Abstract
P58(IPK) may function as an ER molecular chaperone to maintain protein folding homeostasis during Unfolded Protein Responses (UPR). P58(IPK) contains nine TPR motifs and a C-terminal J-domain within its primary sequence. To investigate the mechanism how P58(IPK) functions to promote protein folding within ER, we have determined the crystal structure of P58(IPK) TPR fragment to 2.5Å resolution by SAD method. The crystal structure of P58(IPK) revealed three domains (I, II and III) with similar folds and each domain contains three TPR motifs. An In vitro ELISA assay indicates that P58(IPK) acts as a molecular chaperone by interacting with the misfolded proteins such as Luciferase and Rhodanese. The P58(IPK) structure reveals a conserved hydrophobic patch located in domain I that may be involved in binding the misfolded polypeptides. Structure-based mutagenesis for the conserved hydrophobic residues located in domain I significantly reduced the molecular chaperone activity of P58(IPK).
Introduction
Endoplasmic reticulum (ER) stress leads to protein overloading and protein misfolding within the ER lumen, which induces the so-called unfolded protein response (UPR) 1; 2. Several ER-resident stress sensor proteins such as IRE1, PERK and ATF-6 function to transduce the ER stress signals from ER lumen to nuclei to trigger UPR. In the normal conditions, the ER luminal domains of these sensor proteins are bound by the ER molecular chaperone BiP and these interactions inhibit the UPR signaling. In the stressed conditions, accumulated misfolded protein in the ER lumen competes for BiP and the release of BiP from the ER sensor proteins initiates the UPR signaling 3; 4; 5. The UPR can lower the ER stress burden by regulating a number of transcription pathways. One major pathway is to reduce the ER protein influx and the second is to promote protein folding and degradation of the misfolded proteins within ER 6; 7; 8. Eventually, the ER stress is cleared by UPR and the homeostasis of the ER can be re-established. When UPR fails to rescue the stressed ER, the cell may go through the apoptosis 9. Malfunction in the UPR pathways contribute to some severe human diseases such as inflammation, diabetes and neurodegeneration 10; 11.
P58(IPK), also known as DnaJC3, is a downstream target of UPR signaling and plays major roles in UPR. P58(IPK) knockout in cultured cells causes elevated amount of misfolded protein within ER and higher level of UPR signaling 12. The mutations within P58(IPK) result in diabetes symptoms in a mouse model 13. P58(IPK) has an N-terminal signal peptide for ER targeting and translocation 14. Recent studies showed that P58(IPK) functions as a molecular chaperone to facilitate misfolded protein folding in association with BiP within ER 14; 15. In the cytosolic side, P58(IPK) was first identified as an inhibitor for PKR, a virally induced eIF2α kinase 16. Later P58(IPK) was also shown to be able to bind and inhibit PERK, the ER stress-inducible eIF2α kinase 17; 18. PKR and PERK function to attenuate protein synthesis by regulating the phosphorylation of eIF2α 19. Therefore, P58(IPK) may be a dual-functioned protein serving as a chaperone in the ER lumen and a regulator of global protein synthesis in the cytosol.
P58(IPK) contains an N-terminal ER-targeting peptide sequence (~30 amino acid residues), nine tetratricopeptide repeats (TPR) and a C-terminal J-domain, as predicted by the primary sequence. The TPR motif was first identified as degenerate 34 amino acid repeat present in a variety of proteins from bacteria to eukaryotes. One TPR motif consists of two anti-parallel α-helices and multiple TPR motifs can stack together to form a TPR domain as protein interaction sites 20. The J-domain was first found within the Hsp40 DnaJ sequence and has become a hallmark for all Hsp40 proteins 21; 22. P58(IPK) may recruit ER Hsp70 BiP and stimulate the BiP ATPase activity for the subsequent protein refolding through the C-terminal J-domain 23. The bulk of P58(IPK) contains nine TPR motifs. It has been suggested that P58(IPK) may function as a molecular chaperone to interact with misfolded proteins 12; 15. It is likely that the P58(IPK) TPR fragment may serve as the binding site for the misfolded protein. A functional homologue of P58(IPK) in yeast, Jem1p, was recently indentified to be involved in ER quality control24.
In this study, we have determined the crystal structure of mouse P58(IPK) TPR fragment (residues 35–393) to 2.5Å. The crystal structure of P58(IPK) TPR fragment revealed three domains (I, II and III) with similar folds. The three domains within P58(IPK) structure are connected in a unique head-to-tail fashion. We have established an ELISA assay demonstrating that P58(IPK) TPR fragment acts as a molecular chaperone to bind misfolded model proteins such as Luciferase and Rhodanese. The P58(IPK) structure reveals a conserved hydrophobic patch located in domain I that may be the binding site for the misfolded polypeptides. Structure-based mutagenesis studies strongly support this hypothesis.
