Abstract
We assess the role of DNA breathing dynamics as a determinant of promoter strength and transcription start site (TSS) location. We compare DNA Langevin dynamic profiles of representative gene promoters, calculated with the extended non-linear PBD model of DNA with experimental data on transcription factor binding and transcriptional activity. Our results demonstrate that DNA dynamic activity at the TSS can be suppressed by mutations that do not affect basal transcription factor binding–DNA contacts. We use this effect to establish the separate contributions of transcription factor binding and DNA dynamics to transcriptional activity. Our results argue against a purely ‘transcription factor-centric’ view of transcription initiation, suggesting that both DNA dynamics and transcription factor binding are necessary conditions for transcription initiation.
INTRODUCTION
RNA polymerases require access to a locally denatured single-strand DNA segment (2,3) at the transcription start site (TSS) in order to initiate transcription. It has been demonstrated that introduction of an artificial bubble at the TSS of a viral promoter, via insertion of a 5-bp mismatched segment, is sufficient for the polymerase to initiate transcription in the absence of basal transcription factors (4,5). Use of a negatively supercoiled DNA template (5–8) can also obviate the requirement by polymerase II for basal transcription factors binding (5–7) and helicase activity (8). It has been suggested that under natural conditions in vivo, formation of the transcriptional bubble is seeded by transient, thermally induced strand separation motions of the DNA double helix, commonly known as DNA breathing (9). To investigate this possibility, we have been studying the sequence dependence of breathing dynamics with the non-linear Peyrard–Bishop–Dauxois model (PBD) of DNA (10,11). In support of a link between spontaneous DNA strand separation and transcription initiation, we found that mammalian promoter sequences frequently exhibit a breathing dynamics maximum (bubble) coinciding with the TSS (4,9). We introduced the use of Langevin molecular dynamic (LMD) simulations and use of three dynamic criteria: bubble length, bubble amplitude and bubble lifetime, which can be extracted from the simulated dynamic trajectories of experimentally identified TSS (4). Bubble length is defined as the number of consecutive base pairs that are simultaneously separated from their hydrogen bond minima by more than a given distance threshold (the bubble amplitude). Simulations of several mammalian core promoters demonstrated that a relatively large (length: ∼10 bp; amplitude A: >1.5 Å) and stable (lifetime τ: > 5 ps) bubble forms frequently at the examined TSS (4). We reported that A/T-rich regions such as TATA boxes exhibit faster, lower amplitude motions than TSS regions (4,12). G/C-rich promoters, however, display less obvious bubble-forming motifs in the simulations (4).
The main source of structural and dynamic heterogeneity in G/C-rich sequences presumably originates from a dramatic difference in the stacking interaction between GG/CC steps on the one hand and CG/CG and GC/GC on the other (1,13,14). However, the original PBD Hamiltonian does not account for the sequence dependence of the stacking potentials and performs poorly at reproducing the melting transitions of G/C-rich DNA. For accurate analysis of G/C-rich DNA, we recently derived an extended PBD (EPBD) Hamiltonian that includes sequence-dependent base-stacking potentials, and calibrated the model with DNA melting studies of short repeats and homopolymers (1). Monte Carlo simulations with the resulting EPBD model faithfully reproduce the melting behavior of highly homogenous and repetitive sequences, e.g. the famous 10° Tm difference between poly(dG).poly(dC) and poly(dGdC) (13). Consistent with such differences in melting behavior and with NMR studies of the millisecond-scale dynamics of G/C-rich DNA (15), the EPBD simulations predict significant heterogeneity in pre-melting (breathing) dynamics of various G/C-rich DNA sequences.
Here we examine the EPBD breathing dynamics of two representative mammalian promoters with high G/C content. We aim to establish whether DNA breathing dynamics profiles at the TSS are merely coincidental, or a necessary factor for transcription. We use EPBD LMDs simulations, gene transcription and gel shift assays to explore the relationship between DNA dynamics, efficiency of transcription and basal transcription factor binding at the core promoter. Our hypothesis is that a TSS-specific dynamic signature is a necessary feature of transcription initiation. As a model system, we chose a fully characterized ‘classical’ promoter, the SCP1 promoter (16) and the CpG island promoter of the mouse thymidylate synthase (TS) (17).
