Abstract
The Escherichia coli McrA protein, a putative C5-methylcytosine/C5-hydroxyl methylcytosine-specific nuclease, binds DNA with symmetrically methylated HpaII sequences (Cm5CGG), but its precise recognition sequence remains undefined. To determine McrA’s binding specificity, we cloned and expressed recombinant McrA with a C-terminal StrepII tag (rMcrA-S) to facilitate protein purification and affinity capture of human DNA fragments with m5C residues. Sequence analysis of a subset of these fragments and electrophoretic mobility shift assays with model methylated and unmethylated oligonucleotides suggest that N(Y > R) m5CGR is the canonical binding site for rMcrA-S. In addition to binding HpaII-methylated double-stranded DNA, rMcrA-S binds DNA containing a single, hemimethylated HpaII site; however, it does not bind if A, C, T or U is placed across from the m5C residue, but does if I is opposite the m5C. These results provide the first systematic analysis of McrA’s in vitro binding specificity.
INTRODUCTION
Wild-type Escherichia coli K-12 strains possess several restriction systems in addition to the classical EcoK hsdR/M/S host-specificity restriction-modification mechanism. One of these, for methylated adenine recognition and restriction (Mrr) has been reported to restrict DNA containing N6-methyladenine and also DNA with C5-methylcytosine residues (m5C) (1–4). Neither system restricts DNA methylated by the resident E. coli enzymes encoded by dam, which methylates the A residue in the sequence GATC, or by dcm, which modifies the internal cytosine in CCWGG (W is A or T) sequences at the C5 position (1,3,5).
DNA containing C5-methylcytosine (m5C) or 5-hydroxymethyl cytosine (Hm5C) is also restricted by the modified cytosine restriction (Mcr) system which is identical to the previously described restricts glucose-less phage (Rgl) restriction system that blocks the growth of T-even phages, but only when they contain Hm5C, i.e. when their Hm5C DNA residues are not glucosylated (6–8). Later work further subdivided the Mcr system into two genetically distinct regions: McrA (equal to RglA) on an easily excisable but defective lambdoid prophage element e14 located at 25 min on the E. coli K-12 chromosome (8–9) and McrB (or RglB) at map position 99 min in a region that includes the EcoK restriction/modification and Mrr systems (2,4,8,10). McrA recognizes DNA containing C5-methylcytosine or C5-hydroxymethylcytosine while McrB also recognizes DNA containing N4-methylcytosine. The mcrB locus encodes two polypeptides McrB and C which together function as a nuclease recognizing in cis two half sites 5′-G/A 5mC (N40–3000) G/A 5mC-3′. Cleavage requires GTP hydrolysis and occurs at a non-fixed distance (∼30 nucleotides) between the two methylated half sites (11,12).
Early studies showed that DNAs methylated by M.HpaII (Cm5CGG), M.Eco1831I (Cm5CSGG where S is C or G) and M.SssI (m5CG) are restricted by the McrA system (2,5,13) and further studies demonstrated that clones expressing the McrA open reading frame conferred both McrA and RglA phenotypes on mcr minus E. coli strains (8). Several indirect studies suggest that McrA is a member of the ßßα-Me finger superfamily of nucleases. Its ßßα-Me finger also contains an HNH motif common to homing endonucleases as well as many restriction and DNA repair enzymes. The core ßßα-Me domain of McrA (residues 159–272 of the 277 amino acid long polypeptide) was modeled by Bujnicki and coworkers using a protein sequence threading approach (14). This region contains three histidine residues (H-228, 252 and 256) predicted to coordinate a Mg2+ ion, as well as four cysteine residues (C-207, 210, 248 and 251) which are thought to form a zinc finger, most likely involved in coordinating Zn2+ or some other divalent metal ion to help stabilize the protein’s structure and/or help catalyze nuclease activity.
