Abstract
Abasic sites represent the most frequent lesion in DNA. Since several events generating abasic sites concern guanines, this damage is particularly important in quadruplex forming G-rich sequences, many of which are believed to be involved in several biological roles. However, the effects of abasic sites in sequences forming quadruplexes have been poorly studied. Here, we investigated the effects of abasic site mimics on structural, thermodynamic and kinetic properties of parallel quadruplexes. Investigation concerned five oligodeoxynucleotides based on the sequence d(TGGGGGT), in which all guanines have been replaced, one at a time, by an abasic site mimic (dS). All sequences preserve their ability to form quadruplexes; however, both spectroscopic and kinetic experiments point to sequence-dependent different effects on the structural flexibility and stability. Sequences d(TSGGGGT) and d(TGGGGST) form quite stable quadruplexes; however, for the other sequences, the introduction of the dS in proximity of the 3′-end decreases the stability more considerably than the 5′-end. Noteworthy, sequence d(TGSGGGT) forms a quadruplex where dS does not hamper the stacking between the G-tetrads adjacent to it. These results strongly argue for the central role of apurinic/apyrimidinic site damages and they encourage the production of further studies to better delineate the consequences of their presence in the biological relevant regions of the genome.
INTRODUCTION
Genome integrity is of vital importance to cellular survival and replication. However, a broad variety of causes, ranging from the effect of genotoxic chemicals to error-prone cellular processes, renders DNA rather vulnerable to damages and mutations (1). The most common types of DNA defects are single base mismatch, abasic site, single base bulges and oxidized bases. Among these, abasic sites [apurinic/apyrimidinic (AP) sites] are expected to be one of the most frequent lesions in DNA. They can arise mainly as a result of two processes: the spontaneous hydrolysis of the N-glycosidic bond (2,3) (generally depurination) or the removal of altered bases by DNA glycosylases (2,4) during the first stage of the base excision repair process (5). In addition to these two causes, damaging chemicals such as free radicals and alkylating agents can promote the release of bases, often by introducing modifications that destabilize the N-glycosidic bond by generating a better leaving group moiety (6,7). The measured spontaneous depurination rate in double-stranded DNA causes the loss of ∼10 000 purines per mammalian cell per day (6,7). Combined with the AP sites produced by DNA glycosylases, that remove the altered bases, the daily amount of generated AP sites is probably much higher. One estimate yielded steady state levels of 50 000–200 000 AP sites per cell in several rat tissues and human liver (8). The formation of altered bases can arise in several ways. For example, reactive oxygen species, the products of normal cellular respiration, can generate a variety of oxidized DNA base damages, including 8-oxo-7,8-dihydroguanine that is frequently used as a biomarker for oxidative DNA damage (9,10). Furthermore, non-enzymatic alkylation from endogenous sources forms cytotoxic and mutagenic products, as 3-alkyladenine and O6-alkylguanine (11,12). Moreover, further methylation, oxidation and deamination processes can produce other types of damaged bases, such as N7-methylguanine, 5,6-dihydroxy-5,6-dihydrothymine (Tg) or uracil (12–14). The accumulation of unrepaired AP sites can be lethal because they hinder DNA replication (2,15). Moreover, even when bypassed by DNA polymerases, AP sites frequently lead to the insertion of mutagenic bases opposite to them (2,15). Also, these lesions are subject to relatively facile β-elimination reaction leading to unprocessed DNA strand breaks (4,16,17). Finally, recent studies have shown that the aldehyde residue formed by an AP site can generate an interstrand cross-link via carbinolamine/imine formation with the exocyclic N2-amino group of a guanine residue on the opposite strand of the double helix (18).
Since many of the events generating AP sites involve guanine residues, the presence of such DNA lesions comes out particularly important in G-rich tracts. These have been observed in critical segments of eukaryotic and prokaryotic genomes, promoter regions, both short microsatellite and longer minisatellite repeats, ribosomal DNAs, as well as telomeres in eukaryotes and immunoglobulin heavy chain switch regions of higher vertebrates. Guanine-rich tracts have the potential to form G-quadruplex structures following transient duplex destabilization, a process that accompanies transcription, replication and recombination. Furthermore, systematic algorithmic searches of bacterial and human genomes for guanine-rich tracts (19–21) have noted that such putative G-quadruplex-forming sequences are prevalent in proto-oncogenes and essentially lacking in tumor suppressor genes (22). Although the above considerations clearly suggest a relationship between AP sites, G-rich sequences and G-quadruplex structures, at the best of our knowledge, only reports concerning quadruplex structures containing AP sites into the loops have appeared in literature so far (23–26).
In an effort to investigate the effects of AP sites in both parallel and antiparallel quadruplex structures, we have undertaken a systematic study concerning the structural, thermodynamic and kinetic features of oligodeoxynucleotides (ODNs) potentially able to form quadruplex structures and containing residues mimicking AP sites. In this frame, we have synthesized five ODNs all based on the parallel quadruplex forming sequence d(TGGGGGT) (Table 1), in which all guanines have been replaced, one at a time, by an AP site mimic. Given the instability of the natural hemiacetal AP site, a tetrahydrofuranyl analog was employed instead (dSpacer, dS, Figure 1). The main structural properties of quadruplexes formed by dSpacer containing ODNs have been investigated by NMR and circular dichroism (CD) spectroscopy. Strands association and dissociation kinetics have been studied as well.
Table 1.
Sequences and apparent melting temperatures (T1/2) of the oligonucleotides studied
| Name | Oligonucleotides |
|
|---|---|---|
| Sequence (5′–3′) | T1/2 (°C) | |
| AQ1 | d(TSGGGGT) | >100 |
| AQ2 | d(TGSGGGT) | 87 |
| AQ3 | d(TGGSGGT) | <25a; 75b |
| AQ4 | d(TGGGSGT) | 63 |
| AQ5 | d(TGGGGST) | >100 |
dS = dSpacer, tetrahydrofuranyl analog (Figure 1).
aEvaluated from the melting curve (1°C/min scan rate) after the annealing procedure.
bEvaluated from the melting curve after 1 week storage at 5°C.
Figure 1.
