Abstract
The adenine and guanine riboswitches regulate gene expression in response to their purine ligand. X-ray structures of the aptamer moiety of these riboswitches are characterized by a compact fold in which the ligand forms a Watson–Crick base pair with residue 65. Phylogenetic analyses revealed a strict restriction at position 39 of the aptamer that prevents the G39–C65 and A39–U65 combinations, and mutational studies indicate that aptamers with these sequence combinations are impaired for ligand binding. In order to investigate the rationale for sequence conservation at residue 39, structural characterization of the U65C mutant from Bacillus subtilis pbuE adenine riboswitch aptamer was undertaken. NMR spectroscopy and X-ray crystallography studies demonstrate that the U65C mutant adopts a compact ligand-free structure, in which G39 occupies the ligand-binding site of purine riboswitch aptamers. These studies present a remarkable example of a mutant RNA aptamer that adopts a native-like fold by means of ligand mimicking and explain why this mutant is impaired for ligand binding. Furthermore, this work provides a specific insight into how the natural sequence has evolved through selection of nucleotide identities that contribute to formation of the ligand-bound state, but ensures that the ligand-free state remains in an active conformation.
INTRODUCTION
As other metabolite-sensing riboswitches, adenine and guanine riboswitches are natural biosensors that modulate gene expression in response to the concentration level of their cognate ligand (1–3). They are positioned in the 5′-UTR of bacterial mRNA and are composed of two overlapping domains: a purine-binding aptamer domain and a downstream expression platform. Binding of the ligand stabilizes the formation of a compact aptamer structure, which prevents the formation of an alternate structure and thereby modulates gene expression.
Depending on the expression platform composition, gene expression control takes place at the levels of transcription or translation, and ligand binding, either positively or negatively, regulates gene expression (1–3). For example, the xpt–pbuX guanine riboswitch (G-riboswitch) from Bacillus subtilis is a genetic switch that turns ‘off’ transcription (1). In the absence of guanine, the RNA forms an antiterminator structure, a default ‘on’ state that allows completion of transcription. Binding of guanine stabilizes the structures of the aptamer and adjacent rho-independent terminator, creating an ‘off’ state that results in repression of transcription. The xpt–pbuX operon encodes genes involved in purine metabolism, and riboswitch control ensures that these genes are not transcribed when the levels of guanine are elevated. By contrast, the pbuE (formerly named ydhL) adenine riboswitch (A-riboswitch) from B. subtilis is a genetic switch that turns ‘on’ transcription (Figure 1a) (2). In the absence of adenine, the expression platform adopts a stable rho-independent terminator stem-loop, a default ‘off’ state that prevents transcription. Binding of adenine stabilizes the aptamer domain at the expense of the terminator stem, and the riboswitch adopts an ‘on’ state that allows transcription. The pbuE gene encodes a purine efflux pump, and adenine sensing by the riboswitch allows gene expression of this pump, presumably to reduce intracellular purine concentrations. Thus, the nature of the expression platforms determines if purine binding will activate or repress the expression of a gene under the control of a purine riboswitch.
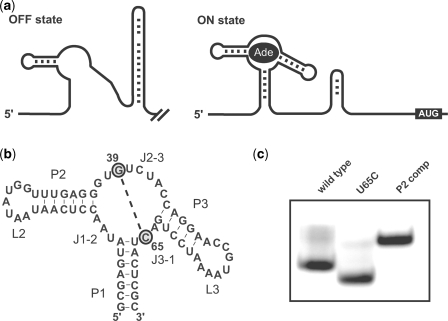
Figure 1.
Formation of a compact structure by the U65C mutant of the A-riboswitch aptamer. (a) Alternative base pairing of the B. subtilis pbuE A-riboswitch associated with the off and on states. The off state is characterized by the presence of a transcription terminator stem-loop, whereas the on state is characterized by the stabilization of the ligand-bound aptamer, which prevents formation of the terminator stem-loop and allows transcription to proceed. (b) Sequence and secondary structure of the U65C mutant A-riboswitch aptamer illustrating the proposed interaction between G39 and C65. (c) Native-gel electrophoresis of the wild-type A-riboswitch aptamer, the U65C mutant and a control loop mutant (P2 comp; see text) in the presence of 1 µM MgCl2.
Although the expression platforms vary, the aptamer domains of the A- and G-riboswitches are structurally very similar (4). The aptamer domains contain several conserved residues and a common three-way junction with two stem-loops (P2 and P3) and a closing stem (P1; Figure 1b) (1,2,5). Several three-dimensional structures determined by X-ray crystallography have revealed a characteristic compact fold. Structures of the xpt–pbuX G-riboswitch aptamer from B. subtilis (or related mutants) were determined in complex with several purine analogs (3,6–11), whereas the add A-riboswitch aptamer from Vibrio vulnificus was determined in complex with adenine (3). The key determinants of the purine aptamer architecture are a loop–loop interaction between L2 and L3, and a compact core that encloses the ligand through intricate hydrogen bonding and stacking interactions. At the center of the core, purine ligands form a riboswitch-specific Watson–Crick base pair with a key residue (guanine-C74 or adenine-U65) and participate in other hydrogen bonds with J2–3 residues [U38 and U42 according to the A-riboswitch numbering (12,13) used here] to constitute a base quadruple. This purine-bound base quadruple is stabilized on both sides by the stacking of two base triples, and together these five-staggered base layers compose the core of the aptamer.