While McrA is predicted to function as a nuclease and induces the DNA damage response (an index of cleavage activity) when a suitable substrate is present (15), McrA-mediated nuclease activity in vitro has never been demonstrated and to date the mechanism for McrA’s biological restriction of modified phage or plasmid DNAs is not known. Recently we reported on the cloning, expression, purification and initial characterization of full-length, biologically active rMcrA (16). However, while all attempts to demonstrate that rMcrA is a nuclease acting on m5C-containing DNA were unsuccessful, electrophoretic mobility shift analysis demonstrates that purified rMcrA interacts specifically with DNA fragments containing Cm5CGG sequences. This prompted us to design experiments to determine the spectrum of endogenously methylated McrA targets in human DNA. In order to address this question we successfully employed McrA fused to an eight-amino-acid long StrepII tag (rMcrA-S) to affinity-capture methylated restriction fragments from total human DNA. Standard sequencing of these fragments, together with bisulfite genome-sequencing analysis of their original loci in human DNA revealed more fully the DNA-binding profile of McrA. We also used rMcrA-S in electrophoretic mobility shift assays (EMSAs) with symmetrically methylated, hemimethylated- and non-methylated-double-stranded DNA probes with a canonical HpaII site and various single base-pair permutations flanking the central m5CpG dinucleotide or opposite the m5C residue. Together, these data help define the minimal recognition sequence and base-pairing requirements for McrA’s interaction with DNA.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were purchased from Integrated DNA Technologies. We dissolved individual oligonucleotides with and without m5C in 10 mM Tris–HCl, 0.01 mM EDTA buffer, pH 7.5, and, as needed, annealed them at 36 µM with their complements, in 1 × One-Phor-All Buffer (10 mM Tris–Acetate pH 7.5, 10 mM Mg–Acetate, 50 mM K–Acetate) (GE Healthcare) by heating for 2 min at 98°C, then slow cooling to room temperature. With 10% PAGE analysis, we verified their conversion to duplexes, leaving only minimal amounts (<5%) of residual single-stranded oligonucleotides. Supplementary Tables S2a and b list the oligonucleotides used for this study and give the sequences of their resulting fully base-paired ds-cassettes. Supplementary Table S3 shows the oligonucleotide cassettes used to determine the binding of rMcrA-S to ds-cassettes with a mismatched base opposite m5C. Oligonucleotides 1–6 were synthesized with m5C at various positions, while the remaining ones were methylated post-synthesis, when needed, using the ds-specific CpG methylase (M.SssI) after annealing to form duplexes, as described above. A typical reaction (20 µl) included 180 pMol duplex DNA in 1× SssI buffer (10 mM Tris–HCl, 50 mM NaCl, 10 mM MgCl2 and 1 mM dithiothreitol) supplemented with 160 µM S-adenosylmethionine (SAM). Reactions were started by adding eight units of M.SssI enzyme and incubating the mix overnight at 37°C, then spiking with additional SAM for a final concentration of 320 µM, and finally incubating the samples at 37°C for another 2 h. The M.SssI was inactivated by heating at 65°C for 20 min. To check that methylation was complete, we included as a control a duplex containing a single HpaII (CCGG) site, and no other CpG dinucleotides. We then checked that the DNA was protected against digestion by HpaII (methyl sensitive), but was susceptible to digestion by the methyl-insensitive isoschizomer MspI. For each assay, 2 µl of methylation reaction was added to 10 mM Bis–Tris–Propane–HCl, 10 mM MgCl2, 1 mM dithiothreitol supplemented with 100 µg/ml BSA, followed by 10 units of either HpaII or MspI enzyme. Reactions were incubated at 37°C for 2 h before being analyzed by agarose-gel electrophoresis. Gels were stained with ethidium bromide and the DNA bands visualized under UV light.
Strains and enzymes
Escherichia coli Top10 (Invitrogen) was used for DNA manipulation and plasmid preparations. Strain BL21-AI was obtained from Invitrogen. All enzymes, unless otherwise stated, were purchased from New England Biolabs.
rMcrA-S
The recombinant McrA coding sequences used here with either an eight amino-acid N terminal or a C terminal StrepII tag (17) (underlined) (MASWSHPQFEKGA-start of McrA or end of McrA–SAWSHPQFEK, respectively) were constructed by PCR amplification from a wild-type, untagged mcrA clone. We used primers that appended the StrepII tag and unique restriction sites to aid in cloning on the ends of the PCR product. After restriction-enzyme digestion and gel-purification, the amplicons were cloned into pET28. Details and clones are available on request. We verified the accuracy of the clones by DNA sequencing and then moved them into the expression host BL21(DE3)/pRIL; the recombinant proteins were expressed following autoinduction at 20°C as previously described (16). The departure here was that after SP-Sepharose Fast Flow (Pharmacia Biotech) chromatography, we further purified the recombinant proteins by binding them to Strep Tactin® SpinPrepTM filters (Novagen), followed by elution with LEW buffer (50 mM NaPO4, pH 8.0; 300 mM NaCl) containing 10 mM biotin. The purified proteins were stored at 4°C. Preliminary studies (data not shown) indicated that the N-terminal tagged protein (rS-McrA) was markedly less efficient in binding HpaII-methylated DNA fragments to streptavidin-coated magnetic beads (Nanolink) than was its C-terminal analogue (rMcrA-S); therefore, the latter was utilized in all remaining experiments.
rMcrA-S affinity enrichment of DNA fragments containing m5CpG
Human genomic A549 DNA (lung carcinoma) was digested exhaustively with MseI whose recognition sites (TTAA) rarely occur in GC-rich regions, so that most regions with a high density of m5CpG dinucleotides were left intact. The digested DNA was phenol-extracted, precipitated with ethanol, and then dissolved in 10 mM Tris–HCl, 0.01 mM EDTA (TEsl), pH 8.0. Approximately 750 ng of fragmented DNA was incubated for 20 min at RT with ∼7 nmol of rMcrA-S in 200 µl LEW buffer supplemented with 100 µg/ml BSA and 250 ng sonicated E. coli ER2925 (dam−, dcm−) DNA as the carrier. Then we added a 25 µl bed-volume of Nanolink magnetic strepavidin beads, prewashed twice in 1 × LEW + 100 µg/ml BSA, and incubated the materials at RT for 1 h with gentle mixing to capture the rMcrA-S/A549 DNA complexes. The unbound fraction was removed and the beads were washed 3 × with 100 µl 1 × HEPES + 250 mM NaCl; 3 × with 1 × HEPES + 700 mM NaCl; and, 2× with 50 µl 1 × HEPES + 250 mM NaCl. The beads with the bound DNA fragments next were washed in 50 µl 1 × Quick Ligase Buffer (NEB) [66 mM Tris–HCl, 10 mM MgCl2, 1 mM dithiothreitol, 1mM ATP and 7.5% (W/V) polyethylene glycol (PEG 6000)] and resuspended in 50 µl 1× Quick Ligase Buffer.
