Abstract
Non-coding RNAs (ncRNAs) with gene regulatory functions are starting to be seen as a common feature of mammalian gene regulation with the discovery that most of the transcriptome is ncRNA. The prototype has long been the Xist ncRNA, which induces X-chromosome inactivation in female cells. However, a new paradigm is emerging – the silencing of imprinted gene clusters by long ncRNAs. Here, we review models by which imprinted ncRNAs could function. We argue that an Xist-like model is only one of many possible solutions and that imprinted ncRNAs could provide the better model for understanding the function of the new class of ncRNAs associated with non-imprinted mammalian genes.
Genomic imprinting: an epigenetic gene regulatory mechanism
Mammals are diploid organisms carrying two alleles of each autosomal gene, one inherited from the mother and one from the father. Whereas in most cases both parental alleles are expressed equally, a subset of genes show genomic imprinting, in which expression is restricted by an epigenetic mechanism (see Glossary) to either the maternal or the paternal allele. Genomic imprinting is generally conserved in mice and humans, but because of the ease of genetic manipulations in mice more details are known for mouse imprinted genes, on which we focus here.
About 80 mouse genes, most of which are grouped in clusters containing 3–15 genes, are subject to imprinting [1,2]. The genes contained in an imprinted cluster are mainly protein-coding genes; however, one gene always encodes a noncoding RNA (ncRNA). Imprinted expression of genes in a cluster is controlled by one cis-acting imprint control element (ICE) [3,4]. Importantly, it is the ICE and not the imprinted genes themselves that carries parental information in the form of a DNA methylation imprint, which is acquired during male or female gametogenesis and maintained only on one parental allele after fertilization when the embryo is diploid. Six well-characterized clusters named after their founding imprinted gene are known, of which two – Igf2 (insulin-like growth factor 2) and Dlk (delta-like 1 homologue) – are paternally imprinted (i.e. modified by a DNA methylation imprint acquired in the male gamete), and four – Igf2r (insulin-like growth factor 2 receptor), Kcnq1 (potassium voltage-gated channel, KQT-like subfamily, member 1), Gnas (guanine nucleotide binding protein a stimulating) and PWS-AS (Prader–Willi and Angelman syndromes; this cluster is named after the human syndromes associated with its deletion) – are maternally imprinted (i.e. modified by a DNA methylation imprint acquired in the female gamete [5]).
Significantly, the parental chromosome carrying the unmethylated ICE is the one that expresses the ncRNA, indicating that the ICE is a positive regulator of ncRNA expression [3]. The position of the ICE varies with respect to the ncRNA promoter: in some clusters it lies a few kilobase pairs upstream of the ncRNA promoter [6,7]; in one it lies within the ncRNA transcription unit [8-9] and in others it lies at the promoter [10-12]. These variations indicate that the ICE might function in different ways to activate ncRNA expression. In all six imprinted clusters, expression of the ncRNA correlates with the repression in cis of some or all of the imprinted protein-coding genes [13,14]. This has given rise to the expectation that expression of the ncRNA would silence the multiple mRNA genes in an imprinted cluster. To date, only three imprinted ncRNAs have been functionally tested (Figure 1 and Box 1). Of these, the H19 (hepatic cDNA clone 19) ncRNA in the Igf2 cluster has no major silencing role [15]. By contrast, the Air (antisense Igf2r RNA) ncRNA in the Igf2r cluster and the Kcnq1ot1 (Kcnq1 overlapping transcript 1) ncRNA in the Kcnq1 cluster have both been shown to have a major silencing role by experiments that truncated these ncRNAs to 3% of their length [16,17]. Although the Air, Kcnq1ot1 and H19 ncRNAs are positively regulated by the ICE and share reciprocal expression with the imprinted mRNA genes in the cluster, they have several differences (Box 2).
Figure 1.