The main difference between the A- and G-riboswitch aptamers is the residue at position 65, which dictates binding specificity. The G-riboswitch aptamer binds guanine and hypoxanthine with Kd values of 5 nM and 50 nM, respectively, and does not bind adenine [Kd > 300 000 nM (1)]. Likewise, the A-riboswitch aptamer binds 2,6-diaminopurine and adenine with Kd values of ∼10 nM and ∼300 nM, respectively, and does not bind guanine [Kd > 10 000 nM (2)]. Conversions of a guanine-sensing to an adenine-sensing riboswitch aptamer have been achieved by replacing C65 with a U (2,5,7,8). In contrast, a similar conversion of the B. subtilis pbuE adenine-sensing to a guanine-sensing riboswitch aptamer by a U65C mutation resulted in an aptamer impaired for purine binding (adenine, 2-aminopurine and guanine) (5). A functional correlation between positions 39 and 65 helped rationalize these results (5). Indeed, phylogenetic and mutational studies demonstrated that only G, C and U at position 39 are compatible with adenine binding to the A-riboswitch aptamer, whereas only A, C and U at position 39 are compatible with guanine binding to the G-riboswitch aptamer (5,14). Thus, the A39–U65 and G39–C65 sequence combinations are not compatible with ligand binding, possibly due to an interaction between these residues (5,14). To gain more insight into the functional correlation between residues 39 and 65, the B. subtilis pbuE A-riboswitch aptamer and the related U65C mutant (Figure 1b) were structurally characterized. Interestingly, it was found that the G39–C65 sequence combination in the U65C mutant induces a ligand-free compact structure that mimics the purine-bound structures of the wild-type A/G-riboswitch aptamers.
MATERIALS AND METHODS
Native gel electrophoresis
The [5′-32P]-labeled A-riboswitch aptamer and mutants (final concentration < 1 nM) were incubated for 5 min at 25°C in 90 mM Tris, 89 mM boric acid, pH 8.5 (TB buffer) and 1 µM MgCl2. The RNAs were loaded on a 10% acrylamide:bisacrylamide (29:1) gel in TB buffer with 1 µM MgCl2 and were allowed to migrate at room temperature with continuous circulation of the running buffer (100 V for 24 h). The dried gel was exposed to a Phosphorimager screen.
Preparation of RNA for NMR studies
Double-stranded PCR fragments coding for RSA–VS and RSA(U65C)–VS and flanked by a T7 promoter were inserted into the pTZ19R-derived pTR-4 vector (15) to generate the plasmids pRSA–VS and pRSA(U65C)–VS, respectively. The DNA sequences of the transcribed regions of these plasmids were verified. The RSA–VS and RSA(U65C)–VS RNAs are precursors of the wild-type A-riboswitch aptamer (RSA; Supplementary Figure S1a) and the related U65C mutant [RSA(U65C); Supplementary Figure S1b], respectively, that contain a Varkud Satellite (VS) ribozyme substrate at their 3′-end (15). The 15N-labeled RSA–VS and RSA(U65C)–VS RNAs were synthesized in vitro using the bacteriophage T7 RNA polymerase, a plasmid template linearized with the AvaI restriction enzyme (New England Biolabs, MA, USA), and 15N-labeled nucleoside triphosphates (16). Following the RNA synthesis, a catalytic amount (∼25 nmol for a 25-ml transcription reaction) of purified trans-cleaving TR–4 VS ribozyme (15,17) was added to the transcription mixture to yield the desired RNAs [15N-labeled RSA and RSA(U65C)] with homogenous 3′-ends. These RNAs were purified by denaturing gel electrophoresis (10% 19:1 acrylamide:bisacrylamide and 7 M urea), followed by anion-exchange HPLC using a Dionex DNA–Pac100 column (9 × 250 mm) heated at 65°C (18). The purified RNAs were concentrated (0.35–1.0 mM) and exchanged with Amicon centrifugal filter devices (Millipore, MA, USA) in NMR buffer [10 mM sodium cacodylate (pH 6.5), 50 mM KCl, 0.05 mM NaN3, 5 mM MgCl2 in 90% H2O/10% D2O]. For spectra collected in the presence of adenine, the NMR buffer was supplemented with 2 mM adenine. Given the low solubility of guanine, the 350 µM sample of the U65C mutant in the presence of guanine was first prepared in NMR buffer with 10 µM RNA and 11.4 µM guanine and then concentrated at 15°C using Amicon Ultra-4 (3K) centrifugal filter devices.
NMR spectroscopy
NMR experiments were collected at either 288 K or 298 K on Varian UnityINOVA 500, 600 and 800 MHz NMR spectrometers equipped with pulse-field gradient units and actively shielded z-gradient HCN triple resonance probes. The following NMR experiments were conducted: 1D 15N-decoupled watergate (19), imino-optimized 2D 1H–15N HSQC (20); 2D HNN–COSY (21); and imino-optimized 3D 15N-edited NOESY–HSQC (τm = 120 ms) (22). 1H and 15N chemical shifts were referenced to an external standard of 2,2-dimethyl-2-silapentane-5-sulfonic acid at 0.00 ppm. NMR data were processed using the NMRPipe/NMRDraw package (23) and analyzed with NMRView (24).
Crystallization and X-ray data collection
The RSA(U65C)GU2 (Supplementary Figure S1c) was synthesized and purified as RSA(U65C) using a plasmid template derived from pRSA(U65C)–VS by site-directed mutagenesis. RSA(U65C)GU2 was crystallized by hanging drop vapor diffusion method where 1 µl of RNA (10 mg/ml) in NMR buffer (100% H2O/0% D2O) was mixed to 2 µl of reservoir solution containing 2 mM spermine, 32% MPD and 50 mM sodium cacodylate buffer pH 6.5 at 23°C. Native diffraction data from one crystal were collected to 2.8 Å resolution at a wavelength of 0.91924 Å on the X29 beamline at the National Synchrotron Light Source, Brookhaven National Laboratory, Upton, NY. Data were processed and scaled with XDS and XSCALE (25). The crystal belongs to space group C2 with two molecules per asymmetric unit and 65% solvent.
X-ray structure determination and refinement
The X-ray structure was determined by molecular replacement method using PHASER (26) from the CCP4 suite (27) and the structure of the add A-riboswitch aptamer from V. vulnificus (PDB code 1Y26) as the search model (3). The initial structure was rebuilt using COOT (28) and refined with PHENIX (29) and CNS (30). The R-factor of the final structure is 0.227 (Rfree = 0.277), using all data from 26.0 to 2.8 Å resolution. The structure contains residues 5–73 for chain A and chain B, 9 Mg2+ ions (only divalent metal ion present in the crystallization conditions), 2 Br− ions (the crystal was soaked in a KBr solution prior to data collection) and 88 water molecules. Ions were added on the basis of analysis of the 2Fo−Fc and the difference Fo−Fc electron density maps, the size of the metal ions potentially present in the crystal and by comparison with other A/G-riboswitch structures. Data collection and refinement statistics are summarized in Table 1. Since both structures in the asymmetric unit are very similar, only chain A is shown here. The r.m.s. deviation of NCS-related subunits is 1.39 Å (residues 6–72). All r.m.s. deviations were calculated with LSQKAB from the CCP4 suite (27), and figures were prepared with PyMOL.
