis. The first clues to the in vivo function of SIRT6 came from analysis of SIRT6 deficiency in mice. SIRT6 knockout mouse cells exhibit DNA damage hypersensitivity and genomic instability, and SIRT6-deficient mice develop a striking degenerative and metabolic phenotype with symptoms suggestive of premature aging (Mostoslavsky et al., 2006). SIRT6 was also found to fractionate with chromatin biochemically, suggesting that it might have a chromatin-regulatory function (Mostoslavsky et al., 2006).
However, direct evidence for a physiologic enzymatic activity of SIRT6 at chromatin was lacking. Initial studies did not detect NAD+-dependent deacetylase activity for SIRT6 on several histone substrates. Instead, SIRT6 was observed to promote ADP-ribosylation, an alternative NAD+-dependent reaction observed for some sirtuins (Liszt et al., 2005; Mostoslavsky et al., 2006), but the physiological importance of this activity remains to be determined.
Recently, we discovered that SIRT6 is indeed an NAD+-dependent histone deacetylase, but because it is highly site-specific, this activity had been difficult to observe. We showed that SIRT6 has specificity for deacetylating lysine 9 of histone H3 (H3K9Ac)†, and we identified functions for this activity in maintaining telomere integrity (Michishita et al., 2008) and in negatively regulating aging-associated NF-κB-dependent gene expression programs (Kawahara et al., 2009). We also showed that SIRT6 is required for efficient DNA double-strand break repair in the context of chromatin, though the specific role of histone deacetylation by SIRT6 in this context remains to be clarified (McCord et al., 2009). More recently, we and others (Michishita et al., 2009; Yang et al., 2009) have shown that SIRT6 has a second substrate, lysine 56 of histone H3 (H3K56), and our study (Michishita et al., 2009) demonstrated that SIRT6 is critical for maintaining dynamic changes in H3K56 acetylation levels at telomeres over the cell cycle.
Despite these important cellular and physiologic functions, the basic molecular mechanisms of SIRT6 enzymatic activity—including the mechanistic and regulatory roles of specific SIRT6 sequences—remain poorly understood. Sirtuin proteins share a phylogenetically conserved central ‘sirtuin domain,’ generally thought to comprise an enzymatic core. Eukaryotic genomes typically encode multiple Sir2 family members, and these proteins contain variable N-and C-terminal extensions flanking the ∼270-amino acid sirtuin core domain (Frye, 1999; Frye, 2000). S. cerevisiae harbors several Sir2 family members, and studies of yeast Sir2 proteins have shown that, in addition to residues within the central core domain, regions outside the catalytic core play important roles in acetyl-lysine and NAD+ binding (Zhao et al., 2003), sub-nuclear distribution (Cockell et al., 2000), and interaction with cellular binding partners (Cuperus et al., 2000). In mammals, nuclear localization signals, nuclear export signals, and mitochondrial localization signals have been identified on the N-terminal extensions of SIRT1 (Tanno et al., 2007), SIRT2 (North and Verdin, 2007a), and SIRT3 and SIRT4 (Haigis et al., 2006; Onyango et al., 2002; Scher et al., 2007; Schwer et al., 2002), respectively, but evidence for other functions of the N- and C-terminal extensions of the mammalian SIRTs is relatively limited, and in the case of SIRT6, completely lacking.
Here, we have conducted a functional analysis of the N- and C-terminal extensions of SIRT6, as well as the role of a conserved catalytic residue in the central sirtuin domain of SIRT6. We show that the C-terminal extension (CTE) of SIRT6 is critical for proper sub-cellular targeting and that the N-terminal extension (NTE) contributes to efficient chromatin association and intrinsic catalytic activity. Surprisingly, a catalytically inactive SIRT6 point mutant is defective in chromatin localization, providing the first example in which the catalytic activity of a mammalian sirtuin is required for the protein’s stable association with chromatin.
2. Materials and methods
2.1. Constructs
To generate vectors for transient transfection, a FLAG tag was inserted downstream of EGFP in pEGFP-N2 (Clontech, to generate pEGFP-N2-FLAG) or was inserted in place of EGFP in pEGFP-N2 (to generate pN2-CT-FLAG). Full-length SIRT6 and SIRT6 deletion mutants were generated by PCR and cloned into these modified pEGFP-N2 vectors as EcoRI-BamHI cassettes, generating C-terminally tagged fusion proteins. For stable transduction and bacterial expression, PCR-generated SIRT6 deletion mutants were cloned as AscI-PacI cassettes into pWZL-3FLAG-hygro and pGEX-6P3 (GE Healthcare), respectively, each containing an altered multiple cloning site (harboring AscI and PacI sites) and generating N-terminally tagged fusion proteins. SIRT6-Δcore contains an Ala-Ala-Ala linker (and a NotI site) between the N- and C-terminal portions of the protein.
2.2. Antibodies
The antibodies used were as follows: anti-FLAG M2 (Sigma-Aldrich), anti-β-tubulin (Millipore), anti-H3K9Ac (Sigma-Aldrich, Abcam), anti-H3K56 (Epitomics), anti-H3 (Abcam). SIRT6 antibodies have been previously described (Michishita et al., 2005).
2.3. Cell culture, transfection, and retroviral transduction
293T and U2OS cells were maintained in DMEM with 10% FBS. 293T cells were transiently transfected using the TransIT-293 transfection reagent (Mirus), according to the manufacturer’s instructions. Cells were harvested 40–48 hours post-transfection. For retroviral transduction, 293T cells were co-transfected with pVPack-VSV-G, pVPack-GP (Stratagene), and pWZL-3FLAG-SIRT6 constructs, and viral supernatants were filtered onto target cells after 40 and 64 hours in the presence of 8 µg/ml polybrene. 48 hours after the second infection, cells were selected with hygromycin (250 µg/ml) for 4 days.