Results and Discussion
The P58(IPK) monomer structure
The P58(IPK) (504 a.a.) from Mus musculus contains nine tetratricopeptide repeats (TPR) and a short C-terminal J-domain in its primary sequence. The crystal structure of mouse P58(IPK) TPR fragment (35–393) was determined to 2.5Å resolution by use of SAD method. The resultant electron density map from the SAD phasing was readily traceable (Supplementary Fig. S1). The P58(IPK) forms a monomer in the crystal structure, which is consistent with our previous gel filtration analysis for P58(IPK) TPR domain 25. The final model of P58(IPK) monomer contains residues 35–393 of P58(IPK).
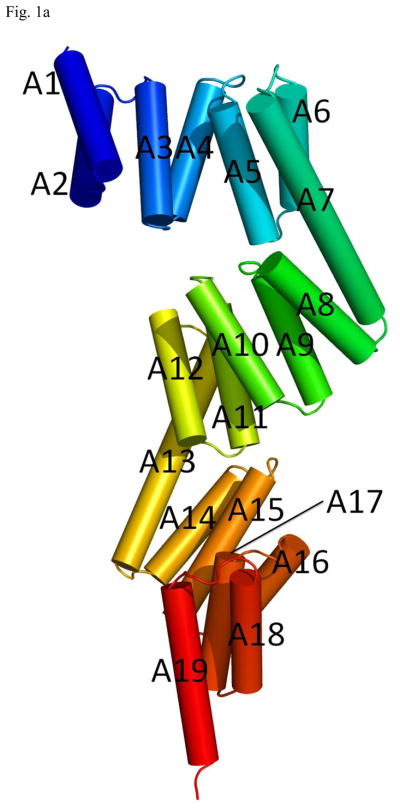
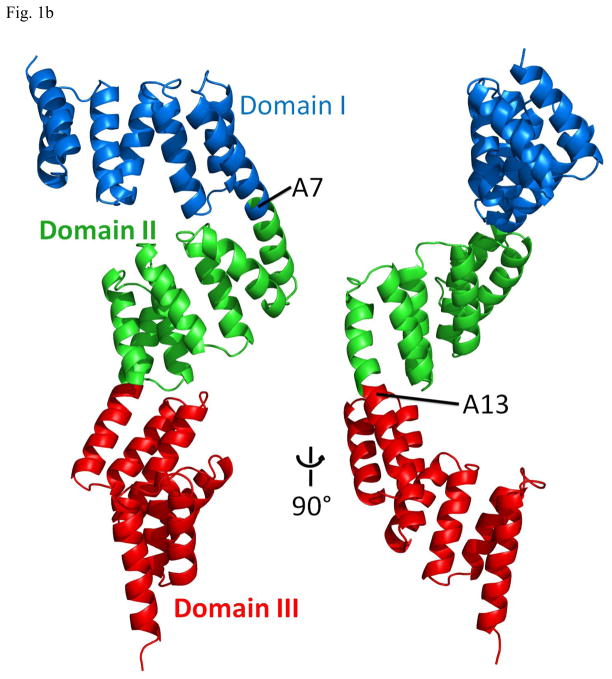
The structure of the P58(IPK) monomer consists of 19 α-helices (A1–A19) and no β-strands (Fig. 1a). A majority of the helices form nine TPR motifs (TPR1–TPR9). Three domains (I, II and III) can be identified from the P58(IPK) structure. The domain I covers helices A1–A7, the domain II consists of helices A7 to A13 and the domain III includes helices A13–A19. Domain I is connected to the domain II by the long helix A7 and the domain II is linked with the domain III through the long helix A13 (Fig. 1b). The three domains are organized in a head-to-tail fashion, which provides P58(IPK) TPR fragment an elongated molecular shape with the length of about 100 Å. In the P58(IPK) domain I, the helices A1–A7 are organized in an anti-parallel fashion, where A1 and A2, A3 and A4, A5 and A6 form TPR1, TPR2 and TPR3, respectively. In the domain II, helices A7–A12 form TPR4–TPR6 and in the domain III helices A13–A18 constitute TPR7-TPR9. The domain II and the domain III of P58(IPK) adopt the similar folds as domain I. When domain II and domain III are superimposed with domain I, the rms derivation for the main chain atoms is 2.76Å and 5.48Å respectively (http://www.bioinf.org.uk/profit).