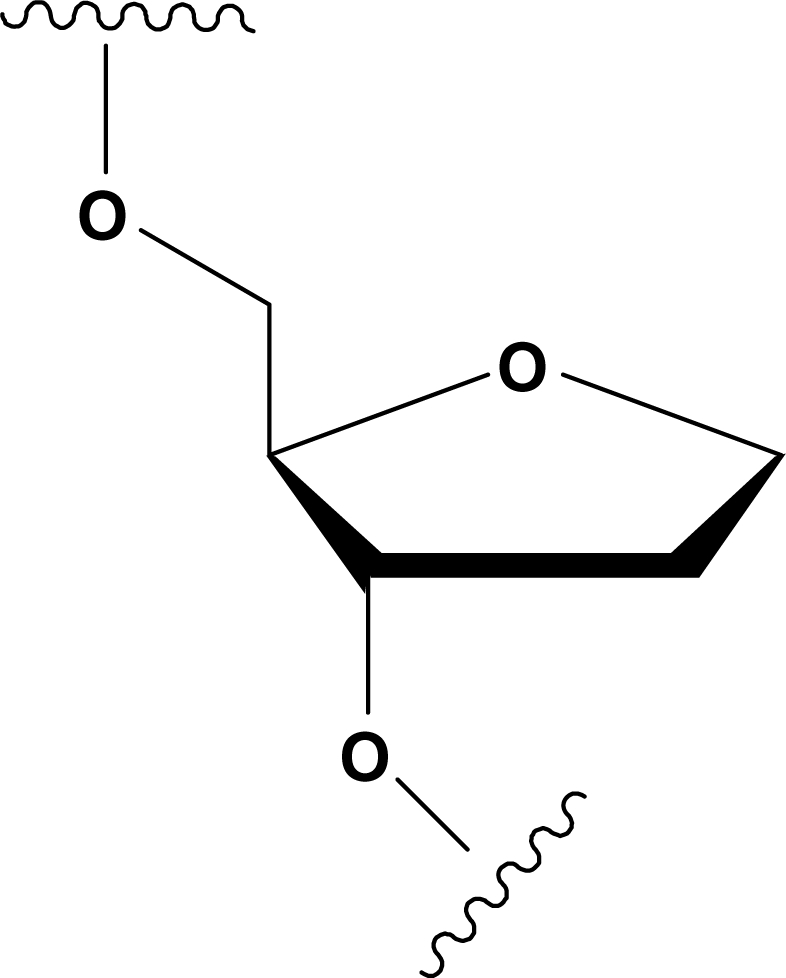
Structure of the tetrahydrofuranyl analog, dSpacer (dS) introduced in ODNs in Table 1 as AP sites mimic.
MATERIALS AND METHODS
Oligonucleotides synthesis and purification
The Oligonucleotides AQ1-AQ5 were synthesized on a Millipore Cyclone Plus DNA synthesizer using solid phase β-cyanoethyl phosphoramidite chemistry at 15 μmol scale. The synthesis were performed by using normal 3′-phosphoramidites and a 5′-dimethoxytrityl-3′-phosphoramidite-1′,2′-dideoxyribose (dSpacer, dS, Link Technologies) for the introduction of an abasic site mimic moiety in each sequence. The oligomers were detached from the support and deprotected by treatment with concentrated aqueous ammonia at 55°C overnight. The combined filtrates and washings were concentrated under reduced pressure, redissolved in H2O, analyzed and purified by high-performance liquid chromatography on a Nucleogel SAX column (Macherey–Nagel, 1000-8/46), using buffer A: 20 mM KH2PO4/K2HPO4 aqueous solution (pH 7.0) containing 20% (v/v) CH3CN and buffer B: 1 M KCl, 20 mM KH2PO4/K2HPO4 aqueous solution (pH 7.0) containing 20% (v/v) CH3CN; a linear gradient from 0 to 100% B for 30 min and flow rate 1 ml/min were used. The fractions of the oligomers were collected and successively desalted by Sep-pak cartridges (C-18). The isolated oligomers proved to be >98% pure by NMR.
NMR
NMR samples were prepared at a concentration of ∼5 mM in 0.6 ml (H2O/D2O 9:1 v/v) buffer solution having 10 mM KH2PO4/K2HPO4, 70 mM KCl and 0.2 mM EDTA (pH 7.0). For D2O experiments, the H2O was replaced with D2O by drying down the sample, lyophilization and redissolution in D2O alone. NMR spectra were recorded with a Varian Unity INOVA 500 MHz spectrometer. 1H chemical shifts were referenced relative to external sodium 2,2-dimethyl-2-silapentane-5-sulfonate. 1D proton spectra of samples in H2O were recorded using pulsed-field gradient WATERGATE (27) for H2O suppression. Phase sensitive NOESY spectra (28) were recorded with mixing times of 180 ms (T = 25°C). Pulsed-field gradient WATERGATE was used for NOESY spectra in H2O with 200 ms mixing times. TOCSY spectra (29) with mixing times of 120 ms were recorded with D2O solutions. NOESY and TOCSY were recorded using a TPPI (30) procedure for quadrature detection. In all 2D experiments, the time domain data consisted of 2048 complex points in t2 and 400–512 fids in t1 dimension. The relaxation delay was kept at 1.2 s for all experiments.
Gel electrophoresis
Modified oligonucleotides were analyzed by non-denaturing PAGE. Samples in the NMR buffer (20 mM KH2PO4, 70 mM KCl and 0.2 mM EDTA, pH = 7) were loaded on a 20% polyacrylamide gel containing Tris–Borate-EDTA (TBE) 2.5× and KCl 50 mM. The run buffer was TBE 1× containing 100 mM KCl. Single-strand samples were obtained by LiOH denaturation. For all samples, a solution of glycerol/TBE 1×, 10 100 mM KCl 2:1 was added just before loading. Electrophoresis was performed at 9.2 V/cm at a temperature close to 5°C. Bands were visualized by UV shadowing.
CD spectroscopy, kinetic and thermal analyses
The quadruplex samples have been prepared by dissolving the lyophilized compound in 10 mM phosphate buffer with 70 mM KCl, 0.2 mM EDTA at pH 7. The solution has been annealed by heating at 95°C for 5 min and slowly cooling to room temperature. The concentration of the dissolved oligonucleotide has been evaluated by UV measurement at 95°C, using as molar extinction coefficient the value calculated by the nearest-neighbour model (31) for the sequence d(TGGGGT). Thermal dissociation and association of the quadruplex sample have been monitored by recording the CD signal at 263 nm as function of the temperature. The molar ellipticity was calculated from the equation [θ] = CD/10 × c × l, where CD is the CD signal, c is the molar concentration of the single strand and l is the path length of the measurement cell in centimetres. The temperature was electronically increased/decreased at scanning rates of 0.5 and 1.0°C/min.
The quadruplexes dissociation kinetics were measured by rapidly increasing the temperature and following the change with time of the CD signal at 263 nm (32). All the experiments were performed at 2.0 × 10−4 M single-strand concentration and in 60–90°C temperature range. In these conditions, the re-association reaction can be neglected. The kinetic constants were obtained by fitting the recorded time-dependent decrease in CD signal with a single exponential function.