Genomic organization of three imprinted mouse clusters containing ncRNAs tested for their silencing function. Only a few genes in each cluster show widespread imprinted expression in extra-embryonic, embryonic and adult tissues, and most genes in a cluster show imprinted expression only in extra-embryonic tissues; thus, the expression pattern in this cell lineage is illustrated. (a) The paternally imprinted Igf2 cluster spans ~80 kb and contains the 2.5-kb H19 ncRNA [68]. Igf2 and H19 show ubiquitous imprinted expression, whereas Ins2 shows imprinted expression only in the extra-embryonic visceral yolk sac. Imprinted expression operates through the ICE, which is itself regulated by a DNA methylation imprint. On the maternal chromosome (pink), the CTCF protein binds to the unmethylated maternal ICE and forms an insulator that blocks the interaction of downstream enhancers to the Igf2 and Ins2 promoters but allows their interaction with the H19 ncRNA promoter. On the paternal chromosome (blue), the DNA methylation imprint blocks CTCF binding. Thus, the enhancers interact with the Igf2 and Ins2 promoters [66]. (b) The maternally imprinted Kcnq1 cluster spans ~800 kb and contains the Kcnq1ot1 ncRNA [16]. The Kcnq1ot1 promoter lies in an antisense orientation in the ICE in intron 10 of the Kcnq1 gene; its full length is unknown, as is whether it overlaps the Kcnq1 promoter. Cdkn1c and the Kcnq1ot1 ncRNA show ubiquitous imprinted expression, whereas the remainder show only limited imprinted expression in the early embryo or in extra-embryonic tissues. Imprinted expression operates through the ICE that contains the Kcnq1ot1 ncRNA promoter. On the paternal chromosome, the Kcnq1ot1 ncRNA is expressed and eight imprinted mRNA promoters are repressed. On the maternal chromosome, the methylated ICE represses the Kcnq1ot1 ncRNA promoter, enabling expression of the eight imprinted mRNAs only on this chromosome [4]. In this cluster, two genes (Nap1/4 and Phemx) show expression from both alleles, although expression of the paternal is less than that of the maternal allele [42]. (c) The maternally imprinted Igf2r cluster spans ~500 kb and contains the 108-kb Air ncRNA [67]. The Air promoter lies in antisense orientation in the ICE in Igf2r intron 2. In this cluster, Igf2r and the Air ncRNA show ubiquitous imprinted expression, whereas the remainder of the genes in the cluster show imprinted expression only in extra-embryonic tissues. Imprinted expression operates through the ICE that contains the Air ncRNA promoter. On the paternal chromosome, the Air ncRNA is expressed and three imprinted mRNA promoters are repressed. On the maternal chromosome, the methylated ICE represses the Air ncRNA promoter, enabling expression of the three imprinted mRNAs only on this chromosome [5]. Gene loci (not to scale) for which the gene name is not given in full are abbreviated as follows: Asc, Ascl2; Cd8, Cd81; Cdk, Cdkn1c; Nap, Nap1/4; Osb, Osbpl5; Phe, Phemx; Phl, Phlda2; Slc, Slc22a18; Trp, Trpm5; Tss, Tssc4.
Box 1. Functional testing of imprinted ncRNAs.
Igf2 cluster
The paternally imprinted Igf2 cluster contains the 2.5-kb spliced H19 ncRNA. Its function was tested by generating a 5-kb deletion of the gene body and promoter [15]. In the absence of H19 expression, the targeted maternal chromosome could maintain silencing of Igf2 in liver, although some loss of silencing was seen in muscle tissue. Ins2 expression was not tested in this experiment. This shows that the H19 ncRNA does not have a major silencing role, despite showing reciprocal parental-specific expression with the silent Igf2 and Ins2 genes. Imprinted expression in this cluster operates through the ICE, which contains a methylation-sensitive insulator that controls the interaction of downstream enhancers with the Igf2 and Ins2 promoters on the two parental alleles. However, the ICE has a dual function because it is also an activator for the H19 ncRNA promoter [6].
Kcnq1 cluster
The maternally imprinted Kcnq1 cluster contains the long Kcnq1ot1 ncRNA. Its function was tested by insertion of a poly(A) cassette that truncated it to 1.5 kb [16]. Maternal chromosomes carrying the truncated allele maintained the DNA methylation imprint and showed no changes in imprinted expression. However, paternal chromosomes expressing the truncated ncRNA showed full re-expression of silenced flanking imprinted genes in both embryonic and extraembryonic tissues. Thus, the Kcnq1ot1 ncRNA acts as a bidirectional silencer of multiple imprinted genes.
Igf2r cluster
The maternally imprinted Igf2r cluster contains the 108-kb unspliced Air ncRNA. Its function was tested by insertion of a poly(A) cassette that truncated it to 3 kb [17]. Maternal chromosomes carrying the truncated allele maintained the DNA methylation imprint and showed no changes in imprinted expression. Paternal chromosomes carrying the truncated ncRNA showed full re-expression of silenced flanking imprinted genes in both embryonic and extraembryonic tissues. Thus, the Air ncRNA acts as a bidirectional silencer of multiple imprinted genes.
Box 2. Characteristics of the Air, Kcnq1ot1 and H19 imprinted ncRNAs.
Position and orientation
The Air and Kcnq1ot1 promoters lie in antisense orientation in an intron of one of the imprinted mRNA genes in the cluster. The ncRNA transcript overlaps part of this gene and has potential to form dsRNA with exonic or intronic sequences. The H19 promoter lies in a sense orientation 80 kb downstream from the two imprinted mRNAs genes in this cluster.
Processing
The majority of the 108-kb Air transcripts are unspliced [36]. As a consequence, Air is a repeat-rich ncRNA because it contains the normal genome complement of transposons and other repetitive sequences [67]. By contrast, the H19 ncRNA comprises single-copy sequences and is fully spliced (splicing of the Kcnq1ot1 ncRNA has not been tested).
Transcript size
The Air ncRNA is 108 kb and the Kcnq1ot1 ncRNA is at least 60 kb (but has not been accurately sized); H19 is 2.5 kb [16,67,68].
Cellular localization
The Air ncRNA is nuclear localized, whereas the H19 ncRNA is efficiently exported to the cytoplasm [36] (Kcnq1ot1 has not yet been tested).
Silencing function
The Air and Kcnq1ot1 ncRNAs can silence multiple genes in cis; the H19 ncRNA lacks this function [15-17].