Fig. 1.
Overall structure of P58(IPK) TPR fragment
a) P58(IPK) TPR fragment contains 19 helices (A1–A19). Domain I, II and III consist of A1–A7, A7–A13, A13–A19, respectively. The P58(IPK) TPR fragment are colored in rainbow (blue in N terminus and red in C terminus).
b) P58(IPK) TPR fragment structure in Ribbons representation. Domain I and II are connected by the long helix A7, domain II and III are connected by A13. Domain I, II and III are colored blue, green and red respectively.
A DALI search for structure homologues did not provide any protein structure with an overall topology similar to P58(IPK) TPR fragment (http://www.ebi.ac.uk/dali/). However, several structure homologues were identified for the three individual domains within P58(IPK) from the DALI search. This data suggests that the three domains within P58(IPK) structure are organized in a novel fashion which is not found in any other TPR motif containing structures. In the P58(IPK) crystal structure, the nine TPR motifs (TPR1–9) are not packed into a “super-helix” which has been identified in other protein structures which consist of multiple TPR motifs 26; 27. Instead, three distinct domains (I, II and III) with similar folds stack together in a head-to-tail manner. Domain II rotates about 180° along the long helix A7 from the position of domain I and domain III rotates about 180° along helix A13 from domain II (Fig. 1).
The DALI search reveals that a number of eukaryotic protein crystal structures show significant structural homology to any of the three domains of P58(IPK) structure with the Z-score higher than 15. These structures include TPR1 and TPR2A domains from Hop (Hsp70 and Hsp90 organizing protein) 28; 29, the N-terminal domain of mitochondrial precursor receptor Tom70p 27, the middle part of the O-linked GlcNAc transferase (OGT) 26, the TPR domains from Serine/Threonine Protein Phosphatase 5 (PP5) 20, CHIP (C-terminal of Hsp70 interacting protein) 30, the peroxisomal targeting signal receptor PEX5 31 and the small G protein-binding cytosolic factor p67pho 32. All of these proteins utilize the large groove formed within the TPR motifs as the binding site for protein substrates. For example, the groove formed within the N-terminal TPR motifs of Tom70 can associate with Hsp70/Hsp90 C-terminal EEVD motif primarily via charge-charge interaction 33.
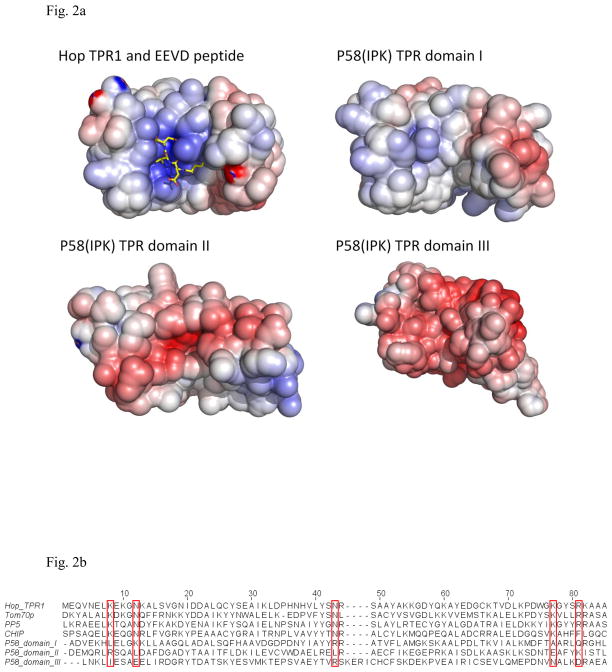
Hop, Tom70p, PP5 and CHIP interact with the C-terminal EEVD motif of Hsp70 or Hsp90 by use of the TPR motifs 20; 27; 33; 34; 35; 36. It has been reported that P58(IPK) may function as a molecular chaperone in pair with ER Hsp70 BiP 14; 15. However, several lines of evidences suggest that P58(IPK) TPR domains do not directly interact with BiP C-terminus. First, BiP does not contain the EEVD motifs in its C-terminus. Second, the EEVD-binding TPR proteins, such as Hop TPR1 domain, are positively charged in their grooves to bind the negatively charged EEVD motif. However, P58(IPK) TPR domain I is neutral in the groove while domain II and domain III of P58(IPK) are negatively charged (Fig. 2a). Third, in Hop TPR1 and Hsp70 C-terminus complex structure, several residues (K8, N12, N43, K73, and R77) are critical for binding the Hsp70 EEVD motif 28. These residues are conserved in the structures of Tom70p, Hop, PP5 and CHIP. However, none of these residues is present in P58(IPK) sequence (Fig. 2b). It is likely that P58(IPK) TPR domain may not associate with BiP via the traditional EEVD motif-TPR interaction. We propose that P58(IPK) may interact with the misfolded proteins by using one of the large grooves located in the domain I, II and III.