Ligation-mediated PCR (LM-PCR)
A MseI-compatible adaptor DNA cassette was formed by annealing two oligonucleotides: MseI Top: 5′-AGCAACTGTGCTATCCGAGGGAT-3′, and MseI Bottom: 5′-TAATCCCTCGGA-3′ and then ligating the product to the MseI-compatible ends of the DNA captured on the strepavidin beads by rMcrA-S. We added 100 pmol of adaptor and 3000 U of T4 DNA ligase to the resuspended magnetic beads in 50 µl 1× Quick Ligation Buffer. The reaction was incubated at 16°C overnight.
The beads were then washed and equilibrated with 100 µl 1 × Thermo Pol Buffer (NEB) (20 mM Tris–HCl pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100). PCR amplification was done in 50 µl NEB Thermo Pol buffer with Taq polymerase using a single unphosphorylated primer: 5′-AGCAACTGTGCTATCCGAGGGAT-3′. The reactions were started by incubating the material at 72°C for 10 min to fill-in the single-stranded regions of the appended cassettes, and then were cycled 14 times at 94°C for 20 s, 68°C for 30 s and 72°C for 2 min 30 s. The amplified products were purified using Qiagen PCR Purification Kit columns (Qiagen), and resuspended in 20 µl of distilled H2O with 50 mM HEPES (pH 7.5). Linkered fragments were cloned using a pSMART GC kit (Lucigen). Individual recombinant clones were sequenced using standard ABI dideoxy sequencing.
Bisulfite sequencing
Genomic A459 DNA was bisulfite-converted according to the manufacturer’s instructions (Qiagen), followed by whole-genome amplification (Qiagen); we omitted the initial denaturing steps since the DNA already was single-stranded after bisulfite conversion. Primers were designed (http://www.urogene.org/methprimer/index1.html) for two regions of the bisulfite-modified genomic DNA. Region C1 was amplified using the forward- and reverse-primers, C1F3: 5′-TGGGGTGTTTTTTTTGTATT-3′ and C1R1: 5′-AAAATCCCACCCTAAACC-3′, respectively, and region C2 with the forward and reverse primers C2F3: 5′-TGGGTTTTGTATAGGTTAAA-3′ and C2R1: 5′-AACAACCAAAAAATTTTCAC-3′, respectively. PCR was done following the manufacturer’s conditions, using NEB Thermo Pol buffer and Taq polymerase supplemented with 5% DMSO (final concentration) and amplified as follows: 94°C for 2 min, two cycles of (94°C for 30 s, 55°C for 30 s, 72°C for 2 min) 2 cycles of (94°C for 20 s, 54°C 30 s, 72°C for 2 min) two cycles of (94°C for 20 s, 53°C for 30 s, 72°C for 2 min) two cycles of (94°C for 20 s, 52°C for 30 s, 72°C for 2 min) and 30 cycles of (94°C for 20 s, 51°C for 30 s, 72°C for 2 min). PCR products were gel purified using PCR purification columns (Qiagen), ligated into pCR4TOPO vector (Invitrogen) via manufacturer’s standard protocol, electroporated into E. coli Top 10 cells, and plated on 2 × YT supplemented with 50 µg kanamycin. We picked individual colonies and grew them in 2 × YT media with 50 µg kanamycin. Plasmids were isolated using alkaline lysis followed by column purification (Fermentas), and sequenced with primers flanking the cloning site. Sequencher software (Gene Codes) was employed to edit the sequences, and CLUSTLW to analyze them.
EMSA binding assays
Reactions (10 µl) typically contained a mixture of 10–25 pMol of various test DNAs, and 250 ng sonicated E. coli ER2925 DNA as a non-methylated, non-specific competitor in 50 mM Tris–HCl, 100 mM NaCl, 1 mM dithiothreitol, 10 mM MgCl2 (NEBuffer No.3), supplemented with 100 µg/ml BSA. We started the reactions adding 25–175 pMol McrA-S; after 45 min at RT, the entire sample was loaded on 10% acrylamide Tris–Acetate/EDTA gel (1 × TAE: 40 mM Tris–acetate, 1 mM disodium EDTA) which was electrophoresed at RT. Thereafter, the gels were stained with ethidium bromide (0.5 µg/ml) at RT and visualized with UV light. Then, we stained the gels with Coomassie Blue to detect rMcrA-S.