2.4. SIRT6-EGFP visualization
293T cells were transiently transfected with pEGFP-N2-FLAG-SIRT6 plasmids in 12-well plates. Twenty-four hours after transfection, live cells were incubated with 1 µg/ml Hoechst 33342 for 2 hours and imaged with a Nikon Eclipse TE300 inverted fluorescence microscope (40×) using Openlab imaging software.
2.5. GST-SIRT6 purification
BL21 E. coli transformed with pGEX-6P3-SIRT6 deletion mutants were induced with 0.1 mM IPTG at 25°C overnight, lysed in E. coli lysis buffer (50 mM Tris pH 7.5, 150–400 mM NaCl, 0.05–0.5% NP40 + 50 µg/ml lysozyme), sonicated with a Branson digital sonicator, bound to Glutathione Sepharose 4B (GE Healthcare), washed twice each with lysis buffer and elution buffer, and eluted in 50–100 mM Tris pH 8 + 15 mM glutathione.
2.6. Extract preparation
For analysis of H3K9Ac levels in SIRT6-overexpressing 293T cells, cells were lysed in their wells in a 1:1 mixture of 2× Laemmli buffer:RIPA (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP40, 1% sodium deoxycholate), and these whole-cell lysates were boiled for 10 minutes. To generate extracts with solubilized histones for histone interaction co-immunoprecipitations (coIPs), cells were lysed in their wells in coIP buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP40, 10% glycerol, 1 mM DTT, 1 mM PMSF, complete protease inhibitor (Roche)), sonicated with a Branson digital sonicator, and clarified at 16,000 g. Extracts were incubated with anti-FLAG M2 agarose (Sigma-Aldrich) for 1 hour at 4°C, washed 3–5 times with binding buffer, and boiled in 2× Laemmli buffer. Biochemical fractionation was performed as previously described (Mendez and Stillman, 2000), and 5% (for FLAG-tagged proteins) or 10% (for endogenous SIRT6) of each fraction was analyzed by western blot.
2.7. In vitro histone deacetylation assays
In vitro deacetylation reactions were performed with 1 µg GST-SIRT6 deletion mutants and 200 ng calf thymus histone H3 (Roche) in NAD+ deacetylation buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10 mM NAD+, 3.3 mM DTT) for 2 hours at 37°C or overnight at 25°C (with indistinguishable results). Histone deacetylation was assessed by western blot with H3K9Ac- or H3K56Ac-specific antibodies.
2.8. Nucleosome-binding gel-shift assays
Mononucleosomes were purified from HeLa cells as described (Shi et al., 2006). 1 µg (Figure 4E and Figure S1) or 400 ng (Figure 4F) mononucleosomes were incubated with 2 µg GST or GST-SIRT6 deletion mutants for 30 minutes at 30°C in mononucleosome binding buffer (20 mM HEPES pH 7.9, 80 mM KCl, 0.1 mM ZnCl2, 0.1% EDTA, 10% glycerol, 0.1% NP40, 0.5 mM DTT) in a total volume of 20 µl. Reactions were fractionated on a 5% non-denaturing TBE gel (BioRad), and mononucleosomal DNA was visualized under UV light after staining with 1 µg/ml ethidium bromide.
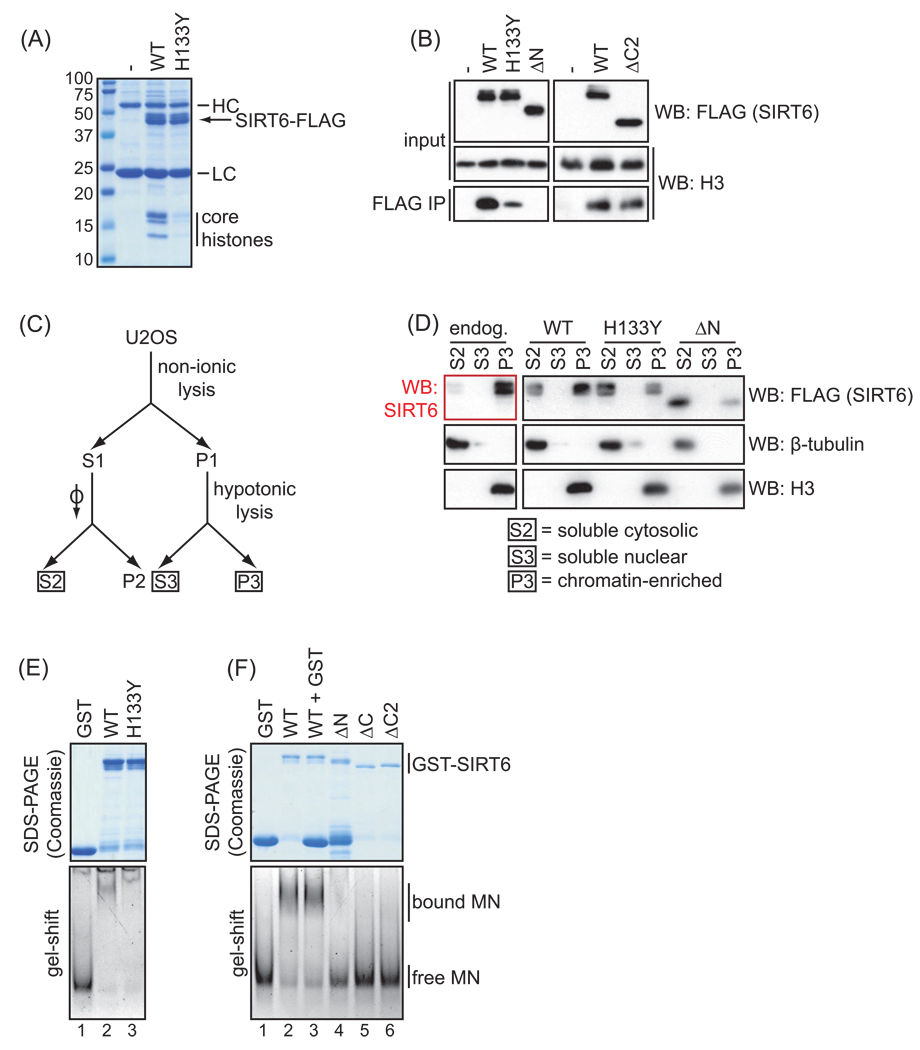
Figure 4.