Fig. 2.
The individual domains within P58(IPK) TPR fragment compared with the TPR domains from other proteins.
a) The surface potential drawings for Hop TPR1 domain (PDB ID 1ELW), P58(IPK) TPR domain I, II and III (Blue for positively-charge regions; White for neutral regions; Red for negatively-charged regions). The groove of Hop TPR1 is positively charged which stabilizes the interaction between Hop TPR1 and EEVD motif of Hsp70 (in yellow stick representation). In contrast, P58(IPK) TPR fragment domain I is neutral in the groove while domain II and III are negatively-charged.
b) Alignment of Hop TPR1, Tom70p, PP5, CHIP and P58(IPK) TPR fragment domain I, II and III. The residues involved in EEVD motif binding are highlighted in the red boxes. These residues are highly conserved in Hop, Tom70p, PP5 and CHIP, but none of them is present in P58(IPK) TPR domain I, II or III.
P58(IPK) TPR fragment functions as a molecular chaperone to bind misfolded proteins
It has been proposed that P58(IPK) may function as a co-chaperone to promote protein folding in co-operation with BiP within ER 14; 15. Here we have established an in vitro ELISA assay to show the BiP independent chaperone activity of P58(IPK) TPR fragment to bind misfolded proteins. P58(IPK) TPR fragment can selectively recognize and bind chemically denatured Luciferase and Rhodanese but not the native proteins (Fig. 3, supplemental Fig. S2). It is highly likely that P58(IPK) TPR fragment interacts directly with the misfolded proteins to prevent the protein from aggregating.
Fig. 3.
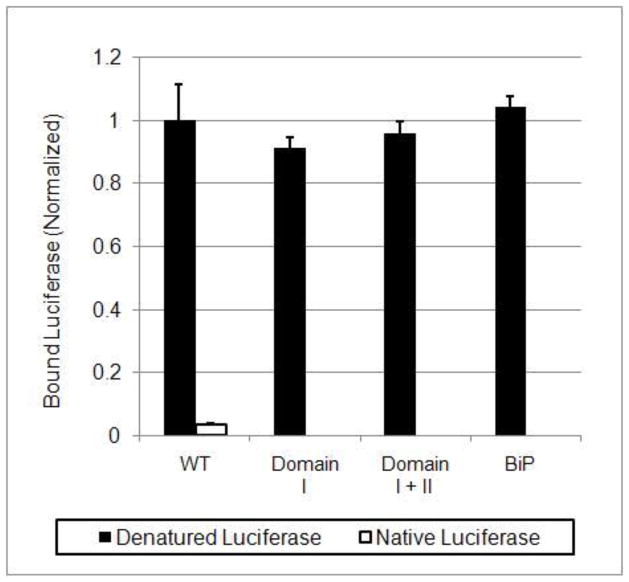
P58(IPK) TPR domain I retains the full molecular chaperone acitvity.
An in vitro ELISA assay was performed to measure the direct binding between P58(IPK) TPR fragment (or its mutant) and the misfolded or native Luciferase (See methods). Denatured or native Luciferase was incubated in the presence of wild type P58(IPK) TPR fragment (residue 33–393, containing domain I, II and III), domain I+II (residue 33–267) or domain I (residue 33–168). The binding between Luciferase and P58(IPK) was monitored by ELISA. The readings are the averaged values of six independent experiments. Hamster Bip with 1mM ATP was used as a positive control.
To investigate which domain is responsible for the molecular chaperone activity of P58(IPK), we have constructed the deletion mutants of P58(IPK) which contains domain I, domain II, domain III, domain I & II, domain II & III. Only domain I, domain I & II can be expressed as soluble protein after extensive trials. The soluble mutants are used in subsequent ELISA assays. The data from the ELISA assay indicates that the domain I by itself retains full activity of the entire P58(IPK) TPR fragment to interact with the misfolded proteins (Fig. 3, Supplemental Fig. S2). The data strongly suggested that the peptide-binding site of P58(IPK) for the misfolded protein is located in domain I.