SCP1 is a single start site promoter artificially constructed from functionally established eukaryotic promoter elements. SCP1, also called the ‘superpromoter’ (16), exhibits one of the strongest known basal activities and is a classical promoter in the sense that it contains the well-known TATA, Initiator (Inr), downstream promoter element (DPE) and motif ten element (MTE) element sequences. It was shown that the Inr, MTE and DPE sequences are sufficient to recruit the TFIID basal transcription factor complex and initiate transcription. This was established by mutations in these promoter elements, correlating TFIID binding with transcriptional activity (16). To separate the effects of transcription factor binding from DNA dynamic properties, we first identified mutations in close proximity to the TSS that do not affect TFIID binding but do change the dynamic signature of DNA. We conducted EPBD simulations on various sequences mutated outside the regions involved in direct contacts with the TFIID complex and chose two of the mutant variants, m1SCP1 and m2SCP1, with silenced and intact TSS dynamics, respectively.
MATERIALS AND METHODS
EPBD computer simulations
Analysis of the dynamic trajectories, bubble probability and bubble lifetime calculations are performed as previously reported (4).
Plasmids
pUC119 plasmid containing the wtSCP1 promoter sequence insert from −36 to +45 (relative to the TSS) (16) was used as a template to construct the m1SCP1 and m2SCP1 mutant variants. Mutations were introduced by QuikChange Mutagenesis Kit (Stratagene) following the protocol of the supplier.
Gel shift reactions
The reactions were assembled as previously described (5,6) TFIID was isolated from HeLa cells (18). TFIIF, TFIIB, TFIIE, TFIIA were purified from Escherichia coli (6). Oligonucleotide probes were labeled with [γ-33P]ATP by T4 polynucleotide kinase (Invitrogen). The sequences of the probes used in the gel shift reactions are as follows: wtSCP1—GGGGCGCGTTCG TCCCAGTCGC GATCG AACACTCGA; m1SCP1—GGGGCGCGTTCGCG CCAGTCGCGG TCGAACGCTCGA; non-specific gel shift reaction competitor—TTCTTCTTCTTCTTCTTCTTCT TCTTCTTCTTCTTCTTC.
Cell transfection
HeLa cells (1 × 105) were transfected with 2 μg of pUC119-SCP1 variants or empty pUC119 plasmid DNA using Magus reagent (BIDMC) and electroporation. Cells were then cultivated in DMEM/10% FBS. RNA was harvested 19 h later and subjected to Q-PCR.
Analysis of promoter transcriptional activity by Q-PCR
Total RNA was extracted from cells by RNeasy® kit (Qiagen), following the manufacturer’s instructions. First-strand cDNA synthesis was performed using RETROscript® (Ambion). PCR was performed using cDNA synthesized from 1.5 μg total RNA in an Mx3000P QPCR system (Stratagene) with the pUC119-specific primers downstream of the SCP1 TSS and SYBR Green PCR Master Mix (Stratagene). The sequences of the primers are as follows: AGCGG ATAACAATTTCACACAGGA and ATCGAACACTCGAGCCGAG. All Q-PCR data are expressed as mean ± SD.
RESULTS
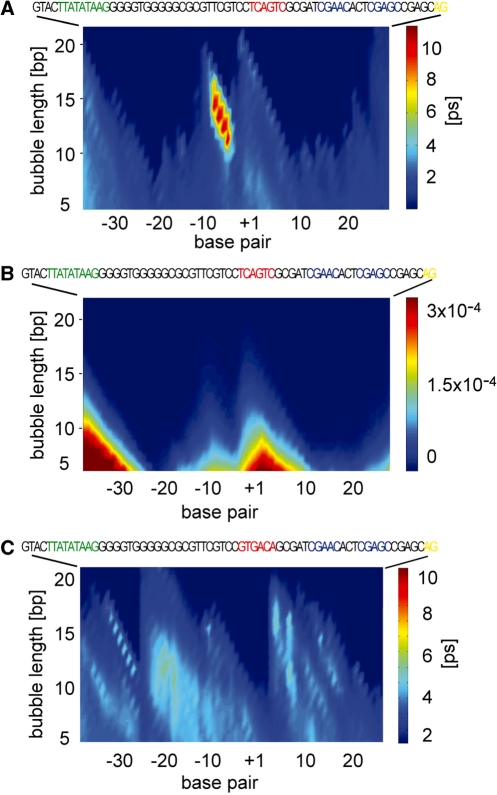
The dynamical trajectories of the SCP1 variants are shown in Figure 1. The wild-type promoter sequence revealed the characteristic dynamic pattern observed previously for other mammalian promoters (4), containing two major sites of dynamic activity, the Inr/TSS region and the TATA region upstream of the TSS. The TSS bubble is distinguished by the longest lifetimes at high amplitudes (Figure 1A). The T/A-rich region, in contrast, exhibits higher bubble probability but lower lifetimes at high bubble amplitudes (Figure 1B). These data closely match our previous observations of long-lived bubbles with high amplitudes at the TSS and short-lived bubbles at TATA and TATA-like sites for several promoters (4).