Table 1.
Data collection and refinement statistics
| RSA(U65C)GU2 | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 124.93, 46.33, 87.24 |
| α, β, γ (°) | 90.00, 120.04, 90.00 |
| Resolution (Å) | 26.04–2.80 (2.93–2.80)a |
| Rsym | 0.130 (0.630) |
| I/σI | 10.5 (3.1) |
| Completeness (%) | 99.1 (99.0) |
| Redundancy | 3.6 (3.0) |
| Refinement | |
| Resolution (Å) | 26.04–2.80 |
| No. of reflections | 10 839 |
| Rwork/Rfree | 0.227/0.277 |
| No. atoms | |
| RNA | 2932 |
| Ions | 11 |
| Water | 88 |
| B-factors | |
| RNA | 30.6 |
| Ions | 22.3 |
| Water | 23.8 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 1.147 |
| Maximum likelihood coordinate error (Å) | 0.39 |
aValues in parentheses are for the highest-resolution shell.
RESULTS
A compact structure for the U65C mutant A-riboswitch aptamer in the absence of ligand
One of the hallmarks of A- and G-riboswitch aptamers is the compactness of their ligand-bound three-dimensional structure. As a result, native-gel electrophoresis can be reliably used to study the effect of mutations on the global architecture of the aptamer, since compact structures migrate faster than more extended ones. More precisely, disruption of the loop–loop interaction by mutation yields slower migrating species compared to the wild-type aptamer (5,13). This is demonstrated here for a P2 loop mutant (Figure 1c; P2 comp), which migrates slower than the wild-type A-riboswitch aptamer (Figure 1c; wild-type). This result was obtained in the absence of ligand and at a low magnesium concentration (1 µM) that is expected to partially destabilize the loop–loop interaction of the wild-type (13,31). Surprisingly, the U65C mutant of the A-riboswitch aptamer migrates faster than the wild-type aptamer under the same conditions (Figure 1c; U65C), suggesting that the U65C mutation helps stabilize the loop–loop interaction. It has been previously shown for both A- and G-riboswitch aptamers that stabilization of the core through ligand binding can promote formation of the distal loop–loop interaction under conditions where this interaction is destabilized (13,31,32). Thus, stabilization of the loop–loop interaction for the U65C mutant most likely results from the organization of a stable core structure.
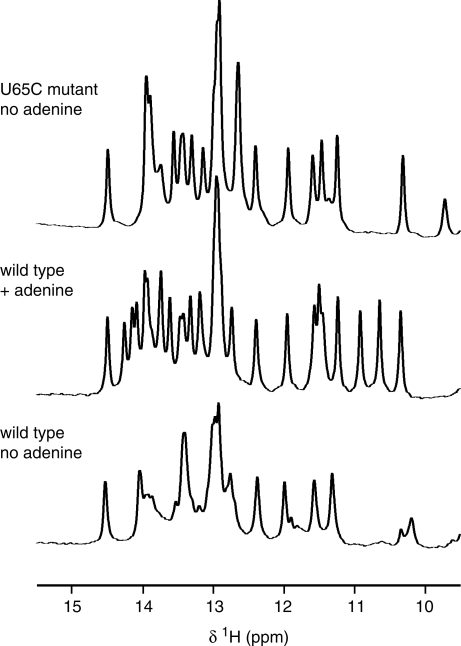
To explore the three-dimensional structure of the U65C mutant, the B. subtilis pbuE A-riboswitch aptamer and the related U65C mutant were investigated by NMR spectroscopy. We exploited the NMR data of imino protons, which can provide a wealth of information on the structure and folding of purine riboswitches (31,33–35). The imino region of the 1D 1H NMR spectrum of the wild-type A-riboswitch aptamer recorded in the absence of ligand shows signals of variable intensities characteristic of folded RNAs containing some dynamic regions (Figure 2). Addition of adenine results in a spectrum with an increased number of well-dispersed signals and more uniform peak intensities (Figure 2), which is more typical for stably folded RNAs. Similar NMR observations have been made when guanine was added to the B. subtilis xpt–pbuX G-riboswitch aptamer (3,34) and when adenine was added to the V. vulnificus add A-riboswitch aptamer (3). The 1D imino 1H NMR spectrum of the U65C mutant also displays a large number of signals with uniform peak intensities, however, in this case, the spectrum was recorded in the absence of ligand (Figure 2). Thus, in agreement with the native gel results, these 1D imino NMR data indicate that the U65C mutation induces a stable aptamer conformation in the absence of ligand.
Figure 2.
Evidence for stable folding of the U65C mutant of the A-riboswitch aptamer in the absence of ligand. The imino region of 1D 1H NMR spectra is shown for the wild-type aptamer in the absence and presence of adenine and for the U65C mutant in the absence of adenine. The spectra were recorded on a 600 MHz NMR spectrometer at 288 K.
Imino NMR spectroscopy of the wild-type B. subtilis pbuE A-riboswitch aptamer
The structure of the wild-type A-riboswitch aptamer bound to adenine was further investigated by heteronuclear NMR methods. In this complex, 29 imino protons were expected to participate in stable base-pairing interactions based on the X-ray structure of the add A-riboswitch aptamer (PDB code 1Y26) (3), and, correspondingly, 29 distinct imino signals were detected in the 2D 1H–15N HSQC spectrum (Figure 3a). Complete imino signal assignment (Supplementary Table S1) was achieved from 2D HNN–COSY (Supplementary Figure S2a) and 3D 15N-edited NOESY–HSQC spectra (Supplementary Table S2). The base-pairing arrangements of residues with detectable imino protons were also derived from these NMR data, and, in certain cases, base stacking could be inferred.