The hydrophobic peptide-binding groove within the domain I of P58(IPK)
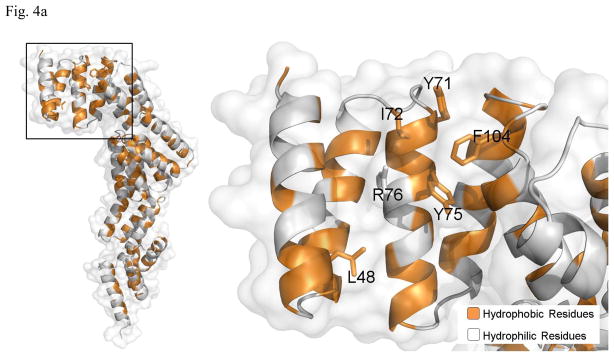
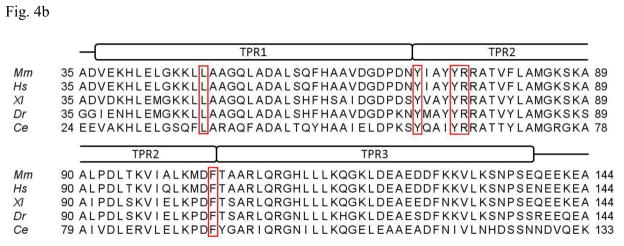
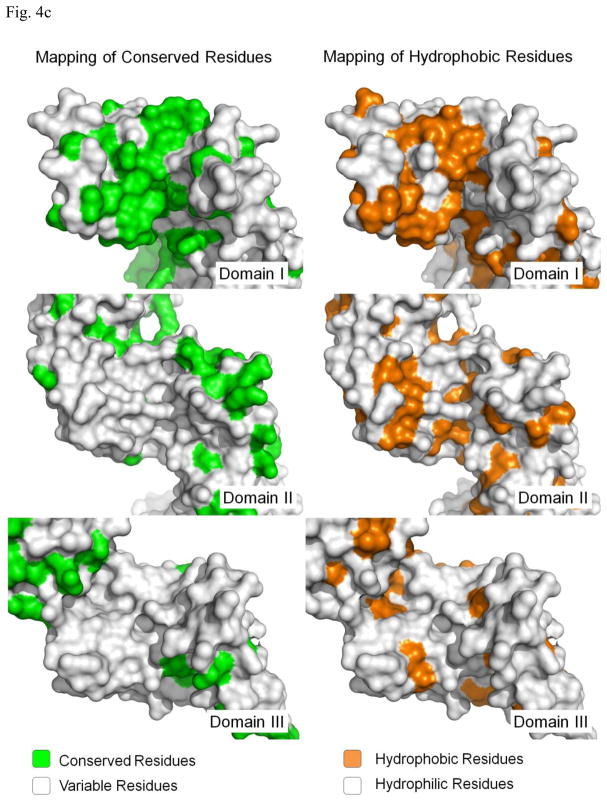
Most molecular chaperones bind non-native polypeptides through hydrophobic interactions. Therefore we examined the molecular surface of P58(IPK) domain I for the possible binding site for the misfolded proteins. A large hydrophobic patch can be identified at the large groove of the domain I. The hydrophobic patch is formed by L48, Y71, I72, Y75, R76 and F104 (Fig. 4a). Sequence alignment of P58(IPK) from different species shows that the hydrophobicity of these residues is well conserved except I72, which indicates that this hydrophobic patch could be a common structural feature for P58(IPK) structures (Fig. 4b). In contrast, the molecular surfaces of the grooves located in P58(IPK) domain II and III are quite hydrophilic and are unlikely to be the binding-sites for the misfolded proteins (Fig. 4c). Moreover, the residues constituting the grooves of domain II and III are not as conserved as those within domain I among the P58(IPK) family members (Fig. 4c). This is consistent with the observation that domain I retains the full chaperone activity of the entire P58(IPK) TPR fragment.
Fig. 4.
Identification of the conserved hydrophobic patch located in the groove of P58(IPK) domain I.
a) In the left panel, the Ribbons are colored according to their hydrophobicity (Gold indicates the hydrophobic regions while white represents hydrophilic regions). In the groove region of domain I, a hydrophobic patch formed by L48, Y71, I72, Y75, R76 and F104 can be identified as the putative peptide-binding site. The right panel represents the magnified version of the black box in the left panel. The hydrophobic residues within the putative peptide-binding site are shown in rod mode and labeled.
b) Alignment of P58(IPK) domain I protein sequences from different species. Domain I consists of three TPR motifs: TPR1, TPR2 and TPR3. The residues constituting the hydrophobic patch are highlighted in the red boxes except I72, which is not 100% conserved. Y71, Y75, R76 and F104 are invariant among all the species aligned. Mm, Mus musculus; Hs, Homo sapiens; Xl, Xenopus laevis; Dr, Danio rerio; Ce, Caenorhabditis elegans.
c) The conservation drawings and hydrophobicity drawings of the grooves within domain I, II and III of P58(IPK) TPR fragment structure. In the conservation drawings, all residues which are conserved among the species aligned in Fig. 4b are mapped onto P58(IPK) structure as green color. In the hydrophobicity drawing, gold represents hydrophobic regions and silver denotes hydrophilic regions. The domain I, II and III are aligned to similar orientations with the groove facing readers.
