Abstract
Despite the identification of H2A.Bbd as a new vertebrate-specific replacement histone variant several years ago, and despite the many in vitro structural characterizations using reconstituted chromatin complexes consisting of this variant, the existence of H2A.Bbd in the cell and its location has remained elusive. Here, we report that the native form of this variant is present in highly advanced spermiogenic fractions of mammalian testis at the time when histones are highly acetylated and being replaced by protamines. It is also present in the nucleosomal chromatin fraction of mature human sperm. The ectopically expressed non-tagged version of the protein is associated with micrococcal nuclease-refractory insoluble fractions of chromatin and in mouse (20T1/2) cell line, H2A.Bbd is enriched at the periphery of chromocenters. The exceedingly rapid evolution of this unique X-chromosome-linked histone variant is shared with other reproductive proteins including those associated with chromatin in the mature sperm (protamines) of many vertebrates. This common rate of evolution provides further support for the functional and structural involvement of this protein in male gametogenesis in mammals.
INTRODUCTION
It will soon be 9 years since the replacement histone variant H2A.Bbd was first identified from an EST database search using nucleotide sequences of members of the human H2A gene (1). Northern blot identified the presence of its mRNA in human testis and PCR of the cDNA obtained from poly(A)+ RNA fraction revealed its occurrence in different human female tissues. Using ectopically expressed myc epitope-tagged and GFP-tagged versions of the protein, it was shown that it was largely excluded from the inactive X-chromosome under these conditions. Furthermore, the ectopically expressed tagged form of the protein co-fractionated in sucrose gradients with the mononucleosome fraction generated from micrococcal nuclease digestion of stably transfected cells (1).
The identification of this new histone H2A variant was followed by many important biochemical studies aimed at the characterization of its role in chromatin organization using mainly in vitro reconstituted systems and cells ectopically expressing tagged forms of the protein (2,3). In this way, it was shown that H2A.Bbd destabilizes the nucleosome core particle (NCP) and it exchanges faster from chromatin than the canonical H2A counterpart (3). The H2A.Bbd-containing reconstituted NCP was shown to have a more relaxed conformation (2–4) and organizes ∼120–130 bp of DNA (2,5), leaving ∼10 bp at the flanking ends of the NCP free from interaction with the histone core octamer (4,5). Also, it was shown that the chromatin remodeling complexes SWI/SNF, ACF and nucleolin are unable to mobilize the variant H2A.Bbd NCP but can assist the process of NCP assembly (6,7) similarly to what has been observed in the case of the histone chaperone NAP-1 (8). From a functional perspective, H2A.Bbd was initially found to co-localize with H4 acetylated at K12 (1) and hence, due to the well-known correlation between histone acetylation and active transcription (9,10), the in vitro work has tried to provide support for an involvement of H2A.Bbd in this process. Indeed, transcription appeared to be more efficient for H2A.Bbd nucleosomal arrays than for conventional H2A arrays (6). The lack of an H2A acidic patch (11) that regulates chromatin compaction (12) in H2A.Bbd has been shown to be structurally responsible for a 3.5- to 5.5-fold increase in transcription in the H2A.Bbd nucleosome arrays (13). However, despite the substantial amount of in vitro work, the histone H2A.Bbd variant in its native form has never been identified. Its physiological role has remained elusive and is still completely unknown.
Here, we report the identification of H2A.Bbd in a native setting and its involvement in mammalian spermiogenesis where the protein is found associated with the highly acetylated H4 chromatin fraction that precedes the replacement of histones by protamines (14,15).
MATERIALS AND METHODS
Phylogenetic analysis
Sequence alignment
Histone H2A.Bbd sequences using in the evolutionary analyses were retrieved through recurrent BLAST searches on GenBank databases including complete genomes from human and mouse. Sequences were edited and aligned based on their amino acid sequences using the BIOEDIT (16) and CLUSTAL_X programs using the default parameters as described elsewhere. The accession umbers of the sequences used in the analyses were: mBbd.1: NM_001102665; mBbd.2: XM_001476045; mBbd.3: XM_88950, XM_911022; mBbd.4: XM_910275; mBbd.5: XM_001472598; hBbd.1: NM_080720, AF254576, NM_001017991; hBbd.2: NM_001017990.