Impaired chromatin association of SIRT6 deletion and point mutants. A. Coomassie stain showing co-immunoprecipitation of core histones with wild-type (WT) but not catalytically inactive (H133Y) SIRT6 protein, from 293T cells transiently transfected with FLAG-tagged SIRT6 proteins or empty vector control (-). HC and LC indicate antibody heavy and light chains present in the IPs. B. Western analysis showing relative co-immunoprecipitation of histone H3 with the indicated mutant SIRT6 proteins compared to WT SIRT6, as in (A). Results are representative of at least 3 independent experiments. C. Schematic of cellular fractionation protocol, based on (Mendez and Stillman, 2000). D. Western analysis showing the relative chromatin association of endogenous SIRT6, or of FLAG-tagged wild-type or mutant SIRT6 proteins stably expressed in U2OS cells, using the biochemical fractionation outlined in (C). β-tubulin and histone H3 westerns provide controls for the cytosolic and chromatin-enriched fractions, respectively. E. In vitro gel-shift assay showing similar nucleosome binding efficiency of the wild-type and catalytically inactive H133Y mutant SIRT6. Top: Coomassie stain of the purified GST-SIRT6 proteins used for the gel-shift assay. Bottom: migration of free or SIRT6-bound mononucleosomes (MN) in nucleosome binding reactions, fractionated on a non-denaturing gel and visualized by ethidium bromide staining of nucleosomal DNA. Incubation with GST alone is shown as a negative control. F. In vitro gel-shift assay as in (E) showing impaired nucleosome binding of N- and C- terminal SIRT6 deletion mutants compared to WT SIRT6. In lane 3, excess GST protein was added to control for the GST degradation product in the ΔN protein sample (lane 4). The slight difference in the nucleosome shift may be due to imperfections in the gel, as this was not observed in other experiments (see Figure S1).
3. Results
3.1. The C terminus of SIRT6 is essential for proper nuclear localization
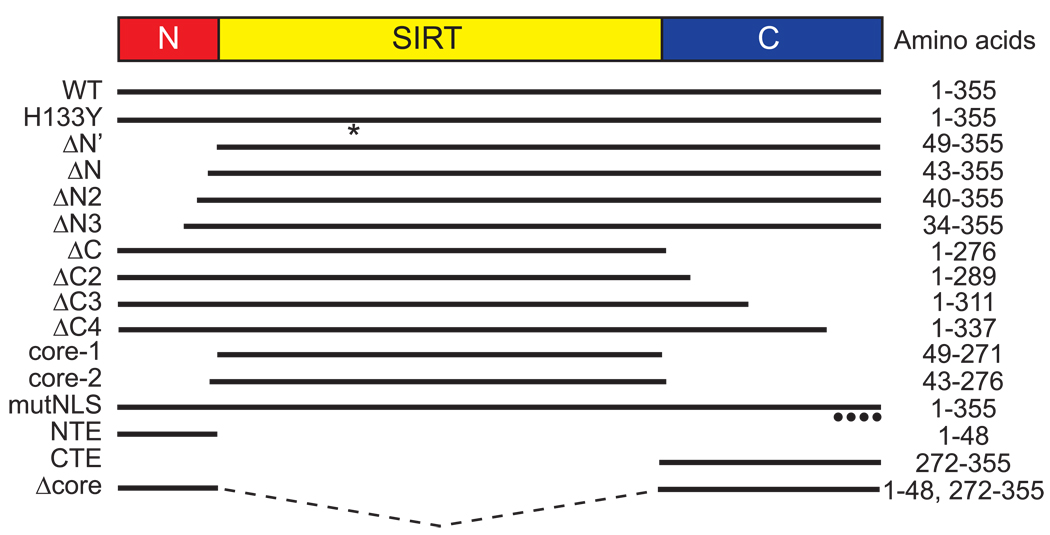
To study potential functions of the N- and C-terminal extensions of human SIRT6, we generated a panel of mutants with progressive deletions from the N and C termini (Figure 1). We also generated mutants consisting of the N-terminal extension alone (NTE), the C-terminal extension alone (CTE), or full-length SIRT6 lacking the catalytic core (Δcore). For comparison, we included the mutant SIRT6-H133Y protein, which harbors a mutation in a highly conserved histidine within the core sirtuin domain of SIRT6; the corresponding mutation abrogates catalytic activity in all sirtuins tested thus far.
Figure 1.
Schematic of SIRT6 deletion and point mutants used in this study. WT, wild-type SIRT6; H133Y, full-length SIRT6 with a catalytic mutation (histidine to tyrosine) at residue 133 (*); mutNLS, K/R-to-A mutations at four residues (••••) in the C terminus of SIRT6. ΔN, ΔC, and ‘core’ mutants are missing the N-terminal extension (NTE), C-terminal extension (CTE), or both the NTE and CTE, respectively. See text for further details.