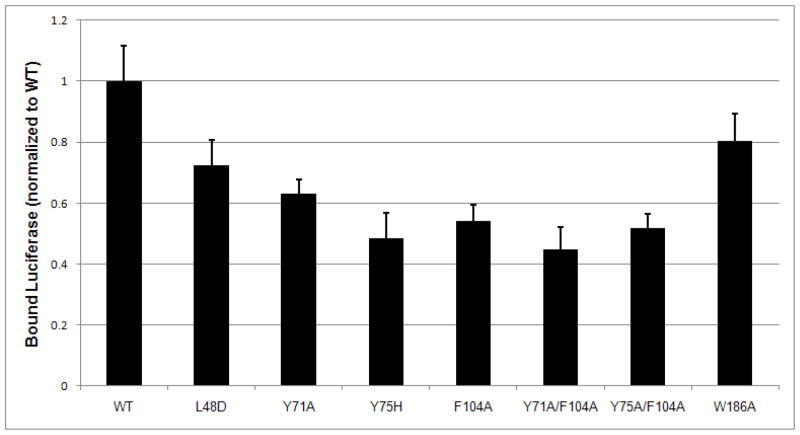
Point mutations (L48D, Y71A, Y75H and F104A) of the hydrophobic patch located at the domain I of P58(IPK) have been performed. The data from the ELISA assay indicated that any mutations for the conserved hydrophobic residues within the patch reduced the binding affinity of P58(IPK) TPR fragment to the misfolded proteins by ~40% (Fig. 5, supplemental Fig. S2). As a negative control, the mutation W186A at the junction of domain I and II did not affect the binding (Fig. 5). The two double mutations (Y71A/F104A and Y75A/F104A) of P58(IPK) also showed ~50% reduction in their binding affinity for misfolded proteins (Fig. 5). The reduction in denatured polypeptide binding ability is similar to the mutants of other ER luminal chaperones, e.g. ERdj3 37. The global structure of P58(IPK) TPR domain is not affected by the double mutations which are evidenced by the CD spectrometry (supplementary Fig. S3). The data strongly suggests that domain I of P58(IPK) recognizes and binds the misfolded proteins through hydrophobic interactions. The large groove formed within domain I of P58(IPK) represents the binding site for the misfolded proteins.
Fig. 5.
P58(IPK) binds the misfolded proteins via the conserved hydrophobic patch within the groove of domain I. The misfolded protein binding assay was performed as in Fig. 3 using Luciferase as the model protein. P58(IPK) TPR fragment (P58 TPR WT, residue 33–393) effectively interacts with the misfolded Luciferase. The mutations for the hydrophobic residues within the groove of domain I significantly reduced the binding ability of P58(IPK) TPR fragment to the misfolded Luciferase. The mutation (W186A) at the junction of domain I and II does not affect the binding.
Working Model of P58(IPK) during UPR
P58(IPK) is over-expressed upon UPR signaling. It functions to alleviate ER stress by promoting protein folding in association with BiP. P58(IPK) consists of an N-terminal TPR containing fragment and a C-terminal J domain. The J domain serves to recruit Hsp70 BiP. However, the function of the P58(IPK) TPR fragment was unspecified. In this study, we demonstrated that the P58(IPK) TPR fragment can function as a molecular chaperone to interact with the misfolded proteins to suppress protein aggregations. The crystal structure of P58(IPK) TPR fragment suggests that P58(IPK) binds misfolded protein via a hydrophobic patch in TPR domain I. Structure-based mutagenesis strongly support this hypothesis.
We proposed the following working model for P58(IPK). In the stressed cells, P58(IPK) can capture and stabilize the misfolded proteins by use of the hydrophobic groove within the domain I. The P58(IPK) C-terminal J domain will then recruit BiP and forms a transient hetero-trimmer of P58(IPK)-BiP-misfolded protein. The misfolded protein will be handed over from P58(IPK) to BiP. Domain II and III within the P58(IPK) structure may facilitate the formation of the hetero-trimmer of P58(IPK)-BiP-misfolded protein and the peptide handover. Once the handover is finished, the complex will be dissociated and P58(IPK) will be released for misfolded protein catching of next cycle.