The Air and the Kcnq1ot1 ncRNAs belong to a growing class of ‘long’ ncRNAs shown to have a cis-restricted gene regulatory role in the mammalian genome. Members of this class include: the Xist (X-inactive specific transcript) and Tsix (X-inactive specific transcript, antisense) ncRNAs that control X-chromosome inactivation [18], and the intergenic ncRNA that might regulate β-globin expression [19]. Apart from these well-known examples, recent studies have shown that the majority of the mammalian transcriptome comprises untranslated transcripts, suggesting that many more ncRNAs might be involved in gene regulation [20]. In this context imprinted ncRNAs seem to be an ideal model to study this new level of gene regulation because they control expression of small groups of flanking genes. Although many features of the silencing effect generated by imprinted ncRNAs are known (Box 3), the actual mechanism is not yet clear. In particular, we still do not know whether imprinted ncRNAs silence through the transcript itself or through the act of transcription. Here we review models whereby ncRNA silencing could operate in different ways: through the ncRNA product, by the act of its transcription, or by regulating higher-order chromatin structure. Most of these proposed models are speculative, but are nevertheless useful at the moment because they might help to design the right strategy to investigate how ncRNAs act as silencers.
Box 3. Silencing features of imprinted ncRNAs.
Silencing is strictly a cis effect on the chromosome expressing the ncRNA.
Silencing extends in both directions relative to the ncRNA transcription start, affecting multiple genes.
Silencing affects genes overlapped by the ncRNA and nonoverlapped genes several hundred kilobase pairs distant.
ncRNA promoters and some mRNAs within the cluster escape silencing.
Silencing is regulated in development and in a tissue-specific manner.
Silencing effects are epigenetic and thus mitotically heritable and reversible.
‘Silencing’ describes repressive effects of imprinted ncRNAs on flanking mRNA genes, including repression that decreases but does not fully silence gene expression.
ncRNA-based silencing models
In this section, we review evidence that the ncRNA product itself is used to silence multiple imprinted genes in cis and discuss RNAi or RNA-directed targeting models.
Is RNAi involved in the silencing of imprinted genes?
RNA interference (RNAi) is a gene-silencing mechanism based on the formation of double-stranded (ds) RNA intermediates that are complementary to the silenced gene (Figure 2a) [20]. The dsRNA intermediates can be used for gene repression in multiple ways – by inhibiting translation, by inducing mRNA degradation or by promoter silencing through heterochromatin formation [21]. An RNAi-based silencing mechanism would be possible for imprinted ncRNAs if they shared sequence homology with silenced imprinted genes. The Air and Kcnq1ot1 ncRNA promoters are present in an antisense orientation inside an intron of one of the silenced genes in their respective clusters. Thus, it is possible that low-level transcription from the ‘silenced’ gene creates a sense-antisense overlap that would enable the formation of dsRNA and attract the RNAi machinery as previously suggested [16,22].
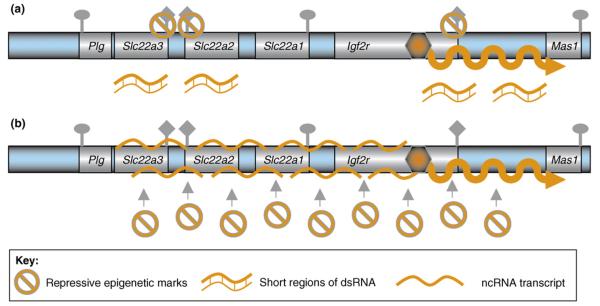
Figure 2.
Gene-silencing models for the imprinted Igf2r cluster based on a role for the ncRNA product. Only the paternal expression pattern in extra-embryonic tissues is shown, in which the Air ncRNA is expressed and Igf2r, Slc22a2 and Slc22a3 are silenced. The remaining genes show tissue-specific silencing in this tissue. (a) RNAi-dependent silencing: the expressed ncRNA triggers RNAi by generating dsRNA, which subsequently induces transcriptional silencing at homologous regions in the cluster. dsRNA might arise either because of the transcriptional overlap with the ncRNA and a protein-coding gene that is found in both the Igf2r and Kcnq1 imprinted clusters, or from intrinsic properties of the ncRNA (i.e. the presence of inverted repeats), or from pairing with RNAs originating from interspersed repeats that are common to introns in all genes in the cluster. The Air ncRNA is unspliced and thus is rich in interspersed repeats. (b) RNA-directed targeting: the expressed ncRNA localizes to the chromosome that expresses it and can spread over the region containing silenced genes. Spreading could be limited by an absence of positive spreading signals on autosomes or be due to some type of insulator that blocks spreading. The ncRNA is subsequently able to recruit epigenetic silencing modifications in the form of DNA methylation, histone modifications or histone variants in a manner similar to that shown for the Xist ncRNA that mediates X-chromosome inactivation.
There are, however, two challenges to the feasibility of this model. First, because the other parental allele is present in an active form in the same nucleus, it is necessary for the RNAi machinery, traditionally perceived as a trans-acting cytoplasmic-based mechanism, to act in cis in the nucleus. An example of cis-restriction of the RNAi machinery has been described in nuclei of fission yeast Schizosaccharomyces pombe. Here a nuclear RNAi effector complex called RNA-induced transcriptional silencing (RITS) is involved in the formation of heterochromatin at the mating-type locus and centromeres [23]. There is also increasing evidence of an RNAi mechanism operating in the mammalian nucleus that can modify transcription and organize centromeric heterochromatin [19,24]. However, so far there is no direct evidence for a functional cis-restricted nuclear RITS complex in mammals [21].