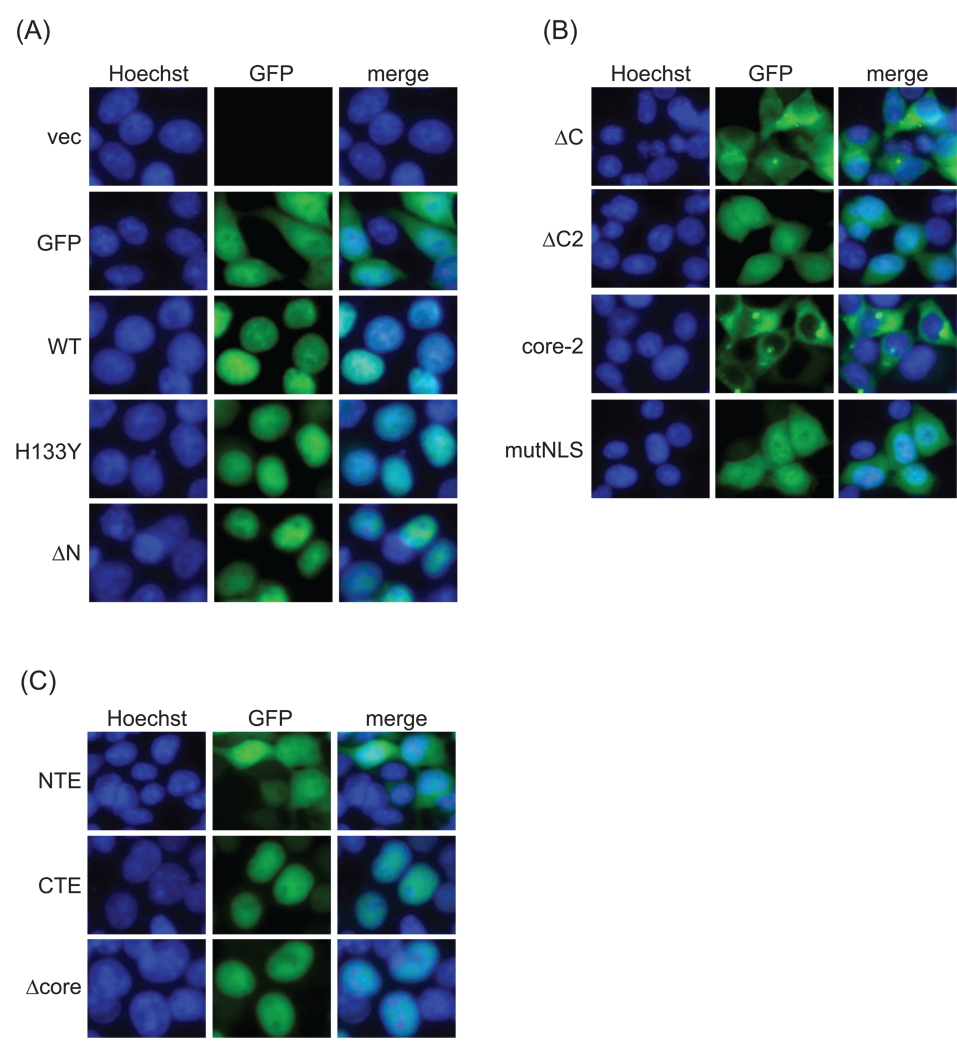
As expected, transiently transfected GFP-tagged wild-type SIRT6 was exclusively nuclear in live 293T cells, as was the catalytically inactive H133Y mutant, whereas the unfused GFP control was detected throughout the nucleus and cytoplasm (Figure 2A). Deleting the NTE of SIRT6 (ΔN) did not significantly disrupt the nuclear localization pattern seen for wild-type SIRT6. Conversely, progressive deletion of the CTE of SIRT6 (ΔC2 and ΔC) resulted in a partial or dramatic mislocalization of SIRT6 from the nucleus to the cytoplasm (Figure 2B). A similar mislocalization of C-terminally truncated SIRT6 proteins was also observed when FLAG-tagged SIRT6 proteins were analyzed by indirect immunofluorescence (data not shown). Intriguingly, the core domain alone (lacking both the N- and C-terminal domains) had a more profound mislocalization than the ΔC mutants (Figure 2B), appearing more substantially excluded from the nucleus. These observations suggest that although the NTE alone is dispensable for proper nuclear localization, it may have a partially synergistic function with the CTE in maintaining proper SIRT6 nuclear localization.
Figure 2.
Sub-cellular localization of wild-type and mutant SIRT6 proteins. Epifluorescence images of live 293T cells transiently transfected with the indicated EGFP-FLAG-tagged SIRT6 deletion and point mutants. Nuclei are co-stained with Hoechst 33342 (40×).
Inspection of the C-terminal sequence of SIRT6 identified a seven-amino acid sequence starting at residue 345—345PKRVKAK351—that resembles a canonical nuclear localization signal (NLS). We mutated the four basic residues (K346, R347, K349, K351) of this putative NLS to alanine in the context of the full-length protein (to generate SIRT6-mutNLS) and examined its localization. GFP-tagged SIRT6-mutNLS was partially mislocalized to the cytoplasm (Figure 2B), and this pattern was indistinguishable from that generated by deleting ∼80 amino acids of the C terminus (ΔC2). These data suggest that the 345PKRVKAK351 sequence at the C terminus of SIRT6 is an NLS essential for proper nuclear localization.
Finally, we analyzed the cellular localization of the isolated CTE and NTE sequences of SIRT6 fused to GFP. The CTE (and Δcore) were sufficient to direct nuclear localization of GFP, generating a pattern indistinguishable from that of full-length SIRT6 fused to GFP (Figure 2C). In contrast, the localization pattern of the NTE was indistinguishable from that of GFP alone (Figure 2C).
Together, our results indicate that the CTE of SIRT6 is necessary and sufficient for proper nuclear localization of SIRT6 and identify an NLS at the very C terminus of SIRT6.