RESULTS
Previously, we reported that purified full-length recombinant McrA (rMcrA) binds to but lacks detectable nuclease activity at methylated HpaII (Cm5CGG) sequences (16) and, as such, might afford utility as a reagent for affinity purification of human DNA fragments containing m5C and possibly Hm5C residues (18,19). The human genome contains ∼2.3-million HpaII sites of which roughly 12% are located in regions with a high density of m5CpG dinucleotides, regions referred to as CpG islands (CGIs) (Supplementary Table S1). The human genome has ∼30 000 CGIs, accounting for ∼10% of the total DNA, about half of which are located near annotated transcription start sites (TSS-CGIs); the remainder are intra- or intergenic (non-TSS). In normal tissues, TSS-CGIs usually are unmethylated but a subset becomes reproducibly methylated in normal cells during imprinting, X-chromosome inactivation, tissue differentiation and in diseased and cancerous cells (20–22). Consequently, there is growing interest in determining global DNA methylation patterns in normal- and abnormal-tissues. Widely used methods for such studies include immunoprecipitation (23); methylation specific PCR (24); m5CpG affinity-capture using m5CpG-binding proteins and related methods (25,26) restriction enzyme-based methods, microarray-based methods or various combinations of these methods (27–29) and bisulfite-based modification followed by single or multi-locus DNA sequencing or more recently by high-throughput, massively parallel sequencing (27).
Since our initial McrA studies used phage T7 DNA methylated in vitro only at HpaII sites, we could not ascertain whether McrA also can bind to, and perhaps be used for affinity purification of DNA fragments with related sequences containing m5CpG’s or, alternatively, if McrA would preferentially enrich a subset of eukaryotic CGIs containing several methylated HpaII sites. A standard technique for converting McrA into such an affinity reagent would be to fuse a (His)6 tag to either end of the protein and then use the tagged protein to generate an affinity matrix for binding DNA fragments containing m5CpG’s. However, rMcrA intrinsically binds to Ni-charged NTA supports, presumably because of interaction with three suitably positioned histidines in its ßßα-Me domain (14,16). Our initial experiments indicated inefficient binding of rMcrA/DNA complexes to Ni-charged NTA magnetic beads, presumably due to steric hindrance. Therefore, we decided to add an eight-amino-acid long StrepII tag (WSHPQFEK) (17) to one end of the protein to foster the affinity capture of methylated DNA fragments independent of the Ni-binding site. In our hands, placing this tag at the C-terminus does not seem to adversely affect the protein’s capture of DNA fragments with methylated-HpaII sequences, whereas its N-terminal tagged counterpart is much less effective (data not shown).
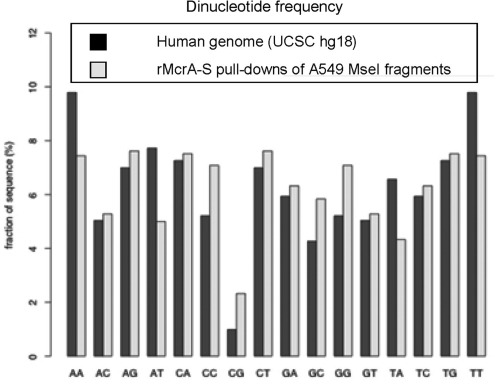
To further resolve McrA’s binding specificity, we initially used rMcrA-S to enrich for methylated sequences from an MseI digest of genomic human DNA because it would contain m5CpG dinucleotides in all possible contexts, and further, MseI leaves CGIs mostly intact (25). In addition, this way we could ascertain whether rMcrA-S preferentially captures CGIs with methylated HpaII sites. Accordingly, we washed bound fragments obtained by rMcrA-S affinity purification with high-ionic-strength buffer, and then LM-PCR-amplified, cloned, and sequenced a subset of the clones to determine if they all contained HpaII sites, and if any originated from chromosome regions defined as being a CGI. We found that most clones from a library of non-size-selected amplicons were not from CGIs, but from regions containing repetitive sequences; i.e. regions known to contain m5CpGs. Some minor enrichment for CGI fragments was noted when the amplified DNA was gel-purified and fragments between 0.5- and 2-kb were used to prepare the library (viz. the size range expected for CGIs in the digest). While many clones in both libraries contained one or more HpaII sites, some had no HpaII sites although they invariably contained several CpG dinucleotides. Overall, we found ∼2–3-fold increase in CpG dinucleotides in these libraries over the starting DNA (Figure 1).
Figure 1.
CpG ratios in the human genome versus rMcrA-s pull-downs. Dinucleotide frequencies compiled for both strands of 33 unique rMcrA-S pull-down fragments (11 400-nt total), compared with total-genome frequencies compiled from UCSC hg18. (http://genome.ucsc.edu/cgi-bin/hgTracks?hgsid=136554479&chromInfoPage=).
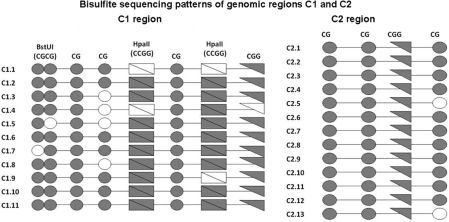
After we located all affinity captured sequences in the human genome using the UCSC Genome Browser (March, 2006 assembly, http://genome.ucsc.edu/), we chose two unique regions to study further: ‘C1’, within Ch. 8q24.3, containing two HpaII sites, one CGG sequence and five other CpG dinucleotides; and, ‘C2’, within Ch. 18q11.2, lacking HpaII sites but having a single CGG sequence plus three other CpGs within the MseI fragment. We next determined the methylation status of these sites in vivo via sodium-bisulfite sequencing. Briefly, total genomic A549 DNA was bisulfite-modified, amplified by a whole-genome amplification assay, and then the C1 and C2 regions were amplified using bisulfite-specific primers C1F3, C1R1 and C2F3, C2R1, respectively. Standard sequencing methods allowed us to deduce the genomic methylation pattern in 12 clones from each region.