The second challenge to an RNAi model is to consider how it can induce silencing of nonoverlapped genes that are located hundreds of kilobase pairs away on either side of the imprinted ncRNA. One possibility is that silencing occurs in two steps [22]. The first step would involve silencing of the overlapped gene by an RNAi mechanism. The second step would involve spreading the silent state to cover the domain of imprinted genes. However, silencing at the Igf2r cluster does not support a two-step silencing model, because silencing of nonoverlapped imprinted genes was shown to be independent of previous silencing of the overlapped gene [25]. As a second possibility, imprinted ncRNAs could contain short repetitive sequences that would form local hairpin structures that trigger the RNAi machinery [26]. Alternatively the ncRNA could form dsRNA with opposite-sense transcripts arising from within its transcribed region. Interspersed repeats could serve this function because they contain sequences reported to function as promoters [27]. To target the RNAi machinery, all imprinted genes in a cluster would need to share sequence similarities, a feature that has not been identified so far. However, repetitive elements, which make up 50% of the mouse genome and are abundant in ncRNAs and introns of mRNA genes, could serve as such common target regions.
Although small ncRNAs have been identified in imprinted gene clusters, there is no evidence that they are involved in imprinted gene silencing [28]. Nor are there supportive data from mice carrying mutations in the RNAi machinery [29]. However, definitive experiments have not yet been performed and despite a lack of supportive data, RNAi involvement cannot yet be excluded from imprinted gene silencing.
ncRNA-directed targeting: similarities to Xist function?
The first mammalian ncRNA shown to have a gene silencing function was the Xist ncRNA, which inactivates one of the two X chromosomes in female mammals [30]. The overall model of Xist function is that of RNA-directed targeting. In this model the Xist ncRNA is expressed and coats in cis the inactive X chromosome; this correlates with the attraction of multiple repressive layers of histone modifications, histone variants and DNA methylation that result in chromosomal inactivation [31]. This model is most easily shown for the imprinted Igf2r cluster, which only contains three imprinted protein-coding genes (Figure 2b), but is equally applicable to the more complex Kcnq1 cluster. The obvious parallel between genomic imprinting and X-chromosome inactivation, namely that both result in homologous sequences behaving differently in the same nucleus, led to an early suggestion that these were related phenomena [32]. More recently, the demonstration that functional cis-acting long ncRNAs lie at the heart of the silencing process for X-inactivation and for some imprinted clusters, has led to the suggestion that both processes share a common origin [33,34]. However, despite these strong similarities there are obvious differences, the most significant being that genomic imprinting affects gene clusters containing 3–15 genes, whereas X-chromosome inactivation affects the whole of the 180-Mbp X chromosome. In addition, although the existence of an imprinted form of X-inactivation in marsupials and in the extraembryonic tissues of mice initially strengthened the parallel between these two processes, it is likely that imprinted X-inactivation in marsupials occurs independently of a Xist homologous ncRNA [35]. Thus, the possibility needs to be considered that X-chromosome inactivation and genomic imprinting act through different mechanisms.
One indication that these mechanisms could differ is the structure of the ncRNA at the heart of each process. The Xist ncRNA is structurally dissimilar to the known functional imprinted ncRNAs Air and Kcnq1ot1; Air is mainly unspliced whereas the spliced form of Xist confers its silencing function [36,37]. Surprisingly, Xist is structurally similar to the nonfunctional H19 ncRNA in that both contain large exons and small introns, the opposite pattern to that displayed by mammalian protein-coding genes [38]. Xist is part of a genomic region containing multiple ncRNAs, including the Tsix ncRNA that regulates Xist expression in cis [39]. Tsix is similar to functional imprinted ncRNAs; it is antisense, overlaps the Xist promoter and makes a partly unspliced 40-kb repeat-rich ncRNA, and thus might offer a better parallel for functional imprinted ncRNAs [36,40].
Do imprinted ncRNAs function in a manner similar to Xist or Tsix? We do not know enough about the Tsix silencing mechanism to make a comparison beyond the correlation between Tsix expression and silencing of the cis-linked Xist promoter [39]. Functional similarity to the Xist ncRNA would require the demonstration that imprinted ncRNAs coat the region containing silenced genes and recruit silencing factors. Several imprinted clusters have been shown to contain repressive histone modifications that correlate with ncRNA expression but causality has not been demonstrated [41-44]. Thus, despite some similarities between these two silencing systems, further work is needed to test if X-chromosome inactivation is recapitulated in miniature at imprinted gene clusters.
Transcription-based silencing models
In this section, we consider the possibility that ncRNA transcription per se leads to silencing of multiple flanking mRNA genes. In these models the ncRNA product is dispensable. An early transcription-based model, named ‘expression-competition’, proposed that ncRNA and mRNA promoters could compete in cis for a common regulatory element [45]. However, the demonstration that truncated forms of the Air and Kcnq1ot1 ncRNAs can be fully expressed in cis with target mRNA genes, argues that imprinted mRNA and ncRNA promoters either do not compete, or are regulated by different cis-regulatory elements [16,17].