Association of single strands into a well-defined quadruplex structure was monitored for each sequence in the same buffer used for the melting experiments. The experiments were carried out at a 1 × 10−4 M single-strand concentration. The samples were incubated at 95°C for 5 min to allow for the quadruplex dissociation. The structural transition from the single strands to the quadruplex structure was monitored in a temperature range 0–20°C by recording the CD spectra as a function of time. Assuming that the dissociation is negligible, the global process allowing the formation of the quadruplex structure, starting from the single strands, could be written as follows:
| 1 |
The change of the concentration of single strand is defined, through the kinetic equation, in the following way:
| 2 |
where v is representative of the association rate; [S] and [Q] are the concentration of the single strand and quadruplex, respectively; k is the kinetic association constant and n is the order of the association reaction. Equation (2) could be solved with respect to the single-strand concentration and integrated. The resulting equation describes the change of the single-strand concentration with the time:
| 3 |
where [S]t is the single-strand concentration at time t, [S]0 is the initial single-strand concentration, n is the order of the reaction with respect to the single-strand concentration and kon is the rate constant for the single-strand disappearance. As previously described (33), the changes in CD spectra at a given time t can be related with the single-strand concentration according to the equation:
| 4 |
where  and
and 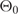 corresponds to the ellipticity of the final and initial state, respectively.
corresponds to the ellipticity of the final and initial state, respectively. 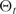 is the ellipticity at time t and
is the ellipticity at time t and 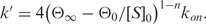
The association rate constants and the reaction orders were obtained by fitting the ellipticity at 263 nm as a function of time by means of Equation (2) using ORIGIN 7.5 software.
The association/dissociation constants have been obtained at different temperatures in order to build the Arrhenius plot. The following form of the Arrhenius equation has been used to interpolate the values of the kinetic constants at different temperatures:
| 5 |
From the Arrhenius analysis, the activation energy and the pre-exponential parameters, of both association and dissociation processes, have been derived.
Molecular modelling
The main conformational features of the quadruplexes AQ1-5 have been explored by means of a molecular modelling study. The AMBER force field using AMBER 99 parameter set was used (34). The initial coordinates for the starting model of the quadruplex [d(TGGGGGT)]4 were taken from the NMR solution structure of the quadruplex [d(TTGGGGT)]4 (Protein Data Bank entry number 139D), choosing randomly one of the four available structures. The initial [d(TGGGGGT)]4 G-quadruplex model was built by replacing the second thymidine residue in each of the four d(TTGGGGT) strands with a guanosine one. The complete structures of quadruplexes were then built using the Biopolymer building tool of Discover by deleting each guanosine residue, one at a time, and replacing it with a dSpacer unit for each strand. The calculations were performed using a distance-dependent macroscopic dielectric constant of 4r, and an infinite cut-off for non-bonded interactions to partially compensate for the lack of solvent used (35). Using the steepest descent followed by quasi-Newton–Raphson method (VA09A), the conformational energy of each complex was minimized until convergence to a RMS gradient of 0.1 kcal/mol Å was reached. Illustrations of structures were generated using the INSIGHT II program, version 2005 (Accelrys, San Diego, CA, USA). All the calculations were performed on a PC running Linux ES 2.6.9.
RESULTS
NMR experiments
All the NMR samples (see ‘Materials and Methods’ section) were heated for 5–10 min at 80°C and slowly cooled down (10–12 h) to room temperature. The solutions were equilibrated at least for 1 week at 4°C and then their 1H-NMR spectra were recorded using pulsed-field gradient WATERGATE for H2O suppression. The achievement of a completed annealing process was guaranteed by the reaching of superimposable 1H-NMR spectra on changing time.
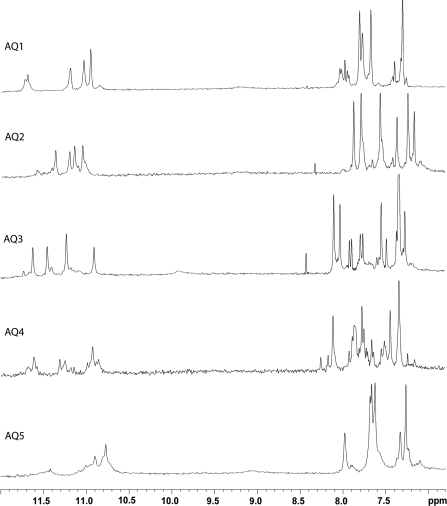
With the exclusion of some weak resonances due to minor conformations also present in solution, the relatively simple appearance of most of the 1D spectra of oligomers (Figure 2) indicates that, in the conditions used here, the modified ODNs form mainly single well-defined hydrogen-bonded conformations, consistent with 4-fold symmetric G-quadruplex structures containing four G-tetrads with all strands equivalent to each other. In fact, all the 1H-NMR spectra of the different samples show four main signals in the region 10.5–12.0 p.p.m., attributable to imino protons involved in Hoogsteen hydrogen bonds of G-quartets, and six main singlets belonging to four guanine H8 and two thymine H6 protons in the aromatic region. On the other hand, it is interesting to note that there are significant differences in the 1D-NMR spectra of the several samples examined, thus suggesting that the introduction of the same AP site mimicking moiety (dSpacer) can produce different effects depending on its position into the sequence.
Figure 2.
Aromatic and imino protons regions of the 1H-NMR spectra of the ODNs (Table 1).
In spite of the quite complicated appearance of 1D NMR spectra showed by some complexes, we were able to perform almost complete resonance assignments of the main species present in solution for all the samples. The resonances assignments have been accomplished on the basis of NOESY and TOCSY spectra, obtained at 500 MHz (see ‘Materials and Methods’ section), following the standard procedures (Supplementary Tables). As reported for other parallel quadruplex structures, the observed NOEs among G-H8 and T-H6 and their own H1′, H2′ and H2′′ ribose protons and the H1′, H2′ and H2′′ protons on the 5′ side suggest that all quadruplexes assume a right-handed helical winding. As for the glycosidic torsion angles, the lack of strong NOEs between G H8 and the H1′ of the same residue, in comparison with those observed between G H8 and its deoxyribose H2′, indicates that all G residues are in an anti-glycosidic conformation in all the complexes.