3.2. The N terminus of SIRT6 is required for H3K9 and H3K56 deacetylation in cells
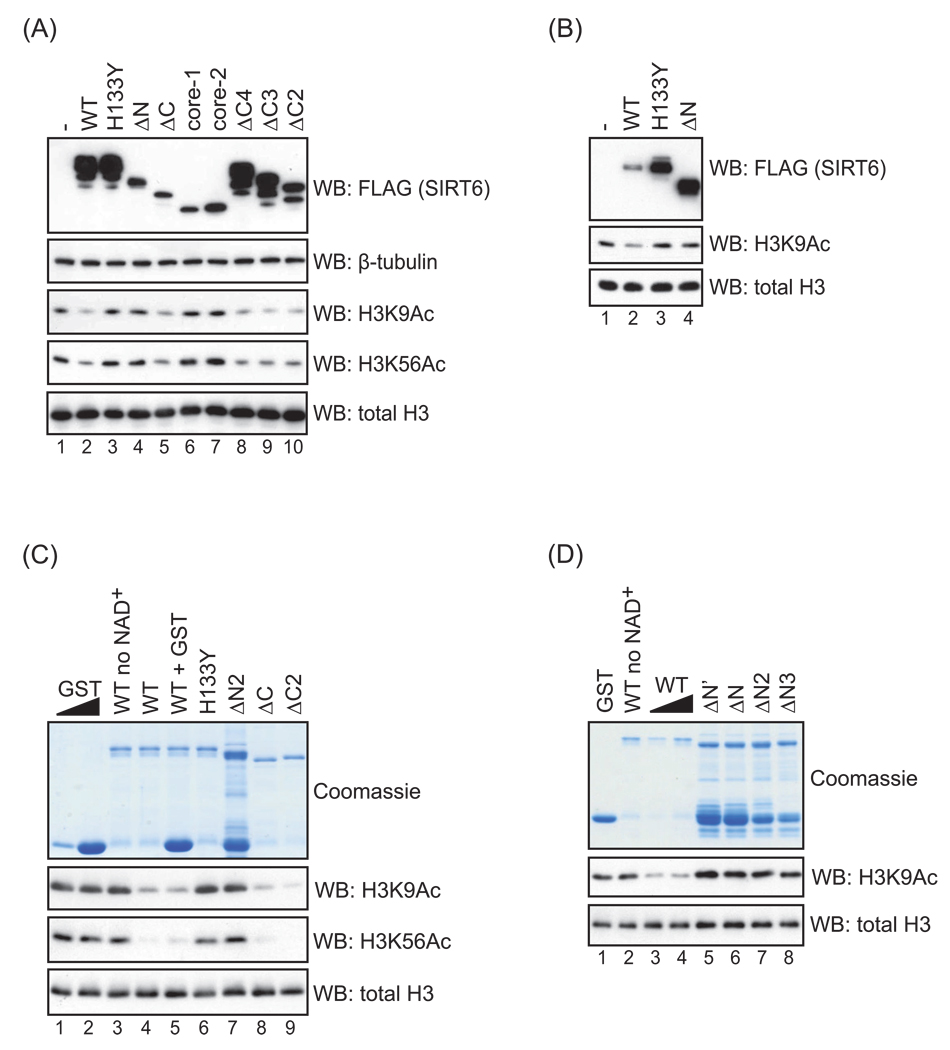
SIRT6 is a relatively site-specific histone deacetylase, for which two substrates have so far been identified, lysines 9 and 56 of histone H3 (H3K9 and H3K56). To determine whether the core sirtuin domain of SIRT6 is sufficient for its deacetylase activity in cells or whether the N and C termini contribute to catalytic activity, we transiently overexpressed a panel of FLAG-tagged SIRT6 deletion mutants (see Figure 1) and assessed global levels of H3K9 and H3K56 acetylation by western analysis. Similar results were obtained for both SIRT6 substrates. As previously shown (Michishita et al., 2008; Michishita et al., 2009; Yang et al., 2009), overexpression of wild-type SIRT6 reduced global H3K9 and H3K56 acetylation levels (but not several other acetylation marks on H3 and H4 [data not shown]), whereas the catalytically inactive H133Y mutant did not (Figure 3A; lanes 1–3).
Figure 3.
Histone deacetylation activity of wild-type and mutant SIRT6 proteins. A–B. Western analysis of H3K9 and H3K56 acetylation levels in 293T cells transiently transfected with FLAG-tagged SIRT6 deletion and point mutants. Quantities of transfected DNA were adjusted to achieve the expression levels shown in (B). Results are representative of at least four (A) or two (B) independent experiments. C–D. Western analysis of H3K9 and H3K56 acetylation levels in in vitro NAD+-dependent deacetylation reactions, using purified GST-SIRT6 deletion and point mutants and purified calf thymus histone H3. (NAD+ was omitted in lanes 3 and 2 of (C) and (D), respectively.) In lane 5, excess GST protein was added to control for the GST degradation product in the ΔN2 protein sample (lane 7). Note the impaired deacetylation for the HY mutant and N-terminally deleted proteins (ΔN’, ΔN, ΔN2, ΔN3), but not the isolated C-terminal deletions (ΔC, ΔC2, ΔC3, ΔC4). We were unable to purify sufficient quantities of the core-1 and core-2 mutants to test their deacetylase activity in vitro. β-tubulin and total histone H3 levels are shown as controls.
Deleting nearly the entire C terminus of SIRT6 (ΔC) did not abrogate the ability of SIRT6 to reduce H3K9 and H3K56 acetylation in cells (Figure 3A, compare lanes 2 and 5). The efficient deacetylation of H3K9 and H3K56 that was observed upon expression of this protein initially seemed surprising, because this ΔC mutant was predominantly cytoplasmic (Figure 2B) and was expressed at lower levels than the exogenous wild-type SIRT6 (Figure 3A). However, in these overexpression experiments, levels of the ΔC protein were substantially higher than endogenous SIRT6, and a considerable fraction of the mutant protein was still present in the nucleus (Figure 2B). In this context, our data suggest that the fraction of the ΔC protein that accesses the nucleus is fully active enzymatically. By deleting fewer amino acids from the C terminus (ΔC2, ΔC3, ΔC4; see Figure 1), we were able to achieve better expression (and less severe mislocalization, Figure 2B and data not shown), and these mutants were as proficient as wild-type SIRT6 in reducing global H3K9 and H3K56 acetylation (Figure 3A, lanes 8–10). Together, these data indicate that when SIRT6 is overexpressed in cells, the CTE is not essential for H3K9 or H3K56 deacetylation, providing evidence that this sequence is dispensable for the intrinsic enzymatic activity of SIRT6.