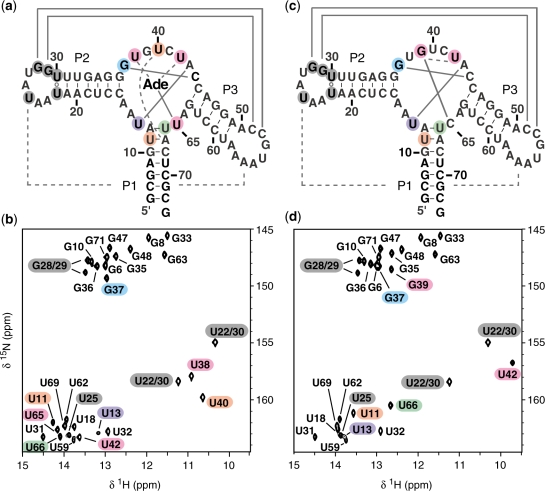
Figure 3.
Sequence and secondary structure of the wild-type (a) and U65C mutant (c) of the B. subtilis pbuE A-riboswitch aptamer. Below, the imino-optimized 2D 1H–15N HSQC spectra (20) of (b) the 15N-labeled wild-type aptamer with unlabeled adenine and (d) the 15N-labeled U65C mutant aptamer. The crosspeaks are annotated by residue types and numbers. Residues in loops L2 and L3 that yield a detectable imino signal in the HSQC spectra are shaded in dark grey, whereas those from the core are shaded in color as in Figure 4. The imino NMR data in (b) and (d) confirm the secondary structures in (a) and (c); canonical (WC/WC G–C and A–U) and non-canonical base pairs are connected by solid lines and open circles, respectively. The imino NMR data also provide evidence for tertiary interactions; canonical and non-canonical base pairs are shown in (a) and (c) by full and dashed lines, respectively. Spectra were recorded on a 600 MHz NMR spectrometer at 288 K.
As expected, 18 imino proton signals were detected for the WC/WC (36) base pairs of the P1, P2 and P3 stems (Figure 3b); the HNN–COSY data confirms the base pairing arrangement and NOEs involving these protons are consistent with a stacked helical structure within each stem (Supplementary Figure S2a and Table S2).
In loops L2 and L3, five imino protons are observed as expected; those from the U22·U30, U25·A56, G28–C52 and G29–C51 base pairs (Figure 3b). The HNN–COSY data confirms the WC/WC G28–C52 and G29–C51 base pairing observed in the X-ray structure, and stacking between these two base pairs is supported from NOEs of G28(H1) and G29(H1) with the same amino protons (not shown). The U25 residue contributes to the single U(N3)–A(N7) correlation in the HNN–COSY spectrum (Supplementary Figure S2a), in agreement with the unique trans WC/Hoogsteen U25·A56 pair observed in the X-ray structure. Formation of the WC/WC U22·U30 base pair that closes loop L2 is supported by an intense NOE signal between the two U imino protons (Supplementary Table S2). Thus, the NMR data clearly indicate that loops L2 and L3 form a stable interaction as described in the X-ray structure of the add A-riboswitch aptamer from V. vulnificus (3).
In the X-ray structure, the adenine-bound core is characterized by the stacking of five successive base layers that involve residues of the three-way junction (J1–2, J2–3 and J3–1) and the two junction-proximal base pairs of P1 (Figure 4a). The expected six imino proton signals from the junction are observed (Figure 3b; U13, G37, U38, U40, U42 and U65), and the NMR data are in agreement with formation of the complex core structure (Figure 4a). All intramolecular NH–N hydrogen bonds in the core were confirmed from the HNN–COSY spectrum (Figure 4a). Since correlations in the HNN–COSY spectra are observed for 15N-labeled nitrogens only, evidence for intermolecular interactions of U38, U42 and U65 with the unlabeled adenine ligand relied exclusively on the observation of NOEs. Both U38(H3) and U42(H3) give a strong NOE to the adenine H9 and both U42(H3) and U65(H3) give a strong NOE to the adenine H2, in agreement with the established pairing for the bound adenine (Figure 4a). Observation of specific NOE signals is also consistent with the stacking of the five base layers at the core of the adenine aptamer (Figure 4a). In summary, the NMR data of the adenine-bound B. subtilis pbuE A-riboswitch aptamer provide high-resolution structural information on the conformation of the stems, loop–loop interaction, and core that is fully compatible with the X-ray structure of a closely related A-riboswitch aptamer (3).
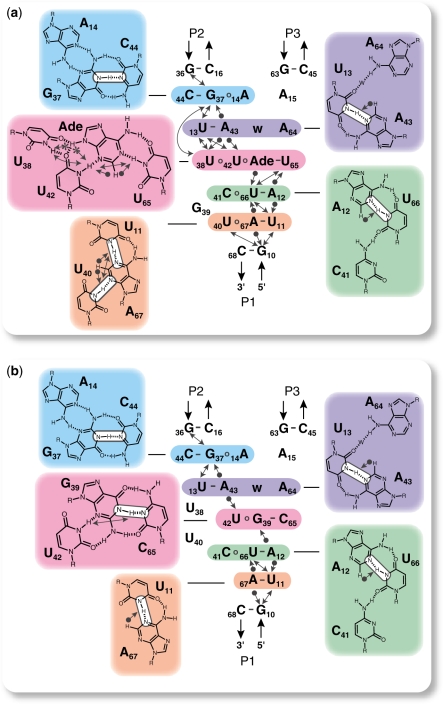
Figure 4.