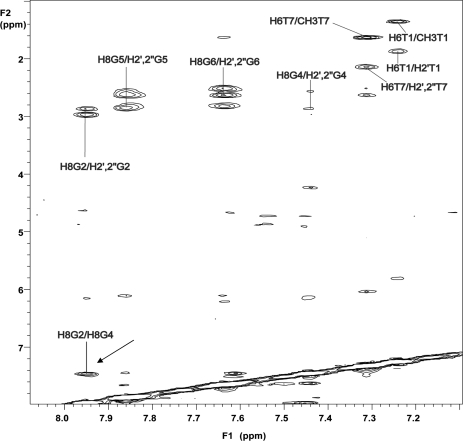
As expected, the normal sequential connectivities path is broken at the dSpacer steps for most of the samples. This datum indicates that the abasic site constitutes a sort of ‘breakpoint’ in the NOE connectivities path along the strands of the complexes, with the added effect of spacing the two resulting tracts of the quadruplexes. However, it is noteworthy that the NOESY spectrum of AQ2 shows the presence of a strong NOE between the aromatic protons of the two dG residues adjacent to the dSpacer unit (Figure 3), thus indicating that, in this case, the two portions of the molecule lie in mutual close proximity, differently from the behaviour of the other quadruplex complexes.
Figure 3.
2D NOESY (500 MHz) region correlating base and H2′/H2′′ sugar protons in AQ2 [d(T1G2S3G4G5G6T7)]4. The arrow indicates a NOE between aromatic protons of G2 and G4.
As far as AQ1 is concerned, the 1D proton spectrum is affected by slight line broadening of the signals and weak splitting of resonances belonging to the G-quartet near to the dSpacer, prompting that the 5′-end of the complex is subjected to a major conformational flexibility.
The insertion of the dSpacer at central position (AQ3), instead, hinders the complete quadruplex association, as demonstrated by the 1D spectrum of the sample, acquired at 25°C after several weeks of annealing at 5°C, as the other quadruplexes. Indeed, this spectrum shows mainly 12 signals in the range between 7.0 and 8.5 p.p.m., and only primarily four imino peaks in the region 10.8–11.8 p.p.m. Raising the temperature to 80°C, the lowest six signals gradually increased in intensity, whereas the other six, along with the four imino peaks, progressively disappeared (data not shown). Thus, at 80°C only six signals were present in the aromatic region of the 1D spectrum, while no imino peak was present. This datum clearly shows that, in this condition, AQ3 is completely unstructured, while at 25°C the quadruplex structure is in equilibrium with its single strand.
The behaviour of AQ4 appears relatively more complicated since its 1D spectrum shows an exceeding number of signals, probably due to the presence of several quadruplex species in solution that has made difficult the complete assignment of the main one. However, in spite of the structural heterogeneity observed, the NOESY spectrum showed rather dispersed cross-peaks that allowed us to assign most of the non-exchangeable protons following the standard procedures (Supplementary Tables), although signals become more weak and less defined for the G-tetrads in proximity of the dSpacer step.
As regards AQ5, its 1D proton spectrum shows that it forms a major quadruplex structure, whose signals are affected by severe line broadening and partial overlapping if compared with the other complexes. This datum suggests that the complex is characterized by a conformational flexibility that becomes more significant for the portions of the molecule close to the dSpacer unit.
Gel electrophoresis
In order to estimate the molecularity of the complexes, we performed a PAGE of all samples compared with the quadruplex [d(TGGGGT)]4 (Supplementary Figure S1). The migration of all non-denaturated samples of the dS-ODNs appears undoubtedly slower than those of the corresponding single strands, clearly showing the presence of multimolecular complexes. Furthermore, their electrophoretic motilities are quite comparable to that of [d(TGGGGT)]4 suggesting the formation of complexes with the same molecularity. More importantly, for all samples, no structures larger than quadruplexes could be detected.
CD spectroscopy, kinetic and thermal analyses
All the sequences investigated in this study show the characteristic CD spectrum of a parallel quadruplex (36) proving their ability to form this structure in the experimental conditions (Supplementary Figures S2, S4A, S5A and S6A). The comparison between the intensity of the molar ellipticity of the CD spectra suggests that the sequences do not form the same amount of structured quadruplex. The molar ellipticity is higher for the quadruplex structures formed by the sequences possessing the dSpacer close to the 5′ terminal region and the signal is weaker for the structures formed by the sequences possessing the dSpacer close to the 3′ terminal region. The thermal dissociation process is kinetically controlled due to the slow rates of both the dissociation and association processes. For this reason, we obtain apparent melting temperatures (T1/2) (37) (Table 1) that are not equilibrium temperatures. Although the kinetic control rules the melting processes, for AQ2 and AQ4, we obtained the enthalpy changes of the quadruplex dissociation processes through the kinetic parameters (Eon, Eoff). Since the sequences show highly different properties, the results for each of them are reported separately.
ODNs AQ1 and AQ5
The thermal analysis carried out on the quadruplex structures formed by these sequences suggests that they possess a higher stability with respect to the other investigated ODNs. Interestingly, the apparent melting temperatures (T1/2), at both, 1°C/min and 0.5°C/min of heating rate, are >100°C and only for the quadruplex structure obtained from the sequence AQ5 partial melting curves have been obtained (Supplementary Figure S3). Both sequences are able to produce very stable quadruplex structures that do not dissociate at high temperature. Interestingly, the samples are still able to reproduce the typical CD spectrum of a parallel quadruplex structure after a long period at 95°C (data not shown). The high thermal stability of the quadruplex structures, obtained with these sequences, prevents us to perform a kinetic characterization of both the association and dissociation processes due to the impossibility to disrupt the starting structure.
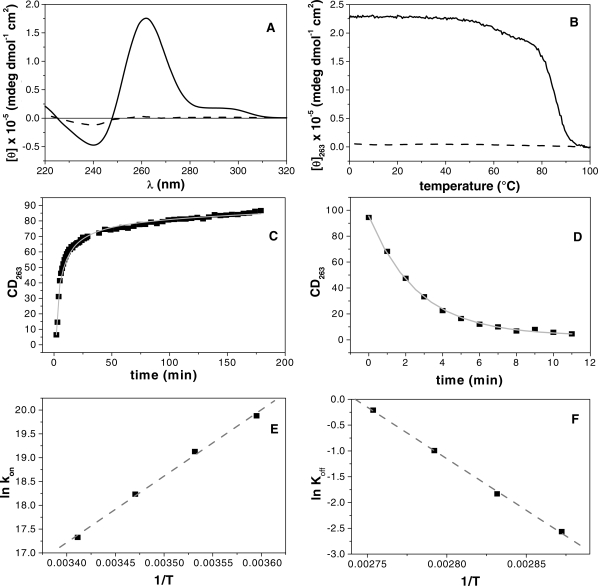
ODN AQ2
In Figure 4B, the melting and annealing CD profiles are shown at the heating and cooling rate of 1.0°C/min and at a strand concentration of 5 × 10−5 M. The quadruplex structure shows a hysteresis between the melting and annealing profiles, thus indicating that the association/dissociation reactions are not at thermodynamic equilibrium (37,38) during the heating and cooling experiments. Interestingly the CD spectrum recorded at 20°C after the annealing shows a very weak intensity, and after 1 day storage at 2°C the CD signal increases its intensity recovering the typical features of a quadruplex structure (Figure 4A). The sample is able to reproduce the typical parallel quadruplex structure CD spectrum after a long period of storage at 2°C, suggesting that the unfolding process is reversible. The association process is strand concentration dependent and it became measurable by using a higher strand concentration (1 × 10−4 M). Particularly, to get a deeper understanding of the latter characteristic we recorded melting/annealing curves at different strand concentrations and at different heating/cooling rates (Supplementary Figure S4A and S4B). Melting and annealing curves have been recorded at three different single strand concentrations (5 × 10−5, 1 × 10−4 and 1 × 10−3 M). All the melting curves show the same middle point temperature, clearly suggesting the monomolecular nature of the dissociation process. On the other hand, the annealing curves change with the increase of the single-strand concentration. Particularly, the higher the concentration of single strand, higher is the fraction of quadruplex structure reformed at the end of the annealing. The latter result is a clear effect related to the molecularity of the association process in which four strands need to interact together to form the quadruplex.