Aligning these sequences showed that 63–100% of all CpG sites in C1 were methylated, and all C1 clones had at least one methylated HpaII sequence or m5CGG sequence (hereafter referred to as a three–fourth HpaII site) (Figure 2a). For the C2 clones, from a region lacking HpaII sites, we found that all 12 clones were methylated at the three–fourth HpaII site (Figure 2b). Ten of them exhibited 100% methylation across all four CpG dinucleotides. These data led us to investigate whether the three–fourth HpaII site is a minimal binding site for rMcrA-S.
Figure 2.
Bisulfite sequencing patterns of two genomic regions (C1 and C2) recovered after rMcrA-S affinity-enrichment from MseI digested A549 DNA. Methylated cytosines in individual clones are depicted as filled symbols and unmethylated cytosines are open symbols. Individual circles represent CpGs, double circles are the BstUI sites (CGCG), triangles are CGG sites, and rectangles are HpaII sites (CCGG).
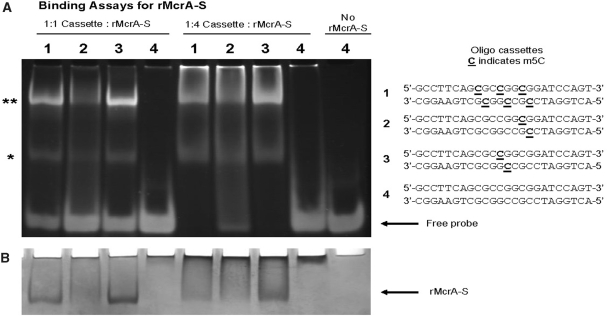
To further define a consensus rMcrA-S DNA recognition site, we turned to EMSA using 24 bp synthetic oligonucleotide cassettes containing one of the following: (i) three m5CpGs including one in an HpaII site (Cm5CGG); (ii) one with a single m5C in a ‘three–fourth HpaII site’ (m5CGG); and (iii) one containing a single m5C in an HpaII site. As a control, we used a ds-cassette with no m5Cs (Supplementary Table S2b). Figure 3 shows that, at the protein/DNA ratios we used, rMcrA-S has high affinity for a ds-cassette containing a single symmetrically methylated HpaII site; however, at higher ratios of protein to DNA, binding to the cassette with a single symmetrically methylated three–fourth HpaII site becomes evident, but not to the unmethylated control cassette (Figure 3a and b). In other experiments we found that binding is independent of Mg++ ions in the buffer (data not shown).
Figure 3.
Binding Assays for rMcrA-S. (A) Assays of electrophoretic mobility shifts with different molar ratios (1 : 1 and 1 : 4—DNA:protein) of rMcrA-S binding unmethylated and symmetrically methylated (m5CpG) containing 24-bp double-stranded oligonucleotides: 1 contains three m5C including one in an HpaII site; 2 contains a single m5C in a ‘three–fourth HpaII site’; 3 contains a single m5C in an HpaII site; 4 has no m5Cs. Molar ratios are based on monomeric McrA (16). Asterisks indicate the positions of McrA-S/DNA complexes. (B) Coomassie Blue staining of EMSA (A) gel.
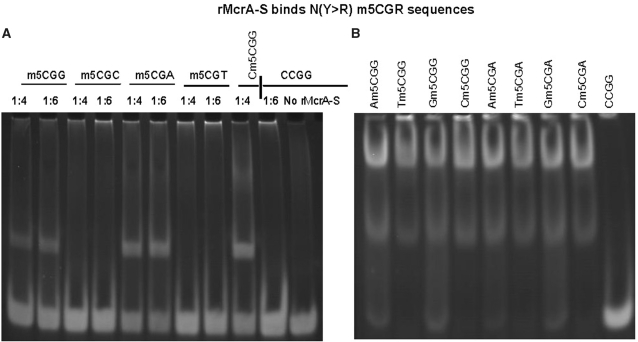
Next, we used complementary synthetic oligonucleotides, annealed and M.SssI methylated in vitro, as described in ‘Materials and Methods’ section to further define rMcrA-S’s binding preference. As Supplementary Table 2b illustrates, these oligonucleotides all contain a single CpG dinucleotide either preceded by D (A, G or T) or followed by H (A, C or T). Figure 4 contains the results of gel-shift assays with these methylated cassettes.
Figure 4.
rMcrA-S binds N(Y > R) m5CGR sequences. (A) EMSA showing rMcrA-S binding 24-bp oligonucleotide cassettes containing a purine following the Cm5CG sequence (lanes 1, 2, 5 and 6) or with a pyrimidine following the Cm5CG (lanes 2, 3, 7 and 8). Lane 9 is a positive control using a cassette with a single symmetrically methylated HpaII site; lanes 10 and 11 are negative controls. The molar ratio of HpaII sites to rMcrA-S for each cassette is 1:4, or 1:6 (ratios based on monomeric rMcrA-S). (B) EMSA showing rMcrA-S binding 24-bp oligonucleotide cassettes containing Nm5CGR sequences. rMcrA-S binds all cassettes with this sequence (lanes 1–8). Lane 9 contains an unmethylated cassette as a negative control. The molar ratio of HpaII sites to rMcrA-S for each reaction is 1:6.