The imino NMR data of the U65C mutant from the B. subtilis pbuE A-riboswitch aptamer are consistent with the formation of a compact core that mimics the adenine-bound wild-type aptamer. (a) Model of the core of the adenine-bound wild-type B. subtilis pbuE A-riboswitch aptamer derived from the imino NMR data and in agreement with the X-ray structure of the V. vulnificus add A-riboswitch (3). (b) Model of the core of the U65C mutant derived from the imino NMR data (see text). For both models, the core consists of a stack of five base layers, and each layer is shaded in a specific color with its pairing arrangement shown on a panel of the same color. Water molecules are denoted as ‘w’. All through-bond 2JNN coupling detected in the 2D HNN–COSY spectra (21) (Supplementary Figure S2) are in agreement with the G/U NH–N hydrogen bonds (depicted by white boxes) shown in these models. All NOEs involving imino protons in the 3D 15N-edited NOESY–HSQC (22) (Supplementary Table S2) are in complete agreement with the base pairing and stacking arrangement depicted in these models. Only the NOEs between two imino protons (lines with two arrowheads), between an adenine H2 and an imino proton (lines with a sphere and an arrowhead) and between the adenine ligand H9 and an imino proton (lines with a star and an arrowhead) were analyzed.
Imino NMR spectroscopy of the U65C mutant A-riboswitch aptamer
Imino NMR data were also obtained to structurally characterize the related U65C mutant. The 2D 1H–15N HSQC spectrum of the U65C mutant (Figure 3d) is similar to that of the adenine-bound wild-type aptamer (Figure 3b), and all but one G imino signal could be easily assigned based on the similarity of imino NMR data (chemical shifts, NH–N hydrogen bonding, and NOE) with the adenine-bound wild-type aptamer (Supplementary Data). G39(H1) was the only suitable candidate for assignment of the extra G imino signal of the U65C mutant (Figure 3d). The HNN–COSY spectrum provides evidence that G39(H1) is hydrogen bonded to a C(N3) (Supplementary Figure S2b), and an NOE between G39(H1) and A12(H2) confirms that G39 resides in the core of the aptamer (Supplementary Table S2). Interestingly, these imino NMR data are in agreement with the formation of a WC/WC G–C base pair involving G39, and a WC/WC G39–C65 base pair (Figure 4b) that mimics the adenine-U65 base pair of the A-riboswitch aptamer (Figure 4a) is considered highly likely based on the NMR data. In addition, since the adenine ligand also participates in a U42·Adenine base pair in the wild-type aptamer (Figure 4a), a similar U42·G39 base pair is proposed for the U65C mutant, although with a hydrogen-bonding pattern that better complements the G39 nucleoside (Figure 4b). Evidence for the proposed U42·G39 interaction is provided by an NOE between the imino protons of U42 and G39 (Figure 4b). Detection of an NOE between the imino proton of U42 and A43(H2) further confirms that U42 occupies similar positions in the U65C mutant and the wild-type aptamer (Figure 4b). Thus, it appears that a U42·G39–C65 base triple in the U65C mutant substitutes for the U42·Adenine–U65 base triple of the A-riboswitch aptamer.
Based on the imino NMR data, a model of the core structure for the U65C mutant was conceived, which consists of a stack of five base layers similar to that of the wild-type aptamer, except for the proposed U42·G39–C65 base triple and the disrupted U40·A67 pair (Figure 4). Interactions involving the two residues adjacent to G39 (U38 and U40) that were observed in the wild-type aptamer appear to be destabilized in the U65C mutant (Figure 4), since these residues did not yield detectable imino signals (Figure 3d). Given that imino protons are normally restricted to G and U residues, structural information on A and C residues cannot be directly extracted from the imino NMR data. Thus, in the model of the U65C mutant core, the tertiary interactions involving A14, C41 and A64 were simply represented as in the wild-type structure (Figure 4a). Analysis of all imino NMR data observed for the core of the U65C mutant aptamer indicates that these data are fully consistent with the proposed model (Figure 4b).
To better compare the structure of the adenine-bound wild-type aptamer with that of the U65C mutant, the chemical-shifts differences of the imino protons and nitrogens between the two RNAs were computed (Supplementary Figure S3). Significant differences in imino chemical shifts (Δ > 0.5 ppm) are observed for all detectable imino protons from the core (U11, U13, G37, U42 and U66). Similarity in imino chemical shifts for other residues (Δ < 0.5 ppm) indicates that the structures of the P1 stem and the P2 and P3 stem-loops are not significantly affected by the U65C mutation. HNN–COSY and NOE data (Supplementary Data) also support formation of the three stems, and indicate that the stable loop–loop interaction is intact in the U65C mutant. Thus, many structural characteristics of the adenine-bound A-riboswitch aptamer are supported by the imino NMR data of the ligand-free U65C mutant.
Ligand binding to the U65C mutant A-riboswitch aptamer
It was previously shown by in-line probing that the U65C mutant is defective for binding to adenine, 2-aminopurine and guanine (5). Given the structural model of the U65C mutant (Figure 4b), it seemed likely that, under appropriate conditions, G39 could transiently dissociate from C65 and allow binding of guanine, but not adenine. The 1D imino 1H NMR spectrum of the U65C mutant (0.35 mM) is not affected by the presence of 2 mM adenine, confirming that the U65C mutant is severely impaired for adenine binding (Supplementary Figure S4). In contrast, the U65C mutant (0.35 mM) is affected by the presence of guanine (∼0.40 mM), yielding 1D imino 1H and 2D 1H–15N HSQC spectra that (Supplementary Figure S4) are very similar to those of the adenine-bound wild-type aptamer (Figures 2 and 3b). Consequently, imino assignment was easily achieved from 2D HNN–COSY and 3D 15N-edited NOESY–HSQC spectra (Supplementary Table S2). The NMR data confirmed that, at high concentrations, the U65C mutant forms a 1:1 complex with guanine and adopts a fold typical of purine-bound A/G-riboswitch aptamers (Supplementary Figure S4).
Crystal structure of the U65C mutant A-riboswitch aptamer
Crystal structure determination of the U65C mutant was pursued to further explore its structure at atomic resolution. Suitable crystals could not be obtained for the U65C mutant sequence characterized by NMR, but a similar sequence containing extra base pairs in the P1 stem [RSA(U65C)GU2; Supplementary Figure S1] yielded crystals diffracting to 2.8 Å that were used for structure determination (Table 1).