Phylogenetic inference
The study of the evolutionary relationships among H2A.Bbd sequences from mouse and human was carried out by reconstructing phylogenetic trees based on nucleotide and protein sequences, using the neighbor-joining tree-building method based on Kimura 2-parameter and Poisson distance matrices, respectively. In order to assess that our results are not dependent on this choice, phylogenetic inference analyses were completed by maximum parsimony trees. We combined the bootstrap and the interior-branch test methods in order to test the reliability of the topologies, assuming values >90% as statistically significant (17). All the molecular evolutionary analyses conducted throughout the present work were performed using the MEGA4 program (18).
Reconstruction of ancestral sequences
Ancestral sequences corresponding to internal nodes of the H2A.Bbd nucleotide phylogeny were estimated by maximum likelihood using the codeml program included in the PAML package (19).
Estimation of evolutionary rates
The rate of H2A.Bbd evolution was estimated by calculating the numbers of amino acid substitutions per site between human and mouse H2A.Bbd proteins taking 112 MYA as a reasonable estimate for human–rodent divergence (20).
PCR
RNA was extracted from mouse testis using RNeasy mini kit (Qiagen). After extraction, cDNAs were synthesized using a Superscript II kit (Invitrogen) following the manufacturer’s procedures, and used as templates for PCR. Genomic DNA was extracted from mouse testis using DNeasy tissue kit (Qiagen). cDNA and genomic DNA were subjected to PCR analysis using the following sets of primers: p5 (5′ primer: TGATCTTTGCAGTGAGCCTG; 3′ primer: GTAGACCTCCAAGTCCAGCG), p4 (5′ primer: ATGACAACACCCCCAGAGAG; 3′ primer: TAGCTGATGATGAGCAGGGG), p(1–2) (5′ primer: GTGGAGCCAAGTCATCCTGT; 3′ primer: AGACAGCCAAGTCCAGCAGT) and p3 (5′ primer: ATCTTTGCTGTGAGCCTGGT; 3′ primer: AAGGCTGGGCAGGACTAACT) of the 5′ and 3′ ends of the coding regions of the corresponding genes (see Figure 1A). Amplification was carried out by 35 cycles of: 95°C for 30 s; 57°C for 30 s; 72°C for 50 s and final extension: 72°C for 7 min. The fragments of the PCR product were cloned into pCR2.1-TOPO vector (Invitrogen) and grown clones were amplified by PCR. The PCR products were analyzed by native (1.5%) agarose and the corresponding gene identification was carried out by digestion with Hpa II (New England Biolads) and analysis on 4% native PAGE in 20 mM sodium acetate, 40 mM Tris–HCl (pH 7.2), 1 mM EDTA buffer. Selected clones from each gene type were also confirmed by the DNA sequencing.
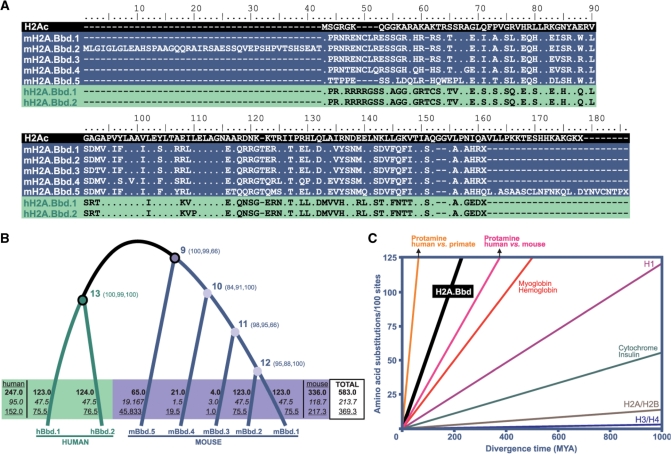
Figure 1.