Figure 1.
LMDs simulations of the collective DNA openings in the SCP1 ‘Super promoter’ predict two major loci of dynamic activity. (A) Average lifetime with length L (vertical axis), beginning at a given nucleotide position (horizontal axis) relative to the TSS (‘+1’). (B) Probability for the formation of bubbles of amplitude >3.5 Å. (C) Average bubble lifetimes of a transcriptionally inactive SCP1 variant mutated at the Inr sequence (16). The SCP1 promoter sequences are shown at the top. Promoter element motifs are indicated with colored boxes: Inr, red; MTE, blue; DPE, yellow.
The dynamic profile of the SCP1 TSS is very similar to other studied promoters despite the fact that its Inr sequence has significantly higher (50%) G/C content than the previously investigated Inr sequences (G/C ∼ 30%) (4). As discussed previously by us (12) and Dornberger et al. (15), this underscores the non-trivial dependence of DNA breathing dynamics on G/C content and the effect of adjacent sequences. Remarkably, the m1SCP1 mutant, which differs from the wild-type sequence by four-point mutations located outside of the TSS, exhibits a dramatically different dynamic profile (Figure 2A). The mutations suppress the dynamic activity of the TSS, clearly silencing the TSS bubble, while preserving the sequence. The second mutant, m2SCP1 displayed a dynamic profile essentially identical to the wild type (Figure 2A). In both mutants, the Inr, MTE and DPE motif sequences were preserved.
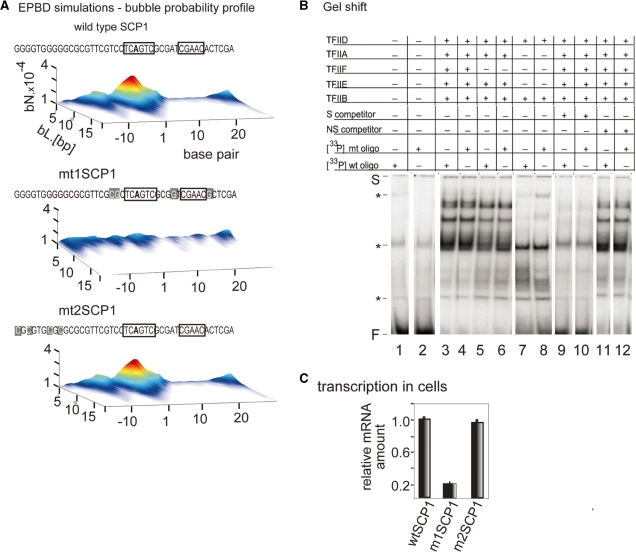
Figure 2.