Figure 4.
ODN AQ2. CD spectroscopy: (A) the panel reports the spectrum at 20°C before heating (solid line) and at 100°C (dashed line). Thermal analysis (B) showing the melting curve (solid line) and the annealing one (dashed line) recorded at a heating and cooling scanning rates of 1°C/min. Association/dissociation kinetic analysis (C and D) of the quadruplex structure following the change in the CD signal at 263 nm; the experimental points are reported as black squares and the interpolating curves as a grey solid line. The panels (E and F) show the Arrhenius plots for the association and the dissociation processes; the experimental values of the kinetic constants (black squares) have been interpolated with the Equation (5) in order to get the activation energies and the pre-exponential values.
In order to shed light on the dissociation and re-association, a kinetic study has been performed by CD, as described in ‘Materials and Methods’ section. Representative molar ellipticity versus time profiles at 20°C (re-association) and 90°C (dissociation) are shown in Figure 4C and D, respectively. The experimental curve in Figure 4C was fitted using the Equation (2). The data for the studied ODNs samples were well fitted by reaction order of 4.0 and the kinetic constants (kon) are reported in the Table 2. The kinetic of quadruplex dissociation was also investigated by performing temperature-jump experiments (32) in which the temperature of the sample was rapidly increased and the rate of quadruplex dissociation was monitored by following the change of CD signal at 263 nm with time. From the kinetic profile recorded at 90°C (Figure 4D), the dissociation rate constant (Table 3) was obtained by fitting the kinetic profile with a single exponential curve. The Arrhenius analysis has been carried out on the kinetic constants in order to get the activation energies and the pre-exponential parameters of both the association and the dissociation process (Tables 2 and 3).
Table 2.
Kinetic parameters for the quadruplexes association
| ODN sequence | T (°C) ±1 | kon (M−3 min−1)a | Eon (kJ mol−1) |
|---|---|---|---|
| AQ2 d(TGSGGGT) | 5 | (4.3 ± 0.4) × 108 | −193 ± 5 |
| 10 | (2.0 ± 0.2) × 108 | ||
| 15 | (8.3 ± 0.8) × 107 | ||
| 20 | (3.8 ± 0.3) × 107 | ||
| AQ4 d(TGGGSGT) | 0 | (1.86 ± 0.2) × 108 | −115 ± 7 |
| 3 | (8.1 ± 0.8) × 107 | ||
| 5 | (3.96 ± 0.4) × 107 | ||
| 7 | (2.27 ± 0.2) × 107 |
All the values have been determined in a 70 mM KCl, 10 mM KH2PO4, pH 7.0 buffer.
akon is defined as d[S4]/dt = −kon[S4]n.
Table 3.
Kinetic parameters for the quadruplexes dissociation
| ODN sequence | T (°C) ±1 | koff × 104 (min−1)a | t1/2 (min)b | Eoff (kJ mol−1) |
|---|---|---|---|---|
| AQ2 d(TGSGGGT) | 75 | 0.08 ± 0.04 | 8.7 | 131 ± 4 |
| 80 | 0.16 ± 0.01 | 4.3 | ||
| 85 | 0.37 ± 0.01 | 1.9 | ||
| 90 | 0.81 ± 0.01 | 0.9 | ||
| AQ4 d(TGGGSGT) | 60 | 0.19 ± 0.04 | 3.6 | 166 ± 6 |
| 63 | 0.30 ± 0.01 | 2.3 | ||
| 65 | 0.40 ± 0.01 | 1.7 | ||
| 67 | 0.50 ± 0.02 | 1.4 |
All the values have been determined in a 70 mM KCl, 10 mM KH2PO4, pH 7.0 buffer.
akoff is defined as d[S]/dt = −d[S4]/dt = koff[S4].
bt1/2 is the half-dissociation time of quadruplex structures defined as 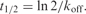
ODN AQ3
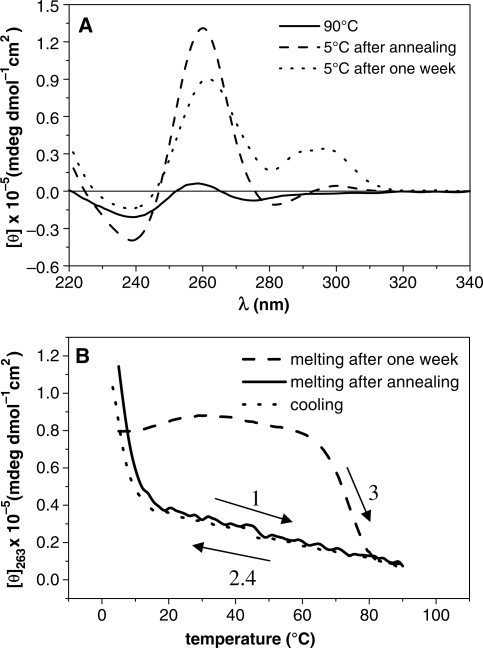
The melting curve, acquired just after annealing and recorded between 5 and 90°C at a strand concentration of 5 × 10−5 M, does not show the sigmoid profile and the annealing curve closely resembles the melting one (Figure 5B). The presence of a very weak hysteresis between the two curves suggests an almost instantaneous reversibility of the dissociation/association process. Because of the absence of the sigmoid shape curves, we could not define a rigorous melting temperature for the studied molecule; however, T1/2 (37) is <25°C, suggesting a low thermal stability of the structured oligonucleotide. Intriguingly, equilibrating the molecule solution at 5°C up to 1 week, the CD spectrum changes. The new CD spectrum exhibits the profile reported in Figure 5A: a weak maximum intensity appears at 290 nm and a maximum intensity at 263 nm decreases suggesting the presence of minor amounts of other quadruplexes. The thermal features have changed too: the new melting curve, sigmoid shaped, displays a higher melting temperature (75°C). Interestingly the annealing curve does not change at all and the higher hysteresis effect clearly proposes that this new system configuration is ruled by a profound kinetic control (36) that it did not allow us to get a deeper characterization of the system.