Under our standardized EMSA conditions, we found that rMcrA-S preferentially shifts cassettes when a purine (R) follows the m5CpG dinucleotide (m5CGR) rather than a pyrimidine (Y) (m5CGY) (Figure 4a). These findings suggest why our previous in vitro studies revealed that McrA did not bind T7 DNA with methylated HhaI sites (Gm5CGC) (16).
We also investigated the importance of cytosine preceding m5CGR. Double-stranded cassettes were designed with a single NCGR site, M.SssI methylated in vitro, and then analyzed for rMcrA-S’s binding by EMSA. rMcrA-S bound all eight cassettes (Figure 4b) but seemingly had a somewhat higher affinity for duplexes with Ym5CGR (Figure 4b, lanes 2, 4, 6 and 8) while shifting duplex cassettes with a Rm5CGR sequence (Figure 4b, lanes 1, 3, 5 and 7). These results are consistent with our initial findings that rMcrA-S can affinity-purify genomic MseI fragments lacking a methylated HpaII site but containing Am5CGG, a three–fourth HpaII site.
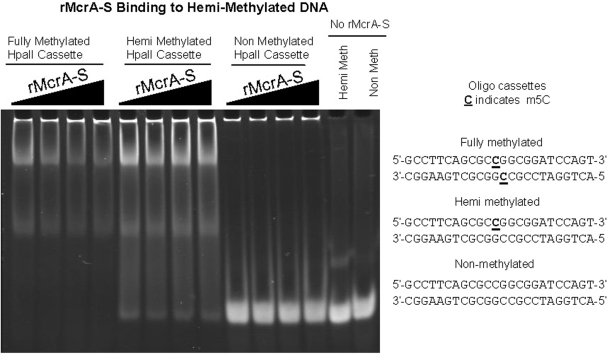
rMcrA-S also binds hemimethylated DNA
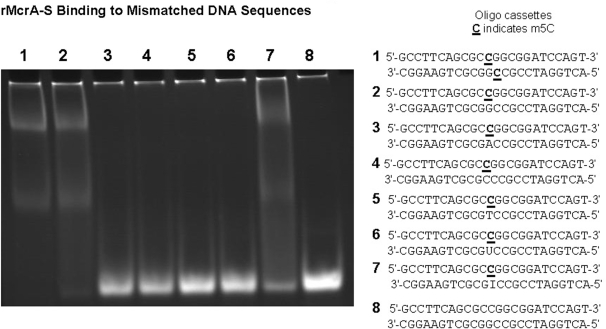
During mammalian DNA replication, CpG dinucleotides in the daughter strand initially are unmethylated until later methylated by the maintenance methyltransferase (30). Therefore, we were interested to learn whether rMcrA-S interacts with a hemimethylated Cm5CGG sequence. For these studies, we annealed an oligonucleotide with and without a single m5C added during synthesis with its methylated- or non-methylated-complement (Supplementary Table S3). As shown in Figure 5, the added rMcrA-S gel shifts only the ds-cassettes with a fully methylated or hemimethylated HpaII site and not the unmethylated cassette. We next tested rMcrA-S’s ability to gel-shift ds-cassettes wherein an A, C, T, U or I residue is placed opposite the m5C (Figure 6). Interestingly, EMSA shifts were observed only when a G or I is opposite the m5C; no shifted complexes are seen with the others.
Figure 5.
EMSA showing rMcrA-S binding to DNA with fully- and hemi-methylated Cm5CGG sites. The molar ratio of HpaII sites to rMcrA-S for each cassette is 1:4, 1:5, 1:6 and 1:7, increasing from left to right. Lanes 1–4 are fully methylated DNA, 5–8 are hemimethylated DNA, and lanes 9–12 are non-methylated DNA. Lanes 11 and 12 are negative controls. The band at the top of the gel is the sonicated non-methylated E. coli competitor DNA.
Figure 6.
rMcrA-S binding to DNA sequences with a mismatch opposite m5C. EMSA showing rMcrA-S binding to a DNA with a symmetrically methylated HpaII site (lane 1), a hemi-methylated HpaII site (lane 2) and an inosine across from the m5C (lane 7). rMcrA-S does not bind if A, C, T or U is opposite the m5C (lanes 3–6, respectively). The molar ratio of DNA to rMcrA-S is 1:6.