EPBD-derived mutations that change bubble probability profile and transcriptional activity while preserving TFIID complex formation at the SCP1 promoter. (A) Bubble probability profiles of the wild-type SCP1 promoter (wtSCP1), m1SCP1, m2SCP1 mutant variants designed to silence transcription activity without affecting protein-binding sites. The probability (z-axis) for the formation of bubbles of amplitude >3.5 Å with length L (y-axis) beginning at a given nucleotide position (x-axis) relative to the TSS (‘+1’). The wtSCP1, m1SCP1 and m2SCP1 sequences are shown at the top. Mutated residues are indicated with gray boxes. Protein binding sites are indicated with black frames. The profile of m2SCP1 is identical to wild type SCP1 as shown at the bottom. (B) Gel shift reactions. Effect of the m1SCP1 mutations on complex formation between TFIID, TFIIB, TFIIF, TFIIE and the Inr promoter fragment. Band shift reactions received a 33P-labeled, double-stranded oligonucleotide (0.12 nM) containing the wild-type (lanes 1, 3, 5, 7, 9 and 11) and the m1SCP1 Inr box sequence (lanes 2, 4, 6, 8, 10 and 12), as indicated in (A). Transcription factors samples are as follows: lanes 1 and 2 bovine serum albumin; lanes 3–12 received equal amounts (in micrograms) of transcription factors as indicated above the lanes. The reactions in lanes 9 and 10 received 3 nM of homologous wild-type cold SCP1 oligonucleotide as a competitor. The reactions in lanes 11 and 12 received 10 nM of unrelated cold oligonucleotide as a competitor. The presence (+) or absence (−) of competitor oligo DNA and basal transcription factors in the reactions is indicated above the lanes. The positions of the gel shift start (S), the free DNA (F) and the non-specific gel shift products (asterisk) are indicated. (C) Transient cell transfection experiments were carried out to measure wtSCP1, m1SCP1 and m2SCP1 promoter activity. The three pUC119-based constructs (16) (2 µg/106 cells) were transfected by electroporation into HeLa cells. Total RNA was extracted from the cells and subject to Q-PCR-based analysis with pUC119 primers to measure cellular level of promoter-specific RNA transcripts. To ensure equal transfection efficiency, DNA instead of RNA was extracted from an aliquot from each reaction, and subjected to Q-PCR with the same primers. Data are expressed as fold induction relative to wtSCP1 mRNA level (on the vertical); all values are normalized to the cellular reference gene ARPO0 mRNA level of expression; reactions were run in triplicate; results were consistent in four independent experiments; error bars, mean ± SD, n = 4. The normalized values are plotted as a bar graph and the identity of the promoter-specific transcripts level is shown below the bars.
The role of the Inr sequence element in transcription is generally attributed to binding of transcriptional initiator factors such as the large TFIID complex (16,19) and/or YY1 (5). These proteins serve to recruit the polymerase and the other basal transcription factors at the TSS (6,8). The m1SCP1 sequence retains the original Inr element, but lacks the characteristic TSS dynamic signature, providing a test case to establish whether binding of the basal transcription factors is sufficient for transcription, or whether the TSS dynamic signature is necessary. To confirm that transcription factor binding was, as intended, unaffected by the m1SCP1 mutations, we performed gel shift assay with TFIID and basal transcription factors (Figure 2B). As positive control, we conducted gel shift reactions with the wtSCP1 promoter oligo fragment. Reactions were assembled with equal protein amounts of TFIID (3 ng/reaction) and TFIIB (2 ng/reaction) alone and together with transcription factors TFIIF (4 ng), TFIIE (3 ng) and TFIIA (3 ng). The results suggest that both the wild-type and the m1SCP1 oligos form nearly identical complexes with TFIID and the tested basal transcription factors. The observed complexes result from sequence-specific recognition, since presence of unlabeled wtSCP1 oligo in the reactions competes equally well for protein binding with both radioactively labeled wtSCP1 and m1SCP1 fragments. To verify the composition of the protein–DNA complexes in crude nuclear extract, we performed gel shift reactions with HeLa extract and anti-TFIIF basal transcription factor-specific antibodies (Supplementary Data). The results suggest that the selected mutations in m1SCP1 only result in suppression of bubble dynamics at the TSS without affecting the binding of the basal pre-initiation transcription complex. The m2SCP1 promoter variant displays both intact dynamics and protein binding (data not shown).
The effect of the mutations on promoter strength was assessed by transiently transfecting wtSCP1, m1SCP1 and m2SCP1 promoter templates in HeLa cells. The transcriptional activity of the promoter variants was determined by measuring the cellular levels of specific RNA transcripts in real-time PCR reactions (Q-PCR). As expected, wtSCP1 and m2SCP1 support high level of transcription in HeLa cells resulting in accumulation of specific RNA (Figure 2C). In comparison, m1SCP1 showed a 4-fold decrease in the level of RNA transcripts. The results of these experiments suggest that suppression of TSS bubble dynamics leads to a decrease in promoter activity, independent of basal transcription factor binding to the core promoter.
To further establish the requirement for strong DNA dynamics in determining a TSS, we conducted EPBD Langevin dynamic simulations on the mouse TS promoter (17,20). The TS promoter is a CpG island promoter that does not contain any of the known elements present in SCP1. It has been suggested that CpG island promoters are commonly associated with constitutively expressed housekeeping genes and may be regulated differently than the other known classes of promoters, such as promoters containing the TATA and Inr elements. TS is a ‘dispersed’ promoter (19,21), by virtue of having multiple TSS dispersed over a 100-bp region (17). This is in contrast to ‘focused’ promoters such as SCP1, which display one or several clearly defined start sites. Most of the TSSs of the TS promoter are known to be regulated by the Ets family of transcription factors in collaboration with Sp1 (20).