Transcriptional interference (TI), where the elongating polymerase from one gene directly interferes with transcription initiation or elongation of an overlapped gene, has been recently reviewed [46]. In one example from human α-thalassemia, a chromosomal deletion juxtaposes two genes and enables transcription from the LUC7L gene to run through the HBA2 (hemoglobin a2) promoter, resulting in silencing and gain of DNA methylation [47]. Silencing by ncRNAs in imprinted gene clusters affects multiple genes, only one of which is overlapped by the ncRNA. Thus, a direct TI mechanism cannot operate in imprinted gene clusters. Instead, we propose that ncRNA transcription leads to TI or to TA (transcriptional activation) of a cis-regulatory element that can regulate all affected mRNA promoters in an imprinted cluster, but, has no effect on the ncRNA promoter. Figure 3 shows three types of cis-regulatory elements (activators, silencers and insulators) that could be affected by TI or TA and induce imprinted silencing. Common to all is the location of the cis-regulatory element within the transcription unit of the ncRNA.
Figure 3.
Transcription-based silencing models that do not require the ncRNA product itself, but only its transcription. (a) Transcription interference of cis-regulatory activator elements: ncRNA transcription displaces binding proteins from an activator that lies in the ncRNA gene body. This activator must be able to activate multiple genes, so is comparable to a locus control region (LCR) or could be considered a ‘domain’ activator. ncRNA transcription interferes with activator function by displacing binding proteins needed for long-range activation of all the imprinted protein-coding genes in the cluster. (b) Transcriptional activation of cis-regulatory silencing elements: ncRNA transcription activates a silencer (located in the ncRNA gene body) by enabling the binding of long-range silencer proteins that repress all imprinted protein-coding genes in the cluster. (c) Transcriptional activation of insulator elements: ncRNA transcription is proposed to activate an insulator or boundary element (located in the ncRNA gene body), which then enables binding of an insulator protein. Implicit in this model is that a domain activator needed for expression of all imprinted genes in the cluster (except the ncRNA) lies on the other side of the insulator element. Formation of the insulator prevents interaction between the domain activator and the imprinted genes.
TI of cis-regulatory activator elements
In this model (Figure 3a), a cis-acting activator element required for expression of multiple genes in the imprinted cluster is located within the ncRNA transcription unit. This element is functional when it interacts with binding proteins, which then leads to gene activation. However, ncRNA transcription perturbs this interaction leading to gene silencing. Although we have no direct knowledge of such a cis-acting activator element we imagine it to be analogous to enhancers or locus control regions that are remote from genes but are involved in recruiting RNA polymerase (RNAP) or chromatin modifying complexes to the promoters they activate [48].
TA of cis-regulatory silencer elements
In this model (Figure 3b), a cis-acting silencer element is located within the ncRNA transcription unit whose interaction with silencer binding proteins requires ncRNA transcription. This could be mediated by ncRNA transcription opening up chromatin to enable silencer proteins to interact with DNA elements. These silencer-binding proteins, similar to the activator-binding proteins described above, are imagined to act on all silenced mRNA genes in an imprinted cluster. Silencer proteins are good candidates for silencing in imprinted gene clusters because some, for example Polycomb complexes that bind locally and induce heterochromatin formation over a broad domain, could fulfill the requirement of repressing multiple promoters in an imprinted cluster [49-51]. A key feature of this model is the use of ncRNA transcription to make DNA accessible to binding proteins. To date there is no direct evidence for this; however, this model was suggested to explain the role of intergenic transcription at the β-globin and the immunoglobulin heavy chain V region multigene clusters [52,53].
TA of cis-regulatory insulator elements
In this model (Figure 3c), a cis-acting insulator element, located within the ncRNA transcription unit, requires ncRNA transcription for interaction with insulator binding proteins. Insulators form structures that block interactions between transcription activators and promoters. Furthermore, because insulators could block multiple activators, they could also operate if they are flanked on both sides by target genes, provided they lie between the promoter and its activator. In the model shown in Figure 3c, the insulator is located within the ncRNA transcription unit, but on one side of all silenced genes. The blocked activator lies upstream of the insulator. Insulators have been found to have important roles in the regulation of some genes [54]. Importantly, the best example of a mammalian insulator is found in the Igf2 cluster (Figure 1). In this cluster, binding of the CCCTC-binding factor (CTCF) protein occurs on the unmethylated ICE, which has recently been shown to be an activator for H19 ncRNA transcription [6]. This provides a tentative link between ncRNA transcription and the binding of insulator proteins.
Higher-order chromatin silencing models
The types of models considered above are to a large extent linear silencing models. However, increasing attention is now being paid to the way that chromosomal spatial organization, which includes three-dimensional structure and position within a nucleus, is used as an expression regulatory mechanism. Two recently developed techniques, termed ‘chromatin-conformation-capture’ (3C) and ‘tagging and recovery of associated proteins’ (RNA-TRAP) have provided evidence that distant enhancers can physically interact with active promoters in the β-globin cluster – the intervening DNA is proposed to form a loop-like structure [55,56]. These higher-order chromatin structures form the basis of the two silencing models considered below.