Rapid H2A.Bbd evolution. (A) Protein sequence alignment of the mouse mH2A.Bbd isotypes compared to a consensus canonical histone H2A (H2Ac) and to human hH2A.Bbd isotypes. (B) Evolutionary pathways leading to the differentiation of mH2A.Bbd and hH2A.Bbd gene and protein lineages. The total number of nucleotide (bold-face), synonymous (italics) and replacement (underlined) changes from the ancestral sequences reconstructed for nodes 9–13 are indicated. Confidence levels for the nodes are indicated in the corresponding internal branches in parentheses. (C) Evolutionary rates (amino acid change per 100 sites) of mammalian H2A.Bbd compared to P1 protamines (31) and with histones (42) and other proteins.
DNA construct and transfection
Total RNA was extracted from mouse tissues and reverse transcriptase-PCR (RT-PCR) was performed as described previously (21). The RT-PCR amplicon was identified to be Mus musculus H2A.Bbd (MmH2A.Bbd) gene isotype 3. The coding region of MmH2A.Bbd was cloned into the pcDNA 3.0 mammalian expression vector (Invitrogen Life Science). The MmH2A.BbD construct was transfected into HeLa cell using Polyfect Transfection Reagent (Qiagen) and cultured at 37°C, 5% CO2 for 24 h. A GFP-version of MmH2A.Bbd isotype 3 was also prepared by cloning its coding region into a pEGFP-N1 vector (Clontech).
Mouse testis cell fractionation
Total cell suspensions were separated by centrifugal elutriation (JE-6B rotor, Beckman Instruments) to obtain fraction 2 enriched in steps 9–12 elongating spermatids (flow rate interval: 12.6–18 ml/min, rotor speed: 3000 r.p.m.), fraction 4 enriched in steps 1–8 round spermatids (9.5–14 ml/min, 2000 r.p.m.) and fraction 6 enriched in pachytene primary spermatocytes (24–30 ml/min, 2000 r.p.m.) (22,23).
Chromatin preparation, histone extraction and purification
The testis or liver tissues from mouse were homogenized using a Kinematica Polytron in 150 mM NaCl, 20 mM Tris–HCl (pH 7.5), 0.1 mM EDTA buffer containing a protease inhibitor mixture (‘Complete’ from Roche Diagnostics at the ratio of one tablet per 100 ml buffer). The homogenate was subsequently processed and digested with micrococcal nuclease (MNase) at 30 U/mg chromatin for the amount of time indicated, to generate S1, SE and P chromatin fractions as described elsewhere (24). Chromatin fractions from native HeLa cells or from HeLa cells ectopically expressing a non-tagged version of mH2A.Bbd isotype 3 (mH2A.Bbd-3) (Figure 1A) were prepared in the same way. Mouse testes nuclei were also extensively digested and the digestion product (mainly mononucleosomes) was processed with a modification of the method described elsewhere (25). Briefly, nuclei were digested with MNase at 100 U/mg of chromatin at 37°C for 30 min. MNase digest reaction was quenched by addition of EGTA to a final concentration of 1 mM. Nuclei were centrifuged at 600g for 10 min and the pellets were resuspended in 20 mM Tris–HCl (pH 7.5), 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EGTA. The resuspended sample was incubated on ice for one and a half hour and then centrifuged at 1000g for 10 min to produce an S supernatant and a P pellet. Histones from the different chromatin fractions were extracted with 1 N HCl (at ∼6 µl/mg of starting tissue), the HCl extracts were precipitated with six volumes of acetone at −20°C and analyzed by SDS–/AU–PAGE or reversed-phase HPLC as described elsewhere (26). Proteins from mature human sperm samples were analyzed as described (27). Alternatively, and in order to visualize the protamines, the mouse testes chromatin fractions were dialyzed against water, lyophilized, pyridylethylated in the presence of 4 M guanidium–HCl, 50 mM Tris–HCl (pH 7.5), 1.25 mM EDTA solution and homogenated. Briefly, 1 μl of β-mercaptoethanol was added to every 250 μl of the guanidinium chloride suspension and incubated for 1.5 h at room temperature in the dark followed by addition of 2 μl of vinyl pyridine and further incubation for 30 min vortexing every 5 min. The sample was then dialyzed, lyophilized and extracted with 1 N HCl as above.