The simulations of the wild-type TS promoter revealed a dynamic activity that is evenly distributed and noticeably weaker than SCP1 (Figure 3A). Conspicuously lacking is the characteristic long-lived bubbles observed for SCP1 (Figure 2A) and other previously studied ‘focused’ promoters (4). This result is again consistent with the notion that a relatively stable, well-defined bubble is required to define a strong, localized TSS.
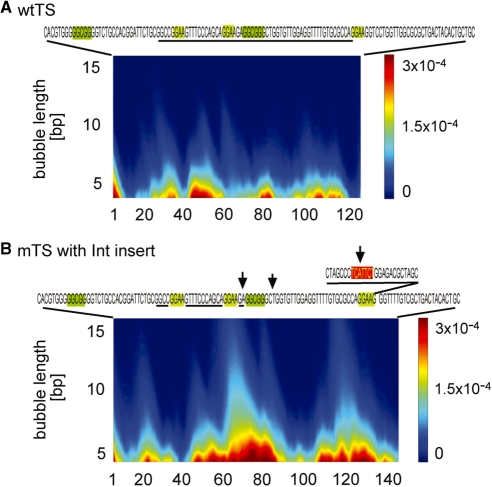
Figure 3.
Changes in the TS promoter bubble probability profile in response to mutations. (A) Wild-type TS promoter (wtTS). The promoter sequence is shown at the top. Experimentally determined window with several TSSs (17) is underlined. (B) Mutant promoter variant (mTS) with altered TSS distribution due to the insertion of an Int-containing sequence (17). The exact position of the insertion is shown above the mTS sequence. The Int is indicated by a red box. The mTS promoter TSS are shown with the arrows above the sequence. The position of Ets and Sp1 transcription factors binding is identical for both promoters, as shown by the light green, respectively, dark green, boxes on the sequence letters. The color scale at the left of the panels presents the probability for the formation of bubbles of amplitude >2.5 Å with length L (vertical axis) beginning at a given nucleotide position (horizontal axis).
It was previously reported (17) that insertion of an Inr sequence in the promoter without changing the dispersed transcriptional window and transcription factors binding sites leads to the appearance of a new strong TSS at the Inr. The transcriptional activity of the wild-type sequence upstream of the insert was altered. The mutant TS exhibited more focused character, with the appearance of another pronounced TSS in addition to the Inr site.
The EPBD simulations of the TS-Inr mutant (Figure 3) revealed a significantly changed dynamic profile, compared with wtTS, with the appearance of a new and strong TSS located within the Inr insert, and a more ‘concentrated’ dynamic activity in the original wild-type sequence upstream of the insert (Figure 3). This dynamic pattern is in striking agreement with the reported (17) pattern of TSSs.
DISCUSSION
The EPBD LMDs simulate DNA transient openings in the picosecond-time scale, a range that is to be expected from the strength of the hydrogen bond potential (1–20 meV) and is consistent with experimental studies of the vibrational frequencies of biomolecules (22–25). While the accuracy of the predicted sequence dependence of DNA breathing frequencies has not been directly verified experimentally, the PBD simulations faithfully reproduce a variety of phenomena directly linked to the hydrogen bond stretching and unstacking (26–29). These include accurate prediction of unzipping force measurements (28), bubble nucleation size (29) and melting profile predictions (1,26,27). Given the striking accuracy of the melting temperature predictions of EPBD, including homopolymers and repeats that are difficult to model (1), we anticipate that the pre-melting dynamic trajectories produced by EPBD simulations are at least qualitatively correct.
The results reported here are consistent with our previous findings that core promoter TSSs exhibit specific breathing dynamic signatures (4). Further, the new data suggest that the observed transient collective openings in the DNA double helix are not simply a coincidental property of the TSS sequence, but actually play an important role in determining the TSS strength and position. Such collective openings may seed the formation of the transcriptional bubble needed for transcription, or they may represent another, yet unknown but important conformational feature of promoter DNA (3). In any event, the data rule out the possibility that the introduced mutations in the SCP1 promoter directly disrupt specific DNA–transcription factor contacts. These mutations were chosen to avoid points of contact with the pre-initiation complex, as identified in thorough DNAse I footprinting studies of SCP1 and SCP1 mutants (16). Our gel shift experiments confirm that transcription factor–DNA interactions are indeed unperturbed.