Figure 5.
ODN AQ3. The panel (A) reports the spectra of the quadruplex structure at 90°C (solid line), at 5°C after annealing by cooling the sample from 90°C to 1°C/min (dashed line) and after 1 week storage at 5°C (dotted line). The difference between the last two spectra at 5°C suggests that a slow conformational rearrangement has occurred. In the panel (B) are represented the melting curves obtained at a heating rate of 1°C/min recorded after the annealing procedure (1) and after 1 week of equilibration at 5°C (3). In both cases, the annealing profiles (2, 4) share high similarities.
ODN AQ4
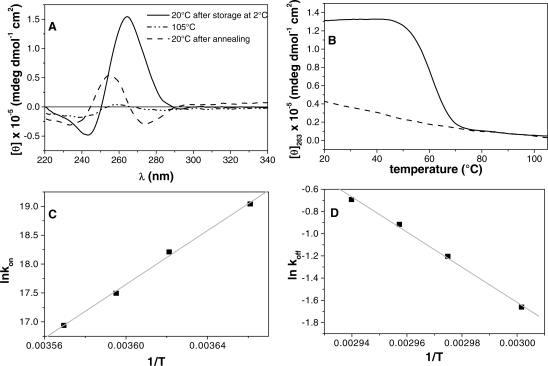
In Figure 6B, the melting and annealing CD profiles are shown at the heating and cooling rate of 1.0°C/min and at a strand concentration of 5 × 10−5 M. The quadruplex structure shows an hysteresis between the melting and annealing profiles (37,38), which indicates that the association/dissociation reaction is not at thermodynamic equilibrium during the heating and cooling experiments. Interestingly the CD spectrum after the annealing shows a very weak intensity, and after 1 day storage at the temperature of 2°C the CD spectrum, recorded at 20°C, increases its intensity recovering the typical features of a quadruplex structure (Figure 6A). The sample is able to reproduce the typical parallel quadruplex structure CD spectrum after a long period of storage at 2°C, suggesting that the unfolding process is reversible. For this sample, as for AQ2, we performed melting/annealing experiments at different single-strand concentration and different scan rates (Supplementary Figure S4C and S4D). Those experiments suggest identical conclusions we got for the AQ2: the dissociation process is monomolecular and the association one possesses a higher molecularity. The dissociation/association process has been studied as we did for the sequence AQ2 and the Arrhenius analysis has been carried out on the kinetic constants in order to get the activation energies and the pre-exponentials parameters of both the association and the dissociation process (Tables 2 and 3).
Figure 6.
ODN AQ4. CD spectroscopy: (A) the panel reports the spectra at 20°C after storage at 2°C and before heating (solid line), at 105°C (dash-dot-dot line) and at 20°C after annealing (dashed line). Thermal analysis (B) showing the melting curve (solid line) and the annealing one (dashed line) obtained with a heating/cooling rate of 1°C/min. The panels (C and D) show the Arrhenius plots for the association and the dissociation processes.
Molecular modelling
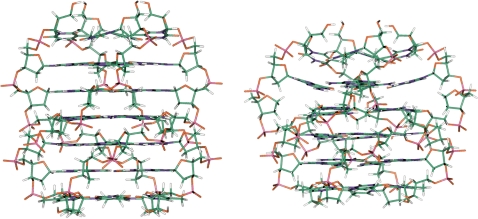
In Figure 7, the molecular models of quadruplexes [d(TGGGGGT)]4 and AQ2 are shown, obtained as described in the ‘Materials and Methods’ section. Apart from the presence of a further G-tetrad, the obtained model of [d(TGGGGGT)]4 resulted very similar to the X-ray structure of the complex [d(TGGGGT)]4 proposed by Phillips et al. (39). On the other hand, the NOE patterns showed by the dS-containing quadruplexes are quite comparable to those exhibited by several parallel quadruplex structures adopting a right-handed helical alignment (included [d(TGGGGT)]4) (40–42), although the pattern is broken by the presence of the dS moiety in most of the modified quadruplexes. These considerations validate the building approach of the model. The molecular model of AQ2 takes into account the NMR data clearly indicating a stacking between the tetrads adjacent to the dS. However, a comparison between the models of the quadruplexes [d(TGGGGGT)]4 and AQ2 suggests significant differences. Particularly, in the AQ2 model the tetrad adjacent to the dS toward the 5′-end appears not completely planar due to a remarkable buckle. Consistently, the T1/2 of AQ2 comes out lower than those of quadruplexes formed by AQ1, AQ5 and TGGGGT in which the G-run is not broken.
Figure 7.
Molecular models of the quadruplexes formed by ODNs TGGGGGT (left) and AQ2 (right). The structures are oriented with the 5′-end upward (carbons, green; nitrogens, blue; oxygens, red; hydrogens, white).
DISCUSSION
The insertion of a dSpacer group mimicking an abasic site in the sequence d(TGGGGT) causes remarkable effects on the kinetics and the thermodynamics of quadruplex formation. In general, the presence of the dSpacer seems to reduce the stability of the resulting quadruplex structure in a site-dependent behavior. The introduction of the dSpacer in the proximity of the 3′ terminal regions decreases the stability in a more dramatic way respect to its presence in the 5′ terminal region. The several modified quadruplexes show different thermal stability and they are all less stable than the natural quadruplex [d(TGGGGT)]4 containing the same number of G-tetrads (33).