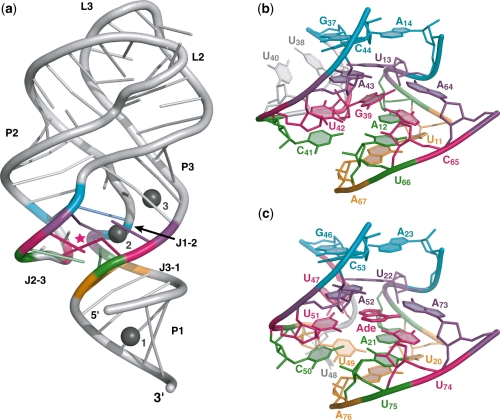
The global scaffold of the ligand-free U65C mutant is typical of purine-bound A/G-riboswitches with its tuning fork-like architecture, loop–loop interaction and compact core (Figure 5a). Structure superpositions show that the global fold of the ligand-free U65C mutant is very similar to that of adenine-bound A-riboswitch and guanine-bound G-riboswitch aptamers (r.m.s. deviations for each superposition are, respectively, 1.14 and 1.15 Å for residues 6–37 and 43–72; Supplementary Figure S5a). Tertiary contacts within the loop–loop interaction of the U65C mutant are identical to those of the guanine-bound B. subtilis xpt–pbuX G-riboswitch aptamer, which carries related L2 and L3 loop sequences (3) (Supplementary Figure S5b). Furthermore, the core structure contains the characteristic five-layer stack (Figure 5b) that mimics the adenine-bound A-riboswitch aptamer (Figure 5c) and corresponds almost exactly to the model derived from the imino NMR data (Figure 4b). The only difference is that the water molecule bridging the U13·A64 interaction in the model (Figure 4b) is missing in the crystal structure of the mutant. However, the presence of a water molecule at this location is variable in A/G-riboswitch aptamers (3) and may depend on the resolution of the X-ray data. All other base pairing interactions within the core that are predicted based on the NMR data (Figure 4b) are observed in the crystal structure of the U65C mutant (Figure 5b).
Figure 5.
Crystal structure of the U65C mutant from the B. subtilis pbuE A-riboswitch aptamer. (a) Ribbon representation of the U65C mutant. G39 is marked by a pink star and the three Mg ions common to both molecules of the asymmetric unit are shown as gray spheres. (b) Stick representation of the U65C mutant core. (c) Stick representation of the core from the crystal structure of the adenine-bound V. vulnificus add A-riboswitch aptamer (PDB code 1Y26) (3) resulting from the superposition with the structure of the U65C mutant. Residues from each base layer of the core are colored as in Figure 4. The residue numbering in (c) is given as in the PDB file.
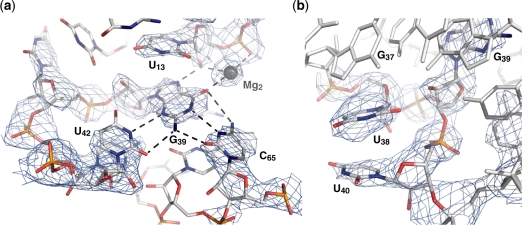
The quality of the electron density map allowed to clearly define the position of G39 in the ligand-binding pocket of the aptamer (Figure 6). Hydrogen bonding within the G39–C65 base pair is typical of WC/WC base pairs, although there is some deviation from planarity, particularly for the G(O6)–C(N4) hydrogen bond (Figure 6a). There is also a significant deviation from planarity for the WC/Sugar Edge U42·G39 base pair, which keeps the U42(O4) away from the ribose of G39, but still allows formation of the U42(N3)–G39(N3) and U42(O2)–G39(N2) hydrogen bonds. Incorporation of G39 in the ligand-binding pocket of the aptamer leads to extrusion of the adjacent residues U38 and U40 from the five-layer stack of the core (Figure 6b). These two residues are not disordered in the structure, but stack on one another near G37 and away from G39. It is clear from the crystal structure that a U42·G39–C65 base triple in the U65C mutant substitutes for the U42·Adenine–U65 base triple of the A-riboswitch aptamer with very minimal structural perturbations.
Figure 6.
Incorporation of G39 in the ligand-binding pocket. (a) Interaction network of G39. The G39 residue forms a WC/WC base pair with C65 and a Sugar-edge/WC base pair with U42. It also interacts with the 2′-OH of U13 and Mg2. (b) Stacking of the U38 and U40 bases on the periphery of the core, near G37 and away from G39. Oxygen, nitrogen, carbon and phosphorus atoms are shown in red, blue, gray and orange, respectively. The Mg ion is shown as a gray sphere and interactions are indicated with dashed lines. The blue electron density represents a 2Fo−Fc omit map contoured at 1σ. The omit map was generated by using the CNS simulated annealing protocol with a sphere of 3 Å around G39 being omitted.
In the structure of the U65C mutant, nine Mg sites and two Br sites are distributed on the two molecules of the asymmetric unit. Although at the resolution of the structure (2.8 Å), it is difficult to precisely define metal binding sites and the exact identity of the metal ion, the three sites that are common to both molecules of the asymmetric unit can be interpreted with more confidence (Figure 5a). One of them (Mg1 in Figure 5a) is a well-defined metal-binding site containing a G·U wobble pair (Supplementary Figure S1) that was rationally incorporated in the P1 stem (37), whereas the remaining two sites (Mg2 and Mg3 in Figure 5a) are associated with the core and correspond to sites previously described for the adenine-bound A-riboswitch aptamer (3). The preservation of the Mg2 site for the U65C mutant is remarkable given that it associates with the U42·G39–C65 base triple (Figure 6a). Thus, the crystal structure presented here demonstrates that the U65C mutant adopts a compact ligand-free structure that essentially mimics the purine-bound structure of purine riboswitch aptamers.