Transcription-based looping model
In this model (Figure 4a), a cis-acting activator required for expression of multiple genes in the imprinted cluster is located within the ncRNA transcription unit. ncRNA transcription induces the formation of a chromatin loop containing the activator, which effectively isolates it and prevents its interaction with promoters. In the absence of activator–promoter interactions the imprinted mRNA genes are silenced. In addition to chromatin looping between enhancers and promoters at the β-globin locus, studies in Saccharomyces cerevisiae have shown that 5′ and 3′ processed ends of RNAPII transcribed genes physically interact by looping [57]. Other experiments, in which a simian virus 40 (SV40) enhancer was separated from its promoter by an inducible loop, support silencing models by which looping can prevent gene activation [58]. A looping model that separates promoters from enhancers has also been proposed for the imprinted Igf2 cluster (Figure 1a) [59]. In the case of Igf2, looping was shown to be dependent on the unmethylated ICE. Together, these data indicate the feasibility of a model whereby ncRNA transcription or transcriptional activation together with 3′-end processing, could induce the formation of a chromatin loop that separates an activator from promoters.
Figure 4.
Higher order chromatin silencing models. (a) Transcription-based looping model: ncRNA transcription and processing is proposed to induce looping of the DNA through interactions between its 5′ and 3′ processed ends. The induced chromatin loop physically isolates a domain activator needed for expression of all imprinted protein-coding genes in the cluster. In this model, the sequence of the ncRNA is not important for silencing function. Thus, this model would also fit the category of transcription-based models. (b) ncRNA-induced compartment model, based on the ncRNA forming a compartment that does not favour mRNA transcription. This could occur for many reasons: for example, the ncRNA physically excludes access to a transcription-compartment, or it could act as a sink to sequester RNA polymerase II, or it could create a subnuclear compartment that favours unspliced, non-exported RNAs.
ncRNA-induced compartment models
There is growing evidence that the mammalian nucleus is organized into distinct subnuclear compartments enriched for activities that transcriptionally or post-transcriptionally regulate gene expression or silencing [60-62]. As one example, shared nuclear-transcription compartments, or ‘transcription factories’, have been identified, that are enriched in active RNA polymerase and expressed genes [63,64]. These expressed genes can come from different chromosomes or from different territories on distant regions of the same chromosome. Together, these data offer support for the notion that the position of expressed and silent genes is organized within a nucleus. Could nuclear organization based on subnuclear compartments regulate imprinted gene expression?
ncRNA-induced compartment models could be based on segregating any transcriptional or post-transcriptional feature needed for expressing mRNA genes. For example, the recent demonstration that the Air ncRNA is inefficiently spliced could indicate that ncRNAs induce subnuclear compartments that inhibit expression of spliced mRNA genes [36]. It is also possible that RNAPII components are altered in transcription compartments containing active ncRNA promoters. Figure 4b shows a model in which ncRNA expression creates a transcription compartment that excludes mRNA processing components, and thus inhibits transcription of cohabiting mRNA genes. We do not have a watertight argument at this time to explain how ncRNA expression could inhibit transcription of mRNA genes in the same transcription compartment. One suggestion, based on their unusual length and tight association to the site of transcription, is that ncRNAs could physically exclude access to this compartment, or act as a sink to sequester RNAPII away from mRNA promoters in this compartment. This type of model predicts a limited number of transcription compartments available for genes in tissues that show imprinted expression, so that the flanking mRNA genes do not have access to other nearby transcription compartments more favorable for mRNA expression. Although this scenario might seem unlikely, because both the ncRNA and the mRNA genes depend on RNAPII and seem to have normal 5′ and 3′ processing, it has recently been shown that the Xist ncRNA induces the formation of an RNAPII-deficient compartment as the first step toward X-chromosome inactivation [65].
Conclusions and future questions
We have considered here imprinted ncRNAs and models by which they could exert cis-silencing effects on small gene domains. It is possible these models do not explain all features of gene silencing in all the imprinted clusters and also possible that combinations of these models operate in more complex imprinted clusters. For example, models need to explain: (i) how full expression of truncated ncRNAs does not induce silencing; (ii) how tissue-specific imprinted expression would operate; (iii) how foreign genes inserted into imprinted clusters are silenced; and (iv) how imprinted ncRNAs escape the silencing they induce. Despite this, the time is right to consider all possible models, because ncRNAs have been shown to be the direct cause of silencing in two imprinted gene clusters, and they might also act in other imprinted and nonimprinted gene clusters [14,16,17]. At this stage of our knowledge, all silencing models – both those considered here and models beyond our conception – are possible, because we lack the answers that would favor one of these models above the others. Box 4 lists several outstanding questions that would aid considerably in deciding which model, if any, fits the situation. In our opinion, the most important question at this time is to know whether ‘transcription is the answer’. An experiment that could shed light on this would be to post-transcriptionally downregulate imprinted ncRNAs and thus separate transcription from its product. Because this experiment might face technical difficulties as a result of their nuclear localization, it is equally important to perform more complete functional and biochemical characterizations of all imprinted ncRNAs – the better we know their characteristics, the better we will understand their mode of silencing.
Box 4. Outstanding issues.
Functional testing and biochemical characterization of imprinted ncRNAs is incomplete (full transcript length, percentage of spliced to unspliced transcripts, percentage of interspersed repeats, transcript stability and steady-state abundance, 5′ and 3′ processing, nuclear or cytoplasmic localization), with some exceptions [16,17,36].
Do imprinted ncRNAs silence through the ncRNA product or through transcription?
Do imprinted ncRNAs coat the chromosomal region containing the silenced mRNA genes?
Do imprinted ncRNAs directly or indirectly induce repressive epigenetic modifications on silenced genes?
Is the full-length imprinted ncRNA transcript needed for its silencing function?