The sperm chromatin was isolated as described elsewhere (28). In brief, semen samples were washed with ice-cold PBS, then cell pellets were treated with PBS containing 0.5% Triton X-100 for 10 min on ice, washed with PBS and resuspended with reaction buffer: 10 mM Tris–HCl (pH 8.0), 2 mM CaCl2, 2 mM MgCl2, 10 mM dithiothreitol. The micrococcal nuclease was added to the suspensions at final concentration of 20 U/mg DNA and samples were incubated for 10 min at 37°C. After centrifugation, the supernatants were discarded and the pellets were resuspended with 10 mM Tris–HCl (pH 8.0), 5 mM EDTA, 10 mM dithiothreitol, extracted on ice for 30 min and supernatants (SE) and the pellet (P) were collected.
Western blot analysis
Western blot analyses of SDS–PAGE of the proteins from the different chromatin and RP-HPLC fractions were carried out using a rabbit polyclonal antiserum elicited against recombinant mouse H2A.Bbd (21) and used at a dilution of 1:1000. Other antibody dilutions used in the western blots were: H4, 1:5000; pan acetylated H4 (Millipore), 1 : 500; and a secondary rabbit horse-radish peroxidase conjugate (Abcam), 1 : 5000. Protein transfer and detection was performed as described (29). Western blot analysis was also used to determine the ratio of histone H2A.Bbd to histone H4 in the mouse spermatogenic fraction 2 obtained by centrifugal elutriation. To this end, the chromosomal proteins from this fraction were run on an SDS–PAGE together with increasing amounts of recombinant mH2A.Bbd and histone H4 followed by a double western using the mouse H2A.Bbd and H4 antibodies described above. The intensity of the different bands was analyzed by densitometry and the relative ratio of H2A.Bbd to H4 in the query sample was determined by interpolation.
Immunofluorescence
Mouse (20T1/2) and human (SK-N-SH) cell lines were grown in DMEM with 10% fetal calf serum, plated on glass cover slips and allowed to attach and grow until between 40% and 80% confluent. They were then co-transfected with GFP-H2A.Bbd using Effectene transfection reagent (Qiagen). The cells were then grown for an additional 16–20 h followed by fixation with 4% paraformaldehyde in PBS. The cells were then mounted in PBS containing 90% glycerol, 0.5 mg/ml para-phenylenediamine and 1 µg/ml DAPI. The cells were then fixed with 4% paraformaldehyde in PBS and imaged by wide-field fluorescence microscopy. The images were collected with a Zeiss Axioplan II fluorescence microscope using a 63 × 1.4 N.A. PlanApo objective and a Photometrics CoolSnap fx 12-bit cooled CCD.
RESULTS AND DISCUSSION
Histone H2A.Bbd is located on the X-chromosome and evolves very quickly under positive Darwinian selection
In order to identify the presence of native H2A.Bbd protein in the cell, we used mouse (Mus musculus) as a biological system. An initial search of the mouse genome indicated that five closely related putative protein isotypes [mBbd.1–5 (Figure 1A)] are encoded by four different genes, all of which are located on the X-chromosome. Isotypes mH2A.Bbd.1 and mH2A.Bbd.2 are encoded by a single gene referred to here as gene 1-2 (see below). This mouse pattern of gene organization contrasts with that of humans where only two protein isotypes, hH2A.Bbd.1 and hH2A.Bbd.2, are present and are encoded by three different genes also located on the X-chromosome. The rapid mode of evolution of these proteins in mouse and human is apparent when analyzing the nucleotide changes from their reconstructed ancestral sequences using methods of maximum likelihood. This reveals that the non-synonymous changes are consistently more frequent than the synonymous changes in all instances (Figure 1B). The non-synonymous variation represents 60–65% of the overall nucleotide variation. This is an intrinsic characteristic of the sexually driven positive Darwinian selection of many genes involved in male reproduction including primate protamines (30). When these results are translated into evolutionary rates (Figure 1C), it appears that H2A.Bbd has a rate of evolution that exceeds that of any other histone including histone H1. Furthermore, the rate of H2A.Bbd evolution is even faster than that of human versus mouse P1 protamines (31). It is noteworthy that the H2A.Bbd gene is encoded on the X-chromosome (Figure 2A), since it has been shown that X-linked sperm proteins evolve at faster rates than their autosomal counterparts (32).