The ODNs AQ1 and AQ5 produce stable parallel quadruplex structures that do not dissociate at the highest temperature reachable. This result suggests that the presence of the dSpacer group before and after the G-tract does not produce strong destabilizing effects on the global quadruplex structure, although, in both cases, the NMR experiments suggest an increased molecular flexibility around the end near the dSpacer. On the other hand, when the dSpacer group is located inside the G-tract (AQ2 and AQ4) the thermal stability decreases and the association/dissociation process becomes measurable. Our kinetic analyses afforded values for the kon that well agree with the ones reported by other authors for [d(TGGGGT)]4 and [d(TGGGGGT)]4 (33,43). Particularly, the kon for the previous quadruplex structures are 3 × 107 at 20°C and 2.3 × 1012 at 3°C. As expected, our data closely resemble the d[(TGGGGT)]4 value suggesting a similar behaviour for AQ2 and AQ4. For both sequences, the kinetic analysis clearly suggests that the association process for the formation of the quadruplex structure possesses an order reaction of four. A possible mechanism could be represented from the following model:
According to this model, in an initial stage there is a quickly pre-equilibrium between the single strands (S) and possible dimeric species (D). After the pre-equilibration step, the dimeric species slowly evolves toward the quadruplex structure (Q). This mechanism agrees with the experimental derived order reaction. Interestingly, an alternative mechanism could be taken into consideration, according to which four separated filaments evolve toward the final quadruplex structure without formation of dimeric species. However, this model would require the interaction of four different strands at the same time and this event is much less probable than the interaction of two single strands followed by the interaction of two dimeric species. Moreover, the mechanism with two states is in agreement with the one reported from Wyatt et al. (44). According to it, the hypothesis of the initial pre-equilibrium step could explain the negative activation energy obtained for the association processes (Table 2). On the other hand, the kinetic analysis showed that the dissociation process has a positive activation energy (Table 3). The difference between the latter values allowed us to estimate the enthalpic changes (44,45) related to the dissociation of the quadruplex structures formed by AQ2 and AQ4: ΔH = 324 kJ/mol and ΔH = 281 kJ/mol, respectively. These values are comparable with the enthalpy change of the dissociation of the parallel quadruplex [d(TGGGGT)]4. Particularly, for this structure the enthalpy change obtained from other authors (33) is centered ∼320 kJ/mol, thus suggesting an individual value per plane of 80 kJ/mol. The enthalpy change for the dissociation of the quadruplex structure formed by ODN AQ2 suggests a conformation in which all the planes in quadruplex structure are formed and stacked each other. Probably, the dSpacer group in that position does not reduce the ability to form stacked G-tetrads in its proximity, in spite of its intrinsic flexibility. This hypothesis is strongly supported by the NMR data. Particularly, the NOESY spectrum (Figure 3) shows a strong NOE between the aromatic protons of the deoxyguanosines closest to the dSpacer moiety, thus clearly indicating that, in this position, the deoxySpacer does not hamper an effective stacking between the adjacent G-tetrads. As a consequence, the dSpacer moiety should be driven out the quadruplex stem toward the outer region, forming a bulge-loop (Figure 7). Some intriguing outcomes are represented by the pre-exponential parameters obtained with the Arrhenius analysis. It is well known that pre-exponential parameter is related to fraction of collisions between reactive molecules that allow the formation of the final product. Considering the dissociation process for AQ2 and AQ4, the pre-exponential parameters are quite close (+53 and +46, respectively) and this is directly related to the fact that the two dissociating quadruplex structures are quite similar. On the other hand, considering the association processes, the two pre-exponential parameters are no longer similar (−30 and −67, respectively). The last result suggests that the single strand of the sequences AQ2 and AQ4 possess different kinetic characteristics; particularly, the presence of the dSpacer close to the 3′ terminal region of the sequence strongly decreases the value of the pre-exponential factor. These results indicate that the presence of the dSpacer in proximity of the 3′ terminal region enhances the configuration entropy of the single strand; more degrees of freedom are accessible to the single strands and this agrees with the reduced stability of the AQ4 quadruplex with respect to its pseudo-symmetric homologous AQ2. The structure deriving from the self-association of the ODN AQ3 is characterized by a melting temperature <25°C and a CD spectrum characteristic of quadruplexes containing all anti-Gs (Figure 5A). On the basis of these results it seems to make sense to think the system is made of a combination of, at least, two different populations: the unstructured species and the structured molecule. The obtained data indicate that the less stable structured molecule gradually evolves in a system composed by a main more stable parallel quadruplex and minor amounts of different quadruplexes, as suggested by the CD spectrum in which signals characteristic of both parallel and antiparallel quadruplexes are present (Figure 5A). Unfortunately, the conversion rate is very low, making unfeasible to plan an appropriate experiment to follow the molecular rearrangement. A qualitative picture of the system produced after annealing could be summarized with the following relation:
where S represents the single-strand species, I stands for the structure with lower stability, Q1 is the more stable parallel structure and Q2 represents the minor amounts of different quadruplexes. The first side of the relation describes an equilibrium that is reached as soon as the annealing process finishes. On the other side, there is the slow process that principally produces a more stable structure. That is the simplest interpretation based on the experimental results and to date we could not find a more rigorous way to estimate the number of species present in the solution. This picture is in agreement with the NMR data. Particularly, the 25°C 1D proton spectrum of the molecule (Figure 2), recorded after a long period at 2°C following the annealing procedure, reveals four well-defined signals in the imino protons region attributable to the presence of a predominant parallel quadruplex conformation and a family of weaker signals related to minor conformations. Moreover, analysing the aromatic protons region at 25°C, we could not exclude the presence of unstructured oligonucleotides too. These data agree with the assumption of the presence of a main structured molecule in solution in equilibrium with the random coil. The whole data suggest that the presence of the dSpacer in the centre of the G-run reduces the ability to form a stable quadruplex structure favouring the formation of a mixture of different conformations. The reason of this dramatic effect is probably related to the uncompensated interruption of the stacking interaction that the dSpacer produces on the molecule.
The presence of the dSpacer in the different positions of the sequence shows quite different effects. Particularly, the formation of stable complexes is not compromised for ODNs AQ1 and AQ5 in which the dSpacer lies in proximity of the 5′- or 3′-end. On the other hand, in the cases of ODNs AQ2, AQ3 and AQ4, the interruption of the G-run by the dSpacer causes a remarkable decrease of the thermal stability, as suggested by a comparison of the T1/2 of the quadruplexes formed by them. This effect is particularly evident when the dSpacer is located in proximity of the 3′ terminal region. In fact, in the case of AQ2, the kinetic and NMR data clearly indicate that the dSpacer does not hinder the stacking between the G-tetrads closest to it, although the molecular model (Figure 7) suggests an increased buckle involving the G-tetrad near the 5′-end. Furthermore, it is noteworthy that the result concerning the Arrhenius analysis on quadruplex AQ4 strongly suggests a non-complete stacking of the G-tetrads, contrary to the behaviour of its pseudo-symmetric counterpart AQ2. The effect of the dSpacer on quadruplexes AQ3 and AQ4, besides to confirm the 3′ effect on stability, clearly points to the inclination to form other types of quadruplex. Although a direct evaluation of the thermal stabilities of quadruplexes AQ1 and AQ5 has not been possible, the 3′ effect is further suggested by a comparison between their imino regions in the 1H NMR spectra, in which the dramatic signals broadening of AQ5 indicates an increased flexibility in the 3′ region, compared with AQ1. Taking into account the results of the present study, it should be quite interesting to investigate the effects of the presence of abasic sites into other types of quadruplex structures, particularly those structures for which a biological role has been ascertained or is strongly suspected as, for example, the telomeric, c-kit and c-myc oncogene promoters quadruplexes. Such investigations are in progress in our laboratories.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian M.U.R.S.T. (P.R.I.N. 2005 and 2006); Regione Campania (L.41, L.5). Funding for open access charge: Dipartimento di Chimica delle Sostanze Naturali Università degli Studi di Napoli Federico II, Napoli, Italy.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to ‘Centro di Servizio Interdipartimentale di Analisi Strumentale’, C.S.I.A.S., for supplying NMR facilities and to ‘Centro Interdipartimentale di Metodologie Chimico-Fisiche’, C.I.M.C.F., for technical support in CD measurements. The authors are also grateful to Luisa Cuorvo, Mirko Ferraiolo, Pasquale Paciello, Annunziata Cummaro and Chesia Ronga for their collaboration.