In contrast to the C-terminal deletion mutants, a SIRT6 mutant lacking the NTE (ΔN)— which was appropriately localized to the nucleus (Figure 2A)—was severely compromised in its ability to reduce global H3K9 and H3K56 acetylation (Figure 3A, lane 4). This held true for more conservative deletions of the N terminus as well (data not shown). Although SIRT6-ΔN is expressed at lower levels than wild-type SIRT6 in this experiment, its expression is comparable to that of the ΔC mutant, which is proficient in deacetylating H3K9 and H3K56. Furthermore, wild-type SIRT6 is able to deacetylate H3K9 when expressed at much lower levels than the ΔN mutant (Figure 3B), arguing that the inability of SIRT6-ΔN to deacetylate H3K9 and H3K56 in cells cannot be attributed to lower levels of expression. Similar to the SIRT6-ΔN mutant, the core of SIRT6 (lacking both the N and C termini) lacked catalytic activity in cells (Figure 3A, lanes 6 and 7). This observation is expected, given that the NTE deletion alone abrogates SIRT6 activity, although the more extensive exclusion of this mutant core protein from the nucleus may also contribute to the observed lack of deacetylase activity in cells. Together, our data indicate that the N-terminal extension of SIRT6 is important for its ability to modulate global levels of H3K9 and H3K56 acetylation in cells.
3.3. The N terminus of SIRT6 is essential for the intrinsic catalytic activity of SIRT6
The inability of the SIRT6 N-terminal deletion mutant (SIRT6-ΔN) to reduce cellular H3K9 and H3K56 acetylation levels could be due either to an intrinsic defect in enzymatic activity or to a failure to properly localize to chromatin substrates or interact with critical binding partners. To directly test whether the NTE of SIRT6 is required for the intrinsic deacetylase activity of SIRT6, we purified GST-tagged wild-type SIRT6, the catalytically inactive H133Y SIRT6, and N-terminally deleted SIRT6 from E. coli and assessed their catalytic activity on purified histone H3 in vitro (Figure 3C). As expected, wild-type SIRT6 deacetylated H3K9 and H3K56 (lane 4), whereas the H133Y mutant lacked activity (lane 6). Because all ΔN mutants were partially degraded to GST during purification, we added an excess of GST to the wild-type reaction to control for any effect of the ΔN degradation products. Wild-type SIRT6 still showed robust deacetylase activity in the presence of excess GST (lane 5). In contrast, the ΔN2 mutant appeared to lack activity in vitro (lane 7), similar to the HY mutant, suggesting that the NTE of SIRT6 is required for the intrinsic H3K9 and H3K56 deacetylation activity of SIRT6. More conservative N-terminal deletion mutants also showed impaired catalytic activity in vitro (Figure 3D; see Figure 1). As expected from the results of overexpression in cells, the ΔC mutants were proficient in deacetylating H3K9 and H3K56 in vitro (Figure 3C, lanes 8–9). From these data, we conclude that sequences in the NTE are essential for the intrinsic deacetylase activity of SIRT6.
3.4. Catalytic and non-core SIRT6 sequences contribute to cellular chromatin association
We next used several different assays to assess the roles of the NTE and CTE of SIRT6 in the interaction of SIRT6 with chromatin. First, we transiently overexpressed FLAG-tagged wild-type, H133Y, and N- and C-terminally truncated SIRT6 and assayed for the presence of core histones in FLAG immunoprecipitates. As previously shown, wild-type SIRT6 readily associates with the core histones in cells, as detected by Coomassie (Figure 4A) and western analysis (Figure 4B); the fact that the four core histones are present in approximately equimolar amounts in the SIRT6 IP argues that this interaction occurs in the context of intact nucleosomes at chromatin (McCord et al., 2009). Intriguingly, co-immunoprecipitation of the core histones with the catalytically inactive H133Y mutant was dramatically reduced compared to wild-type SIRT6, as observed by both Coomassie stain and western blot for histone H3 (Figures 4A and 4B, respectively). This observation suggests that the H133Y mutation not only abrogates the catalytic activity of SIRT6 but also profoundly impacts on the dynamics of SIRT6 association with chromatin in cells.
We next assessed the N- and C-terminal deletion mutants in this chromatin association assay using western analysis for H3, because this method was more sensitive and these mutant proteins were expressed at lower levels than the WT and H133Y SIRT6 proteins. Strikingly, deleting the N terminus of SIRT6 nearly abrogated the ability of SIRT6 to bind histones, an effect even more profound than that observed for the H133Y mutation (Figure 4B). In contrast, deleting the C terminus of SIRT6 had little effect on the interaction with histones in this assay (Figure 4B). Thus, both the NTE and the catalytic activity of SIRT6 appear to be required for its stable association with histones in cells. Consistent with these findings, neither the NTE alone nor the Δcore mutant (containing the NTE and CTE and lacking the core domain; see Figure 1) was able to interact with histones in cells (data not shown), suggesting that multiple regions of SIRT6 are required for its interaction with chromatin.