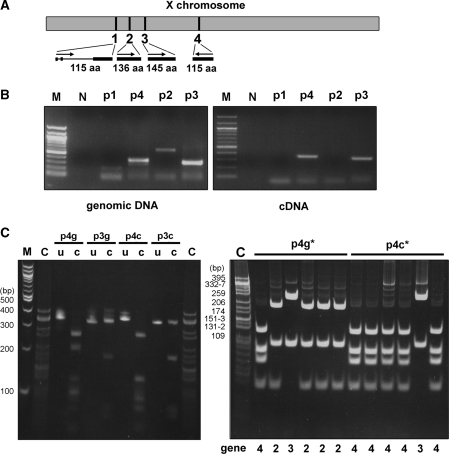
Figure 2.
Transcriptional expression of mouse H2A.Bbd. (A) Schematic representation of the H2A.Bbd genes in the mouse X-chromosome (grey box) and their organization (arrows depict 5′–3′ transcription) and structure (exons, thick line; introns, thin line). (B) PAGE of PCR products from mouse genomic DNA and testis cDNA. The primer set number is indicated on top. (C) The PCR products of the genomic (p3g, p4g) and cDNA (p3c, p4c) amplification were digested with HpaII restriction enzyme (left panel). Alternatively, the PCR products were cloned and the DNA extracted from genomic (3g*) and cDNA (3c*) colonies was digested with HpaII (right panel). The numbers underneath the gel indicate the genes shown in (A). M is a 100 bp DNA ladder and C is a CfoI digest of pBR322 used as markers; u, uncut; c, cut DNA.
Three of the four histone H2A.Bbd genes in mice are differentially expressed in testes
Previous reports had noted enhanced levels of H2A.Bbd mRNA expression in human and mouse testes (1,21). In order to examine this observation, especially considering the evolutionary data described above, we prepared several primers designed to amplify distinct regions within the coding segments of each of the four mouse genes identified in silico as shown in Figure 2A. Each set of primers was able to generate the expected gene fragments using mouse genomic DNA as a template (Figure 2B). However, only the cDNAs corresponding to genes 1-2 and 3 could be amplified from a mouse testis cDNA library, suggesting that only genes 1-2 and 3 are transcribed in mouse testes. Given the intrinsic similarity between the nucleotide sequences of the cDNAs of the different H2A.Bbd isotypes, it was necessary to expand the analysis in order to corroborate this finding. The fragments amplified with the primers for H2A.Bbd-3, H2A.Bbd-4 and H2A.Bbd-1-2 from testes cDNA template were predicted to have 3, 2 and 1 HpaII restriction sites, respectively, and therefore digestion of the PCR products shown in Figure 2B with this enzyme (Figure 2C, left panel) determined that these bands represented a mixed population of PCR amplicons. These amplicons were then cloned and the DNA from the many different colonies obtained was sequenced and digested again with HpaII (Figure 2C, right panel). The amplified fragments from the cDNA library (i.e. p3c*) were able to produce only fragments corresponding to mH2A.Bbd-1-2 and mH2A.Bbd-3, a result that was confirmed by the sequencing analysis. Out of 70 positive colonies obtained from genomic amplification, 43% corresponded to H2A.Bbd-4, 30% to H2A.Bbd-1-2 and 27% to H2A.Bbd-3, and from 30 positive colonies obtained from the testes cDNA library, 80% belonged to H2A.Bbd-3 and 20% to H2A.Bbd-1-2. This indicates that the primers are approximately equally efficient and that H2A.Bbd-3 is expressed at much greater levels than H2A.Bbd-1-2 in testes. The lack of genomic amplification of gene 5 is not surprising as the coding region of this gene is distributed through 3 exons encompassing 20, 100 and 16 amino acids in the N- to C-terminal direction of the protein, with a 5.2 kb intron 2.