DISCUSSION
From these experiments, we conclude that rMcrA-S can bind ds-DNA fragments with a single symmetrically methylated Cm5CGG sequence, or sites where Y precedes, or R follows the methylated central CpG dinucleotide. As shown here, electrophoresis of rMcrA-S/DNA complexes on a polyacrylamide gel typically generates two shifted bands. We believe these may represent mono-dimer equilibria of the complexes that form during EMSA but additional studies will be needed to adequately address this question. Interestingly, our initial studies indicate that rMcrA-S does not demonstrate high affinity to human genomic CGIs with methylated HpaII sites. Further work is needed with CGI-specific- and other types of microarrays to determine if rMcrA-S can be used for high-resolution DNA methylation analysis of tiled CGIs and to verify if McrA is indeed a nuclease in vivo. To date all our tests for double-stranded DNA cleavage by McrA have been negative as have assays for nicking activity on HpaII methylated supercoiled plasmids and for m5C DNA glycosylase activity. In addition, since we see no evidence for cleavage on long T7 DNA fragments with numerous symmetrically methylated HpaII sites, we do not believe that McrA is similar to typeIIS restriction endonuclease that cleave outside or between their recognition sites, as is the case for McrBC.
Presumably, McrA acting simply as a DNA-binding protein could interfere with or ‘biologically restrict’ the maintenance or growth of m5C or Hm5C modified plasmids and phages by interfering with DNA replication (15) since, as shown here, it should bind in vivo to symmetrically methylated as well as hemimethylated sites. Alternatively, binding might not be tightly correlated with cleavage specificity. McrA binding to m5C or Hm5C modified sites might elicit post-translational modification of McrA, or alternatively the synthesis or interaction with some other E. coli protein(s) needed to catalyze cleavage in vivo. The recombinant McrA-S protein described here should be useful in attempts to identify such putative host protein(s) with which it might interact in vivo to a form non-covalent, catalytically active complex by co-chromatography on a Strep-Tactin® affinity matrix (17).
Initial attempts to establish a pET-based McrA plasmid in a Bl21(DE3) host harboring a pACYC-based clone of the HpaII methylase that constitutively expresses M.HpaII [kindly provided by Elisabeth Raleigh (NEB)] were unsuccessful due to lethality caused by leaky expression of the McrA protein. Newer plasmid constructs in which the expression of McrA-S sequence is more tightly repressed (Studier, unpublished) have been introduced into BL21-AI cells along with the M.HpaII plasmid. In these cells the T7 RNA polymerase is under the control of an arabinose promoter and has lower basal expression of T7 RNA polymerase compared to BL21(DE3) cells (31). These new constructs should be useful for understanding the in vivo role of McrA in ‘restricting’ m5C- or Hm5C-containing DNA.
Perhaps the most significant finding of our present study is the discovery that rMcrA-S can bind hemimethylated DNA and that a base mismatch, except for I, opposite the m5C residues abrogates binding. Binding of rMcrA-S to the m5C/I cassette is not totally unanticipated since I opposite a m5C is expected to be less destabilizing than A, C, T or U mismatches which is consistent with the known base-pairing of dC:dI. These results are in contrast to similar studies with (His)6-tagged form of the methyl binding domain (MBD) (residues 77–165) of the mouse MeCP2 protein (32,33). It had been shown previously that the binding of the MBD of MeCP2 requires only one methylated CpG dinucleotide and that the MBD domain is necessary and sufficient for binding of MeCP2 to its target duplex (32,33). However, the MBD of MeCP2 binds equally well to a 27 base long DNA cassette with a symmetrically methylated CpG dinucleotide (m5C/m5C) as to one with a T opposite the m5C (m5C/T) (34). The effects of other mismatches were not reported. The estimated Kd for a hemimethylated CpG duplex is ∼10-fold higher than that of a fully methylated CpG pair, whereas the Kd for an unmethylated duplex is ∼100-fold higher. These data are interpreted by Valinluck et al. (34) as being consistent with MBD binding to its target duplex as a monomer that recognizes both methylated cytosines in a fully methylated duplex. McrA on the other hand appears to need only a single m5CG in a Cm5CGG sequence for efficient binding. In this regard it is quite similar to the SRA (SET- and RING finger-associated) domain of the mouse and human UHRF1 proteins, which recent studies have shown, binds selectively to DNAs with hemimethylated CpG dinucleotides (35–38). These proteins increase their protein–DNA interface by flipping the m5C into a specific protein binding pocket that stabilizes the complex. Similar studies focusing on X-ray crystallization analysis of McrA-S alone and when co-crystallized with methylated and hemimethylated DNA cassettes will be needed to fully understand the molecular basis of its interaction with methylated DNA and whether it also utilizes a base-flipping mechanism to stabilize binding. It also will be interesting to see if McrA can bind DNAs with Hm5C residues and, if binding occurs, whether the differential binding pattern is the same as for DNAs with m5C residues. Preliminary data (JJD) indicate that McrA binds but does not cleave T* DNA containing non-glucosylated Hm5C residues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Laboratory Directed Research and Development Award at Brookhaven National Laboratory; the Low Dose Radiation Research Program of the Office of Biological and Environmental Research program of the US Department of Energy; and by the National Institutes of Health (grant U01-AI56480 to J.J.D.). Funding for open access charge: Office of Biological and Environmental Research program of the U.S. Department of Energy and National Institutes of Health (grant U01-AI56480 to J.J.D.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Barbara Lade, Laura-Li Loffredo and Judi Romeo for technical assistance.