Materials and Methods
Structure determination of mouse P58(IPK) TPR fragment
The cloning, expression and crystallization of mouse P58(IPK) TPR fragment (residues 33–393) have been previously described 25. The gene encoding mouse P58(IPK) TPR fragment (residues 33–393) without the N-terminal ER-targeting signal and the C-terminal J-domain was cloned into pET28b. The recombinant protein from E. coli was purified by use of a metal-chelating column followed by gel filtration column. The P58(IPK) fragment protein was crystallized using hanging drop vapor diffusion method. Large plate-shaped crystals (0.3 × 0.3 × 0.05mm) were obtained using Linbro plates at 4°C. The well solution consisted of 1ml 100mM Hepes buffer (pH 7.0), 15% PEG 5000 MME.
The crystal structure of P58(IPK) TPR fragment was determined using SAD method. The Se-Met P58(IPK) TPR fragment was obtained using standard protocols. The diffraction data for P58(IPK) crystals was collected at SER-CAT beam line 22-ID in APS. The crystal was flash frozen at 100K in a Nitrogen gas stream in the cryoprotectant consisting of 100mM Hepes buffer (pH 7.0), 15% PEG5000 MME and 20% Glycerol. Both of the native and Se-Met P58(IPK) fragment crystals diffracted X-ray to 2.5 Å at SER-CAT. The data were processed using HKL2000 38. The crystals belong to space group of P21 with unit cell parameters of a=84.04Å, b=93.19Å, c=84.41A, α=90.00°, β=119.50°, γ=90.00°. The details of the data set are shown in table I.
Table 1.
Data collection, phasing and refinement statistics for P58(IPK) TPR fragment structure
| Se-Met | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 84.041, 93.186, 84.412 |
| α, β, γ (°) | 90, 119.500, 90 |
|
Peak |
|
| Wavelength(Å) | 0.9789 |
| Resolution (Å) | 2.5 |
| Rsym or Rmerge | 0.060 (0.184) |
| I/sigmaI | 20.7 (4.57) |
| Completeness (%) | 89.4 (89.2) |
| Redundancy | 5.6 (5.3) |
| Refinement | |
| Resolution (Å) | 2.5 |
| No. reflections | 33419 |
| Rwork/Rfree | 23.8/29.6 |
| No. atoms | |
| Protein | 5762 |
| Water | 106 |
| B-factors | |
| Protein | 41.6 |
| Water | 42.9 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.118 |
Highest-resolution shell is shown in parentheses.
Rsym=[ΣhklΣi|Ii−<I>|]/ΣhklΣi <I>
Twelve Selenium atoms were found and phases were calculated by using the program SOLVE 39. The phases from the SAD phasing were further improved by two-fold NCS molecular averaging and solvent flattening by use of the program RESOLVE 40. The resultant electron map had continuous electron density and was readily to interpret. Residues 35–393 of P58(IPK) were modeled into the electron density map with program COOT 41. The model was refined by program CNS 42. Six cycles of positional and annealing refinement were then carried out. Restrained individual B-factor refinement was not performed until the last cycle. After each cycle of refinement, the model was manually rebuilt according to the resultant 2Fo-Fc and Fo-Fc maps. TLS refinement was carried out by use of program Refmac 43. The refinement gave reasonable R.M.S. derivation from the ideal geometry at this resolution (Table I). A Ramachandran plot of the final model was generated by use of program Procheck and revealed that 100% of the residues were in allowed regions.
Misfolded protein binding assay to measure P58(IPK) ’s chaperone activity by ELISA
The binding between P58(IPK) TPR domain and the misfolded polypeptide is examined by ELISA. Wild type or mutant P58(IPK) TPR domains (15ug) are coated on the surface of 96-well plates by incubation for 1 hour at room temperature while rocking. The coated plate is washed with PBS and then blocked using 5% BSA overnight at 4°C. Luciferase (Promega) and Rhodanese (Sigma) are denatured in 8 M Urea in the presence of 1 mM DTT for 1 hour. The denatured polypeptides are diluted 100 fold into each well (0.5ug Luciferase/Rhodanese per well) containing 200ul blocking buffer. After 1 hour binding, the plates are washed with PBST (PBS with 0.05% Tween-20) for 5 times. Polyclonal anti-luciferase antibody from goat (Promega) and polyclonal anit-rhodanese antibody from goat (Santa Cruz) are diluted 2000 fold in PBST and 50 ul of the primary antibodies are added into each well. After incubation at room temperature for 1 hour, the plates are washed with PBST for 5 times. AP-conjugated donkey anti-goat antibody (Promega) is diluted 2000 fold in PBST and added into each well followed by 1 hour incubation at room temperature and washing with PBST for 5 times. The AP substrate is prepared immediately before use and 200 ul of the substrate solution is added into each well. After incubation in dark for 15 min, 50 ul 3M NaOH is added into each well to stop reaction. The plates are read at 405 nm on a multiwell plate reader.