To assay chromatin association using an independent method, we carried out biochemical fractionation of U2OS cells stably expressing the different FLAG-tagged SIRT6 proteins, according to the protocol of Mendez and Stillman (2000). Fractionation into soluble cytosolic, soluble nuclear, and chromatin-enriched fractions (Figure 4C) revealed that wild-type SIRT6 is largely chromatin-enriched under these conditions, as previously shown (Mostoslavsky et al., 2006) (Figure 4D). In contrast, both the catalytically inactive H133Y mutant and the ΔN mutant showed a reduction in the relative amount of SIRT6 present in the chromatin-enriched fraction, further supporting the conclusion that both the NTE and the catalytic activity of SIRT6 contribute to its ability to associate with chromatin in cells.
3.5. The NTE and CTE of SIRT6 are important for nucleosome binding
The inability of the H133Y and ΔN mutants to bind chromatin in cells could be due to an intrinsic defect in nucleosome binding, an inability to interact with chromatin-associated binding partners that propagate SIRT6 along chromatin, or another defect in chromatin targeting. To investigate these models, we assessed in vitro nucleosome binding by incubating the wild-type and mutant GST-SIRT6 proteins with purified HeLa mononucleosomes and measured binding using a gel-shift assay (Figures 4E and 4F). As previously shown (McCord et al., 2009), wild-type SIRT6 binds nucleosomes efficiently, as evidenced by the dramatic shift of nucleosomes to a higher molecular weight. The H133Y mutant induced a similar upward shift of nucleosomes (Figure 4E and Figure S1), suggesting that this mutant is largely proficient at binding nucleosomes in vitro. Although there may be a very subtle difference in the electrophoretic mobility shift for the H133Y mutant compared to wild-type SIRT6, this effect is much less substantial than the defective chromatin association of SIRT6-H133Y observed in cells (Figure 4A–D). Therefore, the impaired chromatin association of the SIRT6-H133Y mutant protein observed in cells is likely not attributable to an intrinsic defect in nucleosome binding. In contrast, the ΔN mutant was completely impaired in the in vitro nucleosome binding assay (Figure 4F, lane 4), suggesting that the NTE of SIRT6 is important for direct binding of SIRT6 to nucleosomes. Surprisingly, although the CTE was dispensable for binding histones in an overexpression context in cells (Figure 4B), deleting the CTE of SIRT6 abrogated nucleosome binding in vitro (Figure 4F, lanes 5 and 6), suggesting that this domain may also impact on the chromatin association of SIRT6 under physiologic conditions.
4. Discussion
SIRT6 is a critical regulator of transcription, genome stability, telomere integrity, and metabolism, but the enzymatic and regulatory roles of specific domains of SIRT6 have not been clearly defined. Here, we describe new functions of the N- and C-terminal extensions of SIRT6 that contribute to sub-cellular localization, catalytic activity, and chromatin binding. Further, we have uncovered an unexpected effect of mutating a highly conserved catalytic residue on the ability of SIRT6 to associate with chromatin.
4.1. Separation of function mutants reveal distinct roles for the NTE and CTE of SIRT6
SIRT proteins share a conserved central ‘sirtuin domain,’ generally thought to be an enzymatic core. In our experiments, the core domain of SIRT6 was not sufficient for proper localization or catalytic activity in cells, indicating that the N- and C-terminal extensions are critical for SIRT6 function. Our data indicate that the NTE and CTE of SIRT6 have several distinct roles: the NTE is required for H3K9 and H3K56 deacetylase activity and chromatin association in vitro and in cells but is largely dispensable for nuclear localization, whereas the CTE is not required for deacetylase activity but is important for nuclear localization. One possible explanation for the effects of deleting the NTE of SIRT6 is that the N terminus of SIRT6 plays a direct role in the chemistry of deacetylation, for example by contributing to binding of substrate or NAD+; such a role has been proposed for the NTE of the S. cerevisiae sirtuin protein Hst2 (Zhao et al., 2003). Alternatively, the NTE could contribute to SIRT6 function by promoting proper folding or protein stability. However, arguing against the possibility that the N-terminally deleted protein is dramatically misfolded are the observations that the mutant protein is appropriately localized to the nucleus in live cells (Figure 2A) and engages efficiently in protein-protein interactions (data not shown).
Intriguingly, an alignment of metazoan SIRT6 homologues reveals that the NTE of SIRT6 is much more strongly conserved than the CTE, which diverges significantly even between the primate and mouse proteins (Figure S2). This is consistent with the critical role of the NTE in SIRT6 catalytic activity and suggests that the CTE of SIRT6 may have diverged to accommodate primate-specific functions of the protein.
Using several independent biochemical assays, we found that both the NTE and CTE of SIRT6 appear to play a role in chromatin association in different in vitro and cellular contexts. The NTE is required for both nucleosome binding in vitro (Figure 4F) and for chromatin association in cells (Figure 4B–D). In contrast, deleting the CTE abrogated nucleosome binding in vitro (Figure 4F) but had a minimal effect on chromatin association in cells under the specific biochemical extraction conditions used (Figure 4B). Thus, the ability to see defects in the chromatin association of the C-terminal deletion mutants depends on the particular assay used. These observations suggest that the CTE may contribute to proper chromatin association of SIRT6 under certain physiologic conditions, but that alternative biochemical extraction conditions may be required to detect such an effect. We note that despite the reduced nucleosome binding of SIRT6-ΔC, this protein was still able to interact sufficiently with its chromatin substrates in cells to efficiently catalyze their deacetylation (Figure 3A). This is not surprising, since enzyme-substrate interactions are frequently very transient. Together, our findings highlight the importance of assaying chromatin association using several independent techniques, as each assay reflects slightly different biochemical and physiological properties.