Is the interspersed repeat content of functional imprinted ncRNAs necessary for silencing?
Can imprinted ncRNAs silence in all stages of development, or is there a developmental window of opportunity for silencing to occur?
Are imprinted ncRNAs localized to subnuclear compartments?
Do imprinted regions form 3D structures that differ between the parental chromosomes?
Acknowledgements
We thank all members of the Barlow laboratory for active discussion of ideas presented here. F.M.P., M.V.K. and D.P.B. are supported by: the European Union FW6 IP ‘HEROIC’ LSHG-CT-2005–018883 and the NoE ‘The Epigenome’ LSHG-CT-2004–053433, the Austrian bm.bmk GEN-AU ‘Epigenetic Plasticity of the Mammalian Genome’ GZ200.141/1-VI/2006 and the Austrian FWF-SFB-17 ‘Modulators of RNA Fate and Function’ SFBF01718(B10). The Barlow laboratory is supported by the Austrian Academy of Science.
Glossary
- Activator
a regulatory DNA element, usually referred to as an enhancer, that can bind to transcription factors and stimulate the transcription of one or more target genes that lie some distance away on the same chromosome [69].
- Chromatin conformation capture (3C)
a technique used to analyze the close physical proximity of two DNA sequences in vivo, e.g. the interaction between an activator and a promoter [55].
- DNA methylation
a heritable, covalent attachment of a methyl group to a DNA base, which in mammals is restricted to the C5 position of cytosine and normally only occurs on cytosines that are part of a CpG dinucleotide [70].
- Epigenetic
describing mitotically heritable but reversible modifications to DNA or chromatin that lead to changes in gene expression in the absence of genetic alterations [70].
- Imprint control element (ICE) or imprint control region (ICR)
a short CpG-rich DNA sequence that is methylated in only one of the two parental gametes and subsequently maintains this methylation imprint only on one parental allele after fertilization when the embryo is diploid. The unmethylated ICE acts as a long-range cis repressor for a cluster of imprinted genes; the methylated ICE lacks repressor function [70].
- Insulator
a regulatory DNA element that binds to specific proteins that block interactions between activators and promoters [54].
- Mating-type locus
a locus in yeast specifying the mating type and flanked by mating type genes that are silent and in a heterochromatic state. Site-specific recombination is used to switch the mating type to the opposite type as a prelude to sexual reproduction [70].
- Polycomb proteins
a class of chromatin-binding proteins originally defined by Drosophila mutants that cause stable and heritable repression of developmentally important gene clusters [70].
- Tagging and recovery of associated proteins (RNA-TRAP)
a technique used to identify chromatin regions lying in close physical proximity to an RNA [56].
- Silencer
a regulatory DNA element that can bind to repressor proteins and repress transcription of one or more target genes that lie some distance away on the same chromosome [69].
- X-chromosome inactivation
an epigenetic process achieving sex chromosome dosage compensation between female mammals that have two X chromosomes and males that have only one. In placental mammals X inactivation is initiated by the Xist ncRNA. In marsupial mammals this process could be initiated independently of Xist expression [70].
References
- 1.Verona RI, et al. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- 2.Barlow DP, Bartolomei MS. Genomic imprinting in mammals. In: Allis CD, et al., editors. Epigenetics. Cold Spring Harbor Laboratory Press; 2006. pp. 357–375. [Google Scholar]
- 3.Spahn L, Barlow DP. An ICE pattern crystallizes. Nat. Genet. 2003;35:11–12. doi: 10.1038/ng0903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis A, Reik W. How imprinting centres work. Cytogenet. Genome Res. 2006;113:81–89. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 5.Regha K, et al. The imprinted mouse Igf2r/Air cluster – a model maternal imprinting system. Cytogenet. Genome Res. 2006;113:165–177. doi: 10.1159/000090829. [DOI] [PubMed] [Google Scholar]
- 6.Thorvaldsen JL, et al. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol. Cell. Biol. 2006;26:1245–1258. doi: 10.1128/MCB.26.4.1245-1258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin SP, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 8.Le Meur E, et al. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev. Biol. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Bielinska B, et al. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet. 2000;25:74–78. doi: 10.1038/75629. [DOI] [PubMed] [Google Scholar]
- 10.Wutz A, et al. Non-imprinted Igf2r expression decreases growth and rescues the Tme mutation in mice. Development. 2001;128:1881–1887. doi: 10.1242/dev.128.10.1881. [DOI] [PubMed] [Google Scholar]
- 11.Fitzpatrick GV, et al. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 12.Williamson CM, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 2006;38:350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- 13.O'Neill MJ. The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum. Mol. Genet. 2005;14(Spec. No 1):R113–120. doi: 10.1093/hmg/ddi108. [DOI] [PubMed] [Google Scholar]
- 14.Pauler FM, Barlow DP. Imprinting mechanisms – it only takes two. Genes Dev. 2006;20:1203–1206. doi: 10.1101/gad.1437306. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt JV, et al. Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9733–9738. doi: 10.1073/pnas.96.17.9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mancini-Dinardo D, et al. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sleutels F, et al. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 18.Chang SC, et al. Mechanisms of X-chromosome inactivation. Front. Biosci. 2006;11:852–866. doi: 10.2741/1842. [DOI] [PubMed] [Google Scholar]