REFERENCES
- 1.Heitman J, Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J. Bacteriol. 1987;169:3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelleher JE, Raleigh EA. A novel activity in Escherichia coli K-12 that directs restriction of DNA modified at CG dinucleotides. J. Bacteriol. 1991;173:5220–5223. doi: 10.1128/jb.173.16.5220-5223.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waite-Rees PA, Keating CJ, Moran LS, Slatko BE, Hornstra LJ, Benner JS. Characterization and expression of the Escherichia coli Mrr restriction system. J. Bacteriol. 1991;173:5207–5219. doi: 10.1128/jb.173.16.5207-5219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kretz PL, Kohler SW, Short JM. Identification and characterization of a gene responsible for inhibiting propagation of methylated DNA sequences in mcrA mcrB1 Escherichia coli strains. J. Bacteriol. 1991;173:4707–4716. doi: 10.1128/jb.173.15.4707-4716.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl Acad. Sci. USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Revel HR. Restriction of nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology. 1967;31:688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- 7.Fleischman RA, Cambell JL, Richardson CC. Modification and restriction of T-even bacteriophages. In vitro degradation of deoxyribonucleic acid containing 5-hydroxymethylctosine. J. Biol. Chem. 1976;251:1561–1570. [PubMed] [Google Scholar]
- 8.Raleigh EA, Trimarchi R, Revel H. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics. 1989;122:279–296. doi: 10.1093/genetics/122.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta P, Casjens S, Krishnaswamy S. Analysis of the lambdoid prophage element e14 in the E. coli K-12 genome. BMC Microbiol. 2004;4:4. doi: 10.1186/1471-2180-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raleigh EA, Murray NE, Revel H, Blumenthal RM, Westaway D, Reith AD, Rigby PW, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988;16:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart FJ, Raleigh EA. Dependence of McrBC cleavage on distance between recognition elements. Biol. Chem. 1998;379:611–616. [PubMed] [Google Scholar]
- 12.Panne D, Raleigh EA, Bickle TA. The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. J. Mol. Biol. 1999;290:49–60. doi: 10.1006/jmbi.1999.2894. [DOI] [PubMed] [Google Scholar]
- 13.Kravetz AN, Zakharova MV, Beletskaya IV, Sineva EV, Denjmuchametov MM, Petrov SI, Glatman LI, Solonin AS. The cleavage sites and localization of genes encoding the restriction endonucleases Eco1831I and EcoHI. Gene. 1993;129:153–154. doi: 10.1016/0378-1119(93)90712-c. [DOI] [PubMed] [Google Scholar]
- 14.Bujnicki JM, Radlinska M, Rychlewski L. Atomic model of the 5-methylcytosine-specific restriction enzyme McrA reveals an atypical zinc finger and structural similarity to betabetaalphaMe endonucleases. Mol. Microbiol. 2000;37:1280–1281. doi: 10.1046/j.1365-2958.2000.02010.x. [DOI] [PubMed] [Google Scholar]
- 15.Anton BP, Raleigh EA. Transposon-mediated linker insertion scanning mutagenesis of the Escherichia coli McrA endonuclease. J. Bacteriol. 2004;186:5699–5707. doi: 10.1128/JB.186.17.5699-5707.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulligan EA, Dunn JJ. Cloning, purification and initial characterization of E. coli McrA, a putative 5-methylcytosine-specific nuclease. Protein Expr. Purif. 2008;62:98–103. doi: 10.1016/j.pep.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt TG, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 18.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 21.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeschnigk M, Martin M, Betzl G, Kalbe A, Sirsch C, Buiting K, Gross S, Fritzilas E, Frey B, Rahmann S, et al. Massive parallel bisulfite sequencing of CG-rich DNA fragments reveals that methylation of many X-chromosomal CpG islands in female blood DNA is incomplete. Hum. Mol. Genet. 2009;18:1439–1448. doi: 10.1093/hmg/ddp054. [DOI] [PubMed] [Google Scholar]
- 23.Wilson IM, Davies JJ, Weber M, Brown CJ, Alvarez CE, MacAulay C, Schubeler D, Lam WL. Epigenomics: mapping the methylome. Cell Cycle. 2006;5:155–158. doi: 10.4161/cc.5.2.2367. [DOI] [PubMed] [Google Scholar]
- 24.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat. Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 26.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab. Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 27.Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 28.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oda M, Glass JL, Thompson RF, Mo Y, Olivier EN, Figueroa ME, Selzer RR, Richmond TA, Zhang X, Dannenberg L, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37:3829–3839. doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolden AH, Ward CA, Nalin CM, Weissbach A. The primary DNA sequence determines in vitro methylation by mammalian DNA methyltransferases. Prog. Nucleic Acid Res. Mol. Biol. 1986;33:231–250. doi: 10.1016/s0079-6603(08)60025-2. [DOI] [PubMed] [Google Scholar]
- 31.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Free A, Wakefield RI, Smith BO, Dryden DT, Barlow PN, Bird AP. DNA recognition by the methyl-CpG binding domain of MeCP2. J. Biol. Chem. 2001;276:3353–3360. doi: 10.1074/jbc.M007224200. [DOI] [PubMed] [Google Scholar]
- 34.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian C, Li S, Jakoncic J, Zeng L, Walsh MJ, Zhou MM. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J. Biol. Chem. 2008;283:34490–34494. doi: 10.1074/jbc.C800169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.