REFERENCES
- 1.Goljer I, Kumar S, Bolton PH. Refined solution structure of a DNA heteroduplex containing an aldehydic abasic site. J. Biol. Chem. 1995;270:22980–22987. doi: 10.1074/jbc.270.39.22980. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA, Preston B. Mutagenesis by apurinic/apyrimidinic sites. Ann. Rev. Genet. 1996;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. DNA repair enzymes. Annu. Rev. Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 4.Weiss B, Grossman L. Phosphodiesterases involved in DNA repair. Adv. Enzymol. Relat. Areas Mol. Biol. 1987;60:1–34. doi: 10.1002/9780470123065.ch1. [DOI] [PubMed] [Google Scholar]
- 5.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 7.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 9.Escodd (European Standards Committee on Oxidative DNA Damage) Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic. Biol. Med. 2003;34:1089–1099. doi: 10.1016/s0891-5849(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 10.Escodd (European Standards Committee on Oxidative DNA Damage) Comparative analysis of baseline 8-oxo-7,8-dihydroguanine in mammalian cell DNA, by different methods in different laboratories: an approach to consensus. Carcinogenesis. 2002;23:2129–2133. doi: 10.1093/carcin/23.12.2129. [DOI] [PubMed] [Google Scholar]
- 11.Schmutte C, Yang AS, Nguyen TT, Beart RW, Jones PA. Mechanisms for the involvement of DNA methylation in colon carcinogenesis. Cancer Res. 1996;56:2375–2381. [PubMed] [Google Scholar]
- 12.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadet J, Berger M, Douki T, Ravanat JL. Oxidative damage to DNA: formation, measurement, and biological significance. Rev. Physiol. Biochem. Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 14.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 15.Bockrath R, Kow YW, Wallace SS. Chemically altered apurinic sites in phi X174 DNA give increased mutagenesis in SOS-induced E. coli. Mutat. Res. 1993;288:207–214. doi: 10.1016/0027-5107(93)90086-u. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Wang Z. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 1999;27:956–962. doi: 10.1093/nar/27.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sud’ina AE, Volkov EM, Oretskaia TS, Degtiarev SKh, Gonchar DA, Kubareva EA. A rapid method for testing the activity of the repair enzyme uracil-DNA-glycosylase. Bioorgan. Khim. (Mosk) 2000;26:442–447. [PubMed] [Google Scholar]
- 18.Dutta S, Chowdhury G, Gates KS. Interstrand cross-links generated by abasic sites in duplex DNA. J. Am. Chem. Soc. 2007;129:1852–1853. doi: 10.1021/ja067294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonacci C, Sheardy RD. The influence of abasic sites on the self-assembly of DNA quadruplexes. Abstracts, 37th Middle Atlantic Regional Meeting of the American Chemical Society; May 22–25; New Brunswick, NJ, USA. 2005. [Google Scholar]
- 24.Cevec M, Plavec J. Role of loop residues and cations on the formation and stability of dimeric DNA G-quadruplexes. Biochemistry. 2005;44:15238–15246. doi: 10.1021/bi0514414. [DOI] [PubMed] [Google Scholar]
- 25.Rachwal PA, Brown T, Fox KR. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007;581:1657–1660. doi: 10.1016/j.febslet.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 26.Rachwal PA, Findlow IS, Werner MJ, Brown T, Fox KR. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007;35:4214–4222. doi: 10.1093/nar/gkm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piotto M, Saudek V, Sklenar VJ. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 28.Jeener J, Meier B, Bachmann HP, Ernst RR. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979;71:4546–4553. [Google Scholar]
- 29.Braunschweiler L, Ernst RR. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 30.Marion D, Wuthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of proton-proton spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- 31.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 32.Merkina EE, Fox KR. Kinetic stability of intermolecular DNA quadruplexes. Biophys. J. 2005;89:365–373. doi: 10.1529/biophysj.105.061259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petraccone L, Pagano B, Esposito V, Randazzo A, Piccialli G, Barone G, Mattia CA, Giancola C. Thermodynamics and kinetics of PNA-DNA quadruplex-forming chimeras. J. Am. Chem. Soc. 2005;127:16215–16223. doi: 10.1021/ja0545923. [DOI] [PubMed] [Google Scholar]
- 34.Cornell WD, Cieplack P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 35.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner PJ. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- 36.Petraccone L, Erra E, Esposito V, Randazzo A, Mayol L, Nasti L, Barone G, Giancola C. Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements. Biochemistry. 2004;43:4877–4884. doi: 10.1021/bi0300985. [DOI] [PubMed] [Google Scholar]
- 37.Mergny JL, De Cian A, Ghelab A, Saccà B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petraccone L, Erra E, Esposito V, Randazzo A, Galeone A, Barone G, Giancola C. Biophysical properties of quadruple helices of modified human telomeric DNA. Biopolymers. 2005;77:75–85. doi: 10.1002/bip.20189. [DOI] [PubMed] [Google Scholar]
- 39.Phillips K, Dauter Z, Murchie AI, Lilley DM, Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J. Mol. Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Patel DJ. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Patel DJ. Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol. 1993;234:1171–1183. doi: 10.1006/jmbi.1993.1668. [DOI] [PubMed] [Google Scholar]
- 42.Aboul-ela F, Murchie AIH, Norman DG, Lilley DMJ. Solution structure of a parallel-stranded tetraplex formed by d(TG4T) in the presence of sodium ions by nuclear magnetic resonance spectroscopy. J. Mol. Biol. 1994;234:458–471. doi: 10.1006/jmbi.1994.1672. [DOI] [PubMed] [Google Scholar]
- 43.Gros J, Rosu F, Amrane S, De Cian A, Gabelica V, Lacroix L, Mergny JL. Guanines are a quartet's; best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyatt JR, Davis PW, Freier SM. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 45.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.