The data indicate that the dynamics at the TSS are as important as the binding of the basal transcriptional factors for determining the transcription initiation strength, and it is thus a ‘localizer’ of the TSS position. Mutations in the MTE and DPE elements that prevent TFIID binding (12) result in decrease in SCP1 activity comparable (∼3- to 4-fold in luciferase reporter assay) with the impact of silencing the transcriptional bubble without perturbing TFIID binding (4-fold in Q-PCR assay). Curiously, a mutation inside the Inr motif of SCP1 was reported to suppress transcription more potently (15-fold in luciferase reporter assay) (16). EPBD dynamic simulations revealed that, in addition to the reported inhibition of TFIID binding, this Inr mutant has significantly reduced dynamic activity (Figure 1C), consistent with our hypothesis that both DNA dynamics and transcription factor binding are determinants of transcription initiation.
The lack of sharply defined TSS-specific dynamics observed in the TS promoter could explain the lack of a ‘focused’ TSS in this gene. Moreover, implanting a segment with localized TSS-like dynamics resulted in the appearance of a new strong TSS coinciding with the location of the dynamic maximum. Surprisingly, changes in the dynamic profile of the original promoter sequence resulting from the insert also reflect the experimentally observed transcriptional activity of this sequence. Considering that the known transcription factor binding sites for the TS promoter were intact in the Inr-insert variant, the reported experimental differences between the TS and the TS-Inr promoters can be readily explained from considerations of DNA dynamics. In contrast, according to a purely ‘transcription factor-centric’ view of transcription initiation, insertion of an Inr segment out of context in terms of transcription factor binding sites may introduce a new start site at the Inr but should not otherwise affect the original TSS distribution.
In conclusion, we propose that transcription factor binding and dynamic activity are both necessary for cellular gene transcription and are interdependent. The EPBD dynamic model appears to be uniquely capable of describing the sequence dependence of DNA dynamic features that are functionally relevant to transcription. In cells, the TSS-specific DNA breathing dynamics are likely to depend not only on DNA sequence, but also to be regulated by transcription factor binding, chromatin and DNA methylation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM073911 to A.U.); National Nuclear Security Administration of the US Department of Energy at Los Alamos National Laboratory (Contract DE-AC52-06NA25396). Funding for open access charge: DE-AC52-06NA25396.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. James Kadonaga for kindly providing the wtSCP1 construct.
REFERENCES
- 1.Alexandrov BS, Gelev V, Monisova Y, Alexandrov LB, Bishop AR, Rasmussen K.Ø., Usheva A. A nonlinear dynamic model of DNA with a sequence-dependent stacking term. Nucleic Acids Res. 2009;37:2405–2410. doi: 10.1093/nar/gkp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 3.Fiedler U, Marc Timmers HT. Peeling by binding or twisting by cranking: models for promoter opening and transcription initiation by RNA polymerase II. Bioessays. 2000;22:316–326. doi: 10.1002/(SICI)1521-1878(200004)22:4<316::AID-BIES2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 4.Alexandrov BS, Gelev V, Yoo SW, Bishop AR, Rasmussen K.Ø., Usheva A. Toward a detailed description of the thermally induced dynamics of the core promoter. PLoS Comput. Biol. 2009;5:e1000313. doi: 10.1371/journal.pcbi.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Usheva A, Shenk T. YY1 transcriptional initiator: protein interactions and association with a DNA site containing unpaired strands. Proc. Natl Acad. Sci. USA. 1996;93:13571–13576. doi: 10.1073/pnas.93.24.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Usheva A, Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 7.Leblanc BP, Benham CJ, Clark DJ. An initiation element in the yeast CUP1 promoter is recognized by RNA polymerase II in the absence of TATA box-binding protein if the DNA is negatively supercoiled. Proc. Natl Acad. Sci. USA. 2000;97:10745–10750. doi: 10.1073/pnas.200365097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 9.Choi CH, Kalosakas G, Rasmussen K.Ø., Hiromura M, Bishop AR, Usheva A. DNA dynamically directs its own transcription initiation. Nucleic Acids Res. 2004;32:1584–1590. doi: 10.1093/nar/gkh335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peyrard M, Bishop AR. Statistical mechanics of a nonlinear model for DNA denaturation. Phys. Rev. Lett. 1989;62:2755–2758. doi: 10.1103/PhysRevLett.62.2755. [DOI] [PubMed] [Google Scholar]