Histone H2A.Bbd is present in cells at advanced stage of spermiogenesis and in human sperm
Since it was clear that mH2A.Bbd isotypes 1-2 and 3 were expressed at the transcriptional level in testes, we decided to look for the protein in testis tissue. Similar protein amounts of HCl nuclear extracts from transfected HeLa cells over-expressing an ectopic version of mH2A.Bbd-3, mouse liver and testes were analyzed by RP-HPLC (Figure 3A and B). The entire volume of each of the HPLC fractions was vacuum-dried and subjected to SDS–PAGE and western blot analysis with a mH2A.Bbd antibody (Figure 3B). As shown in Figure 3B, mH2A.Bbd could only be detected in HeLa cells ectopically expressing the protein and in mouse testes. Furthermore, in the almost identical histone elution profiles (Figure 3A and results not shown), mH2A.Bbd eluted after histone H3. Two bands can be visualized at this position (see arrows in Figure 3B) in mouse testes: an intense one with identical electrophoretic mobility to mH2A.Bbd-3 and one displaying a lower mobility which presumably corresponds to the less expressed H2A.Bbd-1–2.
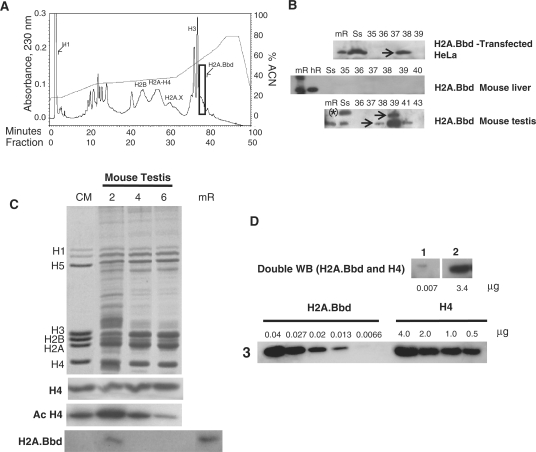
Figure 3.
Identification of native H2A.Bbd in mouse testis. (A) Reversed-phase HPLC of a mouse testes HCl extract. (B) H2A.Bbd, western blot analysis of some of the fractions shown in (A) and of fractions from similar HPLCs carried out with extracts from HeLa cells over-expressing H2A.Bbd-3 or mouse liver. The arrows point to H2A.Bbd bands. The asterisk shows that in addition to Mm.H2A.Bbd isotype 3, the mouse testis starting sample contains an electrophoretic band with reduced electrophoretic mobility that likely corresponds to the larger isoforms indicated in Fig. 2. (C) SDS–PAGE of spermatogenic fractions 2, 4 and 6, and their western blot analysis using H2A.Bbd, pan-acetylated H4 (Ac H4) and H4 antibodies. CM, chicken erythrocyte histones; hR, human recombinant H2A.Bbd; mR, mouse recombinant H2A.Bbd-3; Ss, starting sample. (D) Double western analysis of spermatogenic fraction 2 using H2A.Bbd antibody (panel 1) followed by H4 antibody (panel 2). Panel 3 shows a similar double western analysis of increasing amounts of mouse recombinant H2A.Bbd and H4. The number of micrograms shown in panels 1 and 2 were determined from the best fit linear regression lines obtained from plots of the microgram protein amounts versus the intensity of the electrophoretic bands in 3.