- 19.Haussecker D, Proudfoot NJ. Dicer-dependent turnover of intergenic transcripts from the human beta-globin gene cluster. Mol. Cell. Biol. 2005;25:9724–9733. doi: 10.1128/MCB.25.21.9724-9733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec. No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 21.Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell. 2005;122:13–16. doi: 10.1016/j.cell.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 22.Rougeulle C, Heard E. Antisense RNA in imprinting: spreading silence through Air. Trends Genet. 2002;18:434–437. doi: 10.1016/s0168-9525(02)02749-x. [DOI] [PubMed] [Google Scholar]
- 23.Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 24.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sleutels F, et al. Imprinted silencing of Slc22a2 and Slc22a3 does not need transcriptional overlap between Igf2r and Air. EMBO J. 2003;22:3696–3704. doi: 10.1093/emboj/cdg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 27.van de Lagemaat LN, et al. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Royo H, et al. Small non-coding RNAs and genomic imprinting. Cytogenet. Genome Res. 2006;113:99–108. doi: 10.1159/000090820. [DOI] [PubMed] [Google Scholar]
- 29.Fukasawa M, et al. Genomic imprinting in Dicer1-hypomorphic mice. Cytogenet. Genome Res. 2006;113:138–143. doi: 10.1159/000090825. [DOI] [PubMed] [Google Scholar]
- 30.Marahrens Y, et al. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 31.Thorvaldsen JL, et al. X-tra! X-tra! News from the mouse X chromosome. Dev. Biol. 2006;298:344–353. doi: 10.1016/j.ydbio.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Lyon MF. Epigenetic inheritance in mammals. Trends Genet. 1993;9:123–128. doi: 10.1016/0168-9525(93)90206-w. [DOI] [PubMed] [Google Scholar]
- 33.Lee JT. Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr. Biol. 2003;13:R242–R254. doi: 10.1016/s0960-9822(03)00162-3. [DOI] [PubMed] [Google Scholar]
- 34.Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 2005;6:403–410. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- 35.Duret L, et al. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 36.Seidl CI, et al. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wutz A, et al. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer K, Tilghman SM. Allele-specific gene expression in mammals: the curious case of the imprinted RNAs. Genes Dev. 1994;8:1867–1874. doi: 10.1101/gad.8.16.1867. [DOI] [PubMed] [Google Scholar]
- 39.Luikenhuis S, et al. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol. Cell. Biol. 2001;21:8512–8520. doi: 10.1128/MCB.21.24.8512-8520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa Y, Lee JT. Antisense regulation in X inactivation and autosomal imprinting. Cytogenet. Genome Res. 2002;99:59–65. doi: 10.1159/000071575. [DOI] [PubMed] [Google Scholar]
- 41.Lewis A, et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- 42.Umlauf D, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 43.Fournier C, et al. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 2002;21:6560–6570. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vu TH, et al. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum. Mol. Genet. 2004;13:2233–2245. doi: 10.1093/hmg/ddh244. [DOI] [PubMed] [Google Scholar]
- 45.Barlow DP. Competition – a common motif for the imprinting mechanism? EMBO J. 1997;16:6899–6905. doi: 10.1093/emboj/16.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shearwin KE, et al. Transcriptional interference – a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tufarelli C, et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 48.Dean A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 50.Gaston K, Jayaraman PS. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell. Mol. Life Sci. 2003;60:721–741. doi: 10.1007/s00018-003-2260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiel G, et al. How mammalian transcriptional repressors work. Eur. J. Biochem. 2004;271:2855–2862. doi: 10.1111/j.1432-1033.2004.04174.x. [DOI] [PubMed] [Google Scholar]
- 52.Ashe HL, et al. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolland DJ, et al. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 54.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 55.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat. Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 56.Chakalova L, et al. RNA fluorescence in situ hybridization tagging and recovery of associated proteins to analyze in vivo chromatin interactions. Methods Enzymol. 2004;375:479–493. doi: 10.1016/s0076-6879(03)75029-0. [DOI] [PubMed] [Google Scholar]
- 57.O'Sullivan JM, et al. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 58.Ameres SL, et al. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. EMBO J. 2005;24:358–367. doi: 10.1038/sj.emboj.7600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murrell A, et al. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 60.Foster HA, Bridger JM. The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma. 2005;114:212–229. doi: 10.1007/s00412-005-0016-6. [DOI] [PubMed] [Google Scholar]
- 61.Bartova E, Kozubek S. Nuclear architecture in the light of gene expression and cell differentiation studies. Biol. Cell. 2006;98:323–336. doi: 10.1042/BC20050099. [DOI] [PubMed] [Google Scholar]
- 62.Spector DL. SnapShot: cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 63.Bartlett J, et al. Specialized transcription factories. Biochem. Soc. Symp. 2006;73:67–75. doi: 10.1042/bss0730067. [DOI] [PubMed] [Google Scholar]
- 64.Fraser P. Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 65.Chaumeil J, et al. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gabory A, et al. The H19 gene: regulation and function of a non-coding RNA. Cytogenet. Genome Res. 2006;113:188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 67.Lyle R, et al. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- 68.Pachnis V, et al. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. U. S. A. 1984;81:5523–5527. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maston GA, et al. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–52. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 70.Allis CD, et al. Epigenetics. Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]