DISCUSSION
Multiple sequence alignments of A/G-riboswitches have allowed the identification of conserved residues that play important roles in the structure and function of this class of RNA. For example, it has been demonstrated that residue 65, which is a key determinant of ligand-binding specificity, is completely conserved through evolution, being a C in G-riboswitch aptamers and a U in A-riboswitch aptamers. Multiple sequence alignments can also reveal residues that are not allowed at a given position. It was recently shown that the residue at position 39 is never a G for the G-riboswitch aptamer and never an A for the A-riboswitch aptamer (5,14). It was demonstrated that aptamer mutants that contain the G39–C65 or A39–U65 combination bind their respective exogenous purine ligand with reduced affinity (5,14). Based on these results, it was proposed that an interaction between G39 and C65 is responsible for the impaired ability of the aptamer to bind its ligand (5,14). This hypothesis was tested through a comprehensive structural characterization of the U65C mutant and enabled the discovery that G39 can act as a surrogate ligand in this aptamer mutant.
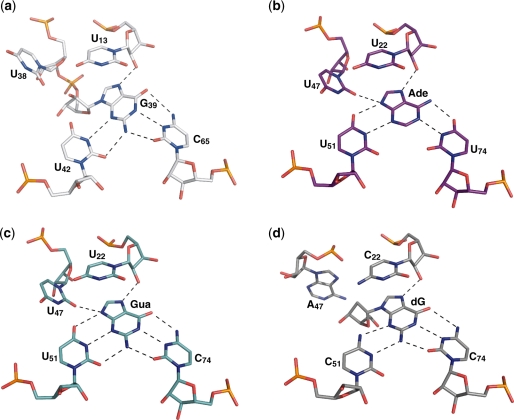
The structural investigations of the U65C mutant B. subtilis pbuE A-riboswitch aptamer presented here clearly demonstrate that formation of a WC/WC G39–C65 base pair yields a compact structure that can no longer accommodate a purine ligand. In the structure of the adenine-bound wild-type aptamer, G39 was extruded from the core and exposed to solvent (3). In the structure of the ligand-free U65C mutant, the overall scaffold, including the loop–loop interaction and the five-layer base stacking at the core are maintained, but G39 is no longer extruded. Instead, G39 occupies the ligand-binding site by forming base-pairing interactions with C65 and U42 (Figure 7a), similarly to the adenine ligand in the A-riboswitch aptamer (Figure 7b) and the guanine ligand in the G-riboswitch aptamer (Figure 7c) (3). Only minimal structural differences are observed between the ligand-free U65C mutant and these purine-bound A/G-riboswitch aptamers (3). First, there is some deviation from planarity for the U42·G39–C65 base triple of the U65C mutant that is not evident for the U42·Adenine–U65 base triple of the adenine-bound A-riboswitch aptamer (U51·Adenine–U74 in Figure 7b) and not as pronounced for the U42·Guanine–C65 base triple of the guanine-bound G-riboswitch aptamer (U51·Guanine–C74 in Figure 7c). Such deviation from planarity in the U65C mutant is largely due to G39, which is oriented slightly differently than the adenine in the ligand-binding site. In the superposition shown in Supplementary Figure S5, G39(O6) and adenine N6 are 1.2 Å apart and there is a ∼20° roll angle between these two bases. The deviation from planarity for the U42·G39–C65 base triple may result from the covalent attachment of the G39 ligand mimic to the RNA aptamer, which evidently makes this residue conformationally more restrained than exogenous purine ligands. A crystal structure of a G-riboswitch aptamer in complex with 2′-deoxyguanosine was recently determined (10), and revealed how this nucleoside is incorporated in the ligand-binding pocket (10). Similarly to G39 in the U65C mutant, the 2′-deoxyguanosine does not permit hydrogen bonding at the N9 position like the adenine and guanine ligands (Figure 7). However, to avoid a steric clash involving its ribose, the 2′-deoxyguanosine forms a shifted base pair with the nucleotide corresponding to U42 (C51; Figure 7d), whereas G39 simply forms a non-planar base pair with U42 (Figure 7a). Thus, in comparison with the 2′-deoxyguanosine, the incorporation of G39 in the ligand-binding pocket is more similar to that of adenine and guanine. Second, the two residues adjacent to G39 (U38 and U40) that stabilize the core via hydrogen bonding of their Watson–Crick edges in the wild-type aptamer fulfill this role somewhat differently in the U65C mutant. The imino proton signals of U38 and U40 were not observed for the U65C mutant, indicating that their Watson–Crick edges no longer participate in stable interactions. The crystal structure confirms that U38 and U40 adopt different conformations in the U65C mutant; their bases stack on one another and their Watson–Crick edges are exposed to the solvent. In-line probing of the U65C mutant aptamer gave significant cleavage products at positions U38 and U40 of the core, and these cleavage products were not observed for the adenine-bound wild-type aptamer (5). Thus, the previously reported in-line probing data of the U65C mutant highlight the subtle but most significant difference between the structure of the ligand-free U65C mutant and that of the adenine-bound A-riboswitch aptamer (5). Overall, the resemblance between these structures is astonishing and reveals how an internal residue can mimic the role of the exogenous ligand in A/G-riboswitch aptamers.
Figure 7.
Ligand-binding pockets in A/G-riboswitch aptamers. Stick representations are shown for (a) the U65C mutant, (b) the V. vulnificus add A-riboswitch aptamer bound to adenine (PDB code 1Y26) (3), (c) the B. subtilis xpt–pbuX G-riboswitch aptamer bound to guanine (PDB code 1Y27) (3) and (d) a mutant of the B. subtilis xpt–pbuX G-riboswitch aptamer bound to 2′-deoxyguanosine (PDB code 3DS7) (10). The residue numbering is given as in the PDB files.
Structural studies of the A- and G-riboswitch aptamers in the absence of ligand but in the presence of magnesium ions indicate that they form partially folded structures. Under those conditions, the loop–loop interaction is formed and the ligand-binding pocket is preorganized with respect to two base triples (C44–G37·A14 and A64·U13–A43), but the J1–2 and J2–3 strands are conformationally dynamic (8,13,32,34). Folding of the A/G-riboswitch aptamer upon ligand binding is thought to proceed via a two-step mechanism, which involves the initial docking of the purine ligand with the pyrimidine residue in J3–1 (U/C65), followed by folding of the J1–2 and J2–3 strands to completely enclose the ligand (8,32). The inherent local flexibility of the core in the absence of ligand plays an important role in the adaptive binding of the A/G-riboswitch aptamer to analogs of guanine and adenine (10,11). However, this local flexibility of the core also has an influence on the folding energy landscape and the thermodynamic equilibrium associated with ligand binding (8).