Structure-based mutagenesis
Deletion mutants of P58(IPK) were generated by polymerase chain reaction (PCR). The resultant deletion mutation constructs include domain I (residues 33–168), domain I+II (residues 33–267). Mis-sense mutagenesis was performed by use of quick-change kit from Stratagene.
Coordinates
The coordinates and structure factors of P58(IPK) TPR fragment have been deposited to Protein Data Bank with an accession number of 3IEG.
Supplementary Material
Acknowledgments
We are grateful to the staff scientists in APS beamline SER-CAT for their help in data collection. This work was supported by grants from NIH (R01 DK56203), NIH (R01 GM080261) and Army Research Office (51894LS) to BDS and DK47119 and ES08681 to DR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–9. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marciniak SJ, Garcia-Bonilla L, Hu J, Harding HP, Ron D. Activation-dependent substrate recruitment by the eukaryotic translation initiation factor 2 kinase PERK. J Cell Biol. 2006;172:201–9. doi: 10.1083/jcb.200508099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang SW, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–7. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 8.Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci. 2007;32:469–76. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–62. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, Oyadomari M, Harding HP, Goodman AG, Harant H, Garrison JL, Taunton J, Katze MG, Ron D. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–39. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 13.Ladiges WC, Knoblaugh SE, Morton JF, Korth MJ, Sopher BL, Baskin CR, MacAuley A, Goodman AG, LeBoeuf RC, Katze MG. Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54:1074–81. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- 14.Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell. 2007;18:3681–91. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–72. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barber GN, Thompson S, Lee TG, Strom T, Jagus R, Darveau A, Katze MG. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci U S A. 1994;91:4278–82. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99:15920–5. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Huizen R, Martindale JL, Gorospe M, Holbrook NJ. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J Biol Chem. 2003;278:15558–64. doi: 10.1074/jbc.M212074200. [DOI] [PubMed] [Google Scholar]
- 19.Ron D, Harding HP. eIF2a phosphorylation in cellular stress-responses and disease. In: Hershey JWBMM, Sonenberg N, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Press; 2007. [Google Scholar]
- 20.Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. Embo J. 1998;17:1192–9. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–81. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 22.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–66. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 23.Melville MW, Tan SL, Wambach M, Song J, Morimoto RI, Katze MG. The cellular inhibitor of the PKR protein kinase, P58(IPK), is an influenza virus-activated co-chaperone that modulates heat shock protein 70 activity. J Biol Chem. 1999;274:3797–803. doi: 10.1074/jbc.274.6.3797. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, Maruyama D, Endo T, Nishikawa S. Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol. 2008;49:1547–62. doi: 10.1093/pcp/pcn119. [DOI] [PubMed] [Google Scholar]
- 25.Tao J, Wu Y, Ron D, Sha B. Preliminary X-ray crystallographic studies of mouse UPR responsive protein P58(IPK) TPR fragment. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:108–10. doi: 10.1107/S1744309108000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–7. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol. 2006;13:589–93. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- 28.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 29.Wu B, Li P, Liu Y, Lou Z, Ding Y, Shu C, Ye S, Bartlam M, Shen B, Rao Z. 3D structure of human FK506-binding protein 52: implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc Natl Acad Sci U S A. 2004;101:8348–53. doi: 10.1073/pnas.0305969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–38. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Gatto GJ, Jr, Geisbrecht BV, Gould SJ, Berg JM. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol. 2000;7:1091–5. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- 32.Grizot S, Fieschi F, Dagher MC, Pebay-Peyroula E. The active N-terminal region of p67phox. Structure at 1.8 A resolution and biochemical characterizations of the A128V mutant implicated in chronic granulomatous disease. J Biol Chem. 2001;276:21627–31. doi: 10.1074/jbc.M100893200. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Qian X, Hu J, Sha B. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J Biol Chem. 2009;284:23852–9. doi: 10.1074/jbc.M109.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demand J, Luders J, Hohfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol. 1998;18:2023–8. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 36.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–6. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 37.Jin Y, Zhuang M, Hendershot LM. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry. 2009;48:41–9. doi: 10.1021/bi8015923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski Z, WM Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55 (Pt 4):849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–72. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54 (Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.