4.2. A link between catalytic activity and chromatin association
Surprisingly, although the catalytically inactive SIRT6-H133Y mutant was able to associate with nucleosomes in vitro (Figure 4E and Figure S1), this mutant was significantly impaired in associating with chromatin in cells (Figure 4A–D). This indicates that the reduced chromatin association of SIRT6-H133Y is not due to an intrinsic defect in nucleosome binding. One model that could account for the link between catalytic activity and chromatin association of SIRT6 is that there may be a feed-forward mechanism in which histone deacetylation by SIRT6 stabilizes SIRT6 occupancy at chromatin or promotes propagation of SIRT6 molecules along chromatin. The latter scenario is analogous to models of heterochromatinization in yeast, in which histone deacetylation by Sir2 generates additional binding sites for the Sir protein complex, mediating spreading of this complex from the telomere toward the centromere and at the silenced mating type locus (Hecht et al., 1995; Hoppe et al., 2002; Rusche et al., 2002). An alternative and not mutually exclusive model is that auto-ADP-ribosylation of SIRT6 (Liszt et al., 2005) may generate a novel interface for chromatin binding; for example, macro domains—present in histone macroH2A—are reported to bind mono-ribosylated proteins (Karras et al., 2005). Such interactions could be important for stabilizing or propagating SIRT6 at chromatin. In the future, it will be interesting to probe these models (for example, by determining whether chemical inhibition of SIRT6 leads to altered chromatin association in cells).
4.3. Mechanistic insights into SIRT6 domains as targets for chemical modulation
In conclusion, our data provide important mechanistic information regarding structural features of SIRT6 that impact on SIRT6 cellular function, and suggest testable models for the regulation of sirtuin enzymes. Our data demonstrating functional roles for the N- and C-terminal extensions of SIRT6 suggest that these domains may serve as sites of regulation, for example, through post-translational modification (North and Verdin, 2007b) or interactions with yet-to-be-identified SIRT6 regulatory factors. Furthermore, because chromatin-modifying enzymes are highly amenable to small molecule regulation, SIRT proteins are actively being pursued as pharmacological targets for metabolic disorders, age-related diseases, and cancer (Lavu et al., 2008; Szczepankiewicz and Ng, 2008; Westphal et al., 2007). In this context, deciphering the structural determinants of SIRT6 enzymatic activity—both in vitro and in cells—should be valuable for improving the design of small-molecule modulators. Our identification of important roles for the N- and C-terminal extensions of SIRT6–along with a surprising new role for a conserved catalytic residue—may inform the development of SIRT6 as a pharmacologic target.
Supplementary Material
In vitro gel-shift assay confirming similar nucleosome binding efficiency of wild-type SIRT6 and the catalytically inactive H133Y SIRT6 mutant. Assay was performed as described in Figure 4E, using purified GST-SIRT6 proteins and purified HeLa mononucleosomes.
Sequence alignment of metazoan SIRT6 homologues, showing the conservation of the N-terminal extension (red) and core (yellow) of SIRT6 and the relative divergence of the C-terminal extension (blue). Black shading indicates residues that differ from the human SIRT6 sequence; asterisk indicates conserved histidine 133; filled circles indicate residues mutated to alanine in SIRT6-mutNLS. Residue 45 (indicated by an arrowhead) is polymorphic (serine/asparagine) in the human sequence. Alignment was compiled using MegAlign (DNASTAR Lasergene).
Acknowledgements
We thank Steven Artandi for modified pWZL-3FLAG and pGEX-6P3 vectors, Jeff Glenn for use of the Nikon Eclipse TE300 inverted fluorescence microscope, and Or Gozani and members of the Gozani and Chua labs for helpful discussions. This work was supported by grants to K.F.C. from the NIH (R01AG028867 and K08AG028961) and Department of Veterans Affairs Merit Review. R.I.T. is funded by a National Defense Science and Engineering Graduate Fellowship, a National Science Foundation Graduate Research Fellowship, and an NIH training grant (1018438-142-PABCA). K.F.C. is a Paul Beeson Scholar and an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The abbreviations used are: H3K9Ac, acetylated lysine 9 on histone H3; H3K56Ac, acetylated lysine 56 on histone H3; NTE, N-terminal extension; CTE, C-terminal extension; IP, immunoprecipitation; NLS, nuclear localization signal.
References
- Cockell MM, Perrod S, Gasser SM. Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics. 2000;154:1069–1083. doi: 10.1093/genetics/154.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus G, Shafaatian R, Shore D. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. Embo J. 2000;19:2641–2651. doi: 10.1093/emboj/19.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem Sci. 2003;28:41–48. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. Embo J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins--novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, Bohr VA, Chua KF. SIRT6 stabilizes DNA-dependent Protein Kinase at chromatin for DNA double-strand break repair. Aging. 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009:8. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007a;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Verdin E. Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J Biol Chem. 2007b;282:19546–19555. doi: 10.1074/jbc.M702990200. [DOI] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepankiewicz BG, Ng PY. Sirtuin modulators: targets for metabolic diseases and beyond. Curr Top Med Chem. 2008;8:1533–1544. doi: 10.2174/156802608786413465. [DOI] [PubMed] [Google Scholar]
- Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- Westphal CH, Dipp MA, Guarente L. A therapeutic role for sirtuins in diseases of aging? Trends Biochem Sci. 2007;32:555–560. doi: 10.1016/j.tibs.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009:8. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Chai X, Clements A, Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Biol. 2003;10:864–871. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vitro gel-shift assay confirming similar nucleosome binding efficiency of wild-type SIRT6 and the catalytically inactive H133Y SIRT6 mutant. Assay was performed as described in Figure 4E, using purified GST-SIRT6 proteins and purified HeLa mononucleosomes.
Sequence alignment of metazoan SIRT6 homologues, showing the conservation of the N-terminal extension (red) and core (yellow) of SIRT6 and the relative divergence of the C-terminal extension (blue). Black shading indicates residues that differ from the human SIRT6 sequence; asterisk indicates conserved histidine 133; filled circles indicate residues mutated to alanine in SIRT6-mutNLS. Residue 45 (indicated by an arrowhead) is polymorphic (serine/asparagine) in the human sequence. Alignment was compiled using MegAlign (DNASTAR Lasergene).