Sequence composition within the core likely evolved to avoid stable ligand-free conformations like that of the U65C mutant, which may be perceived as deep potholes in the energy landscape associated with ligand-induced RNA folding. Based on mutational analysis of the G-riboswitch aptamer, it was previously proposed that a number of nucleotides in the three-way junction may have been selected to maintain the ligand-free RNA in a form that is capable of binding (9). More specifically, it was suggested that A64, A15 and the Y13–R43 base pair are conserved because alternative bases at these positions collapse the binding pocket and are not compatible with ligand binding (9). Sequence conservation at other residues within the A/G-riboswitch aptamer core could also result from specific exclusion of certain residue types. For example, the two residues adjacent to G39 are always a uridine (U38 and U40) in the reported sequences of the A-riboswitch aptamer (5). Mutations of these residues (U38C, U40C and U40G) in the context of the G-riboswitch aptamer reduce binding by 20–100-fold (9). It is plausible that exclusion of certain residue types at U38 and U40 could result from the natural selection against A-riboswitch aptamers with stable ligand-free conformations that can no longer respond appropriately to changes in ligand concentration. In the ligand-bound conformation, these two residues make interactions within the core structure that are likely not crucial for formation of a compact aptamer fold and stable ligand binding. It is evident from the NMR and X-ray studies presented here that the hydrogen-bonding interactions involving U38 and U40 in the adenine-bound A-riboswitch aptamer are disrupted in the U65C mutant, suggesting that these residues are not essential for formation of a stable, compact core. Furthermore, in the crystal structure of a purine riboswitch aptamer bound to 2′-deoxyguanosine (Kd ∼ 60 nM), the aptamer adopts a compact structure similar to the ligand-bound A/G-riboswitch aptamer structures, but residues that correspond to U38, G39 and U40 are extruded from the core (10). It turns out that sequence conservation for many core residues of the A/G-riboswitch aptamer may be strongly governed by the importance of maintaining a ligand-free aptamer structure that does not severely interfere with ligand binding. Considering the evolutionary exclusion of the G39–C65 sequence combination, the structure of the U65C mutant reveals a remarkable example of an RNA aptamer mutant trapped in a stable conformation that impairs its ligand-dependent folding.
Given that the evolutionary-excluded G39–C65 sequence combination impairs ligand binding (5,14), it is highly likely that this combination severely perturbs the proper functioning of the riboswitch in vivo (2). Even though the U65C mutant can still properly bind guanine at high concentration, it may no longer respond appropriately to changes in ligand levels. In this regard, it has been previously shown that natural sequence variants of the G-riboswitch have variable ligand affinities that modulate riboswitch function in vivo (14). Consequently, partial stabilization of ligand-free conformations may represent an effective strategy for fine-tuning the functional properties of the riboswitch. Given that riboswitch aptamers have become attractive targets for the development of novel antibiotics, riboswitch inactivation through stabilization of ligand-free conformations may also represent an interesting avenue for the development of antibacterial agents. In this regard, the crystal structure of the U65C mutant could provide a useful starting point in the search of such compounds.
ACCESSION NUMBER
PDB: 3IVN
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes for Health Research (CIHR) (MOP-82877 to D.A.L and MOP–64341, MOP–86502 to P.L.). The 800 MHz NMR data (15N-edited HSQC–NOESY of the wild-type A-riboswitch aptamer) were recorded at the Québec/Eastern Canada High Field NMR Facility, supported by grants from the Canada Foundation for Innovation, the Quebec Minister of Research, Science and Technology and McGill University. X-ray data were collected at the National Synchrotron Light Source, Brookhaven National Laboratory, which is supported by the United States Department of Energy, Division of Materials Sciences and Division of Chemical Sciences under Contract DE-AC02-98CH10886. Banting and Best Ph.D. scholarship from CIHR and a graduate scholarship from the Université de Montréal (to P.B.). Graduate scholarship from the Université de Montréal (to E.B.). Graduate scholarship from the National Sciences and Engineering Research Council of Canada (to J.-F.L.). D.A.L. is a Chercheur-Boursier Junior II of the Fonds de la Recherche en Santé du Québec and a CIHR New Investigator Scholar. P. L. holds a Canadian Research Chair in Structural Biology of RNA. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEGEMENTS
The authors thank D. Chaussé E. Benoit and P. Lampron for RNA preparation and R. Richter for computer support. They thank B. Golden for advice with RNA crystallization and molecular replacement, J. Sygusch for providing synchrotron access and J. G. Omichinski for critical reading of the manuscript.
REFERENCES
- 1.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 2.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 3.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lescoute A, Westhof E. Riboswitch structures: purine ligands replace tertiary contacts. Chem. Biol. 2005;12:10–13. doi: 10.1016/j.chembiol.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Lemay JF, Lafontaine DA. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:339–350. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J. Am. Chem. Soc. 2006;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J. Mol. Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 13.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rastogi T, Collins RA. Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol. 1998;277:215–224. doi: 10.1006/jmbi.1997.1623. [DOI] [PubMed] [Google Scholar]
- 16.Nikonowicz EP, Sirr A, Legault P, Jucker FM, Baer LM, Pardi A. Preparation of 13C and 15N labelled RNAs for heteronuclear multidimensional NMR studies. Nucleic Acids Res. 1992;20:4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo HCT, Collins RA. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shields TP, Mollova E, Ste Marie L, Hansen MR, Pardi A. High-performance liquid chromatography purification of homogenous-length RNA produced by trans cleavage with a hammerhead ribozyme. RNA. 1999;5:1259–1267. doi: 10.1017/s1355838299990945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piotto M, Saudek V, Skleñár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 20.Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 21.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 22.Zhang O, Kay LE, Olivier JP, Forman-Kay JD. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 23.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 26.McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, et al. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noeske J, Schwalbe H, Wohnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keel AY, Rambo RP, Batey RT, Kieft JS. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.