- 11.Dauxois T, Peyrard M, Bishop AR. Entropy-driven DNA denaturation. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1993;47:R44–R47. doi: 10.1103/physreve.47.r44. [DOI] [PubMed] [Google Scholar]
- 12.Alexandrov BS, Wille LT, Rasmussen K.Ø., Bishop AR, Blagoev KB. Bubble statistics and dynamics in double-stranded DNA. Phys. Rev. E. 2006;74:50901(R). doi: 10.1103/PhysRevE.74.050901. [DOI] [PubMed] [Google Scholar]
- 13.Wells RD, Larson JE, Grant RC, Shortle BE, Cantor CR. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J. Mol. Biol. 1970;54:465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]
- 14.Sponer J, Riley KE, Hobza P. Nature and magnitude of aromatic stacking of nucleic acid bases. Phys. Chem. Chem. Phys. 2008;10:2595–2610. doi: 10.1039/b719370j. [DOI] [PubMed] [Google Scholar]
- 15.Dornberger U, Leijon M, Fritzsche H. High base pair opening rates in tracts of GC base pairs. J. Biol. Chem. 1999;274:6957–6962. doi: 10.1074/jbc.274.11.6957. [DOI] [PubMed] [Google Scholar]
- 16.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat. Methods. 2006;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 17.Geng Y, Johnson LF. Lack of an initiator element is responsible for multiple transcriptional initiation sites of the TATA-less mouse thymidylate synthase promoter. Mol. Cell. Biol. 1993;13:4894–4903. doi: 10.1128/mcb.13.8.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsui TS, Segallg J, Weill PA, Roeder RG. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J. Biol. Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- 19.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Opin. Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudge TL, Johnson LF. Synergistic activation of the TATA-less mouse thymidylate synthase promoter by the Ets transcription factor GABP and Sp1. Exp. Cell Res. 2002;274:45–50. doi: 10.1006/excr.2001.5451. [DOI] [PubMed] [Google Scholar]
- 21.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 22.Urabe H, Sugawara Y, Tsukakoshi M, Kasuya T. Low-frequency Raman spectra of guanosine and nucleotides in ordered states: origin of the lowest-frequency mode. J. Chem. Phys. 1991;95:5519. [Google Scholar]
- 23.Fischer BM, Walther M, Uhd Jepsen P. Far-infrared vibrational modes of DNA components studied by terahertz time-domain spectroscopy. Phys. Med. Biol. 2002;47:3807–3814. doi: 10.1088/0031-9155/47/21/319. [DOI] [PubMed] [Google Scholar]
- 24.Saxena VK, Van Zandt LL. Local modes in a DNA polymer with hydrogen bond defect. Biophys. J. 1994;67:2448–2453. doi: 10.1016/S0006-3495(94)80731-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov BS, Gelev V, Bishop AR, Usheva A, Rasmussen K.Ø. DNA breathing dynamics in the presence of a terahertz field. LA-UR 09-03248, arXiv:0910.5294v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campa A, Giansanti A. Experimental tests of the Peyrard-Bishop model applied to the melting of very short DNA chains. Phys. Rev. E. 1998;58:3585. [Google Scholar]
- 27.Alexandrov BS, Voulgarakis N, Rasmussen K.Ø., Usheva A, Bishop AR. Pre-melting dynamics of DNA and its relation to specific functions. J. Phys. Condens. Matter. 2009;21:034107. doi: 10.1088/0953-8984/21/3/034107. [DOI] [PubMed] [Google Scholar]
- 28.Voulgarakis NK, Redondo A, Bishop AR, Rasmussen K.Ø. Probing the mechanical unzipping of DNA. Phys. Rev. Lett. 2006;96:248101. doi: 10.1103/PhysRevLett.96.248101. [DOI] [PubMed] [Google Scholar]
- 29.Ares S, Voulgarakis NK, Rasmussen K.Ø., Bishop AR. Bubble nucleation and cooperativity in DNA melting. Phys. Rev. Lett. 2005;94:035504. doi: 10.1103/PhysRevLett.94.035504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.