A western blot analysis of mouse testes cells at different stages of spermatogenesis is shown in Figure 3C. The cells were separated by centrifugal elutriation and three fractions, 2, 4 and 6 enriched in elongating spermatids, round spermatids and pachytene spermatocytes, respectively, were collected. The analysis shows that mH2A.Bbd is present in the elongating spermatid fraction (Figure 3C) at a time when histone H4 is maximally acetylated and when histones are being replaced by protamines (14,15). A titration western blot using different amounts of H4 and mH2A.Bbd and their corresponding antibodies (see ‘Materials and Methods’ section) indicates that there are ∼485 mg (3.4/0.007) of histone H4 for every microgram of mH2A.Bbd in this mouse spermatogenic fraction. Upon correction for the difference in molecular mass of these two histones, this corresponds to one molecule of mH2A.Bbd present for every 540 molecules of histone H4 (270 nucleosomes) (Figure 3D). Moreover, when human sperm was digested with micrococcal nuclease under strong reducing conditions (to break the inter-protamine disulfide bridges), H2A.Bbd was found to be present in the histone variant-enriched (33,34), nucleosomally organized (28) complement that coexists with protamines in this sperm (Figure 4A–C). This rather unique nucleosome fraction has been described to be associated with telomeres (35). It has been found to be enriched in gene regulatory regions and CTCF binding sequences (36), and to include genes that are important for embryo development (37).
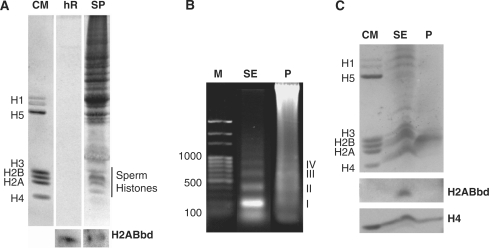
Figure 4.
Presence of Native H2A.Bbd in human sperm. (A) SDS–PAGE of human sperm histones and H2A.Bbd western. Human sperm chromatin fractions obtained from micrococcal nuclease digest are used in (B) and (C). (B) Agarose electrophoresis of the DNA fragments of the chromatin fraction soluble upon digestion (SE) and the insoluble protamine-containing (P) fraction. (C) SDS–PAGE of the proteins associated with the chromatin fractions and the western analysis using mouse H2A.Bbd and histone H4 antibodies. HR, human recombinant H2A.Bbd; M, 100 bp DNA marker (New England Biolabs) DNA marker; P, pellet chromatin fraction (P); SE, chromatin fraction (SE); SP, total human sperm protein. The roman numerals in (E) denote DNA fragments corresponding to mono-, di-, tri- and tetra-nucleosomes, respectively.
Histone H2A.Bbd is found associated with a chromatin insoluble fraction in cells ectopically expressing the native form of the protein and in mouse (20T1/2) cell line, H2A.Bbd is enriched at the periphery of chromocenters
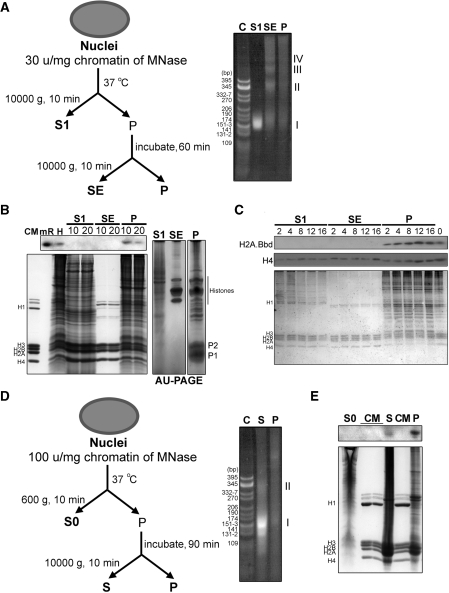
Micrococcal nuclease digestion analysis of chromatin from mouse testes under non-reducing conditions (Figure 5A, B, D and E) and from HeLa cells ectopically over-expressing mH2A.Bbd (Figure 5C) showed that mH2A.Bbd is preferentially present in the insoluble fractions regardless of the extent of digestion (Figure 5D and E). In mouse testes, this is the fraction that is enriched in protamines (Figure 5B). However, in this latter instance, this does not preclude the association of this variant with potential nucleosome structures (see DNA patterns in Figure 5A and E) which most likely are trapped in the insoluble protamine mesh resulting from the protamine P1/P2 (Figure 5B) inter-molecular disulfide bridges (15).
Figure 5.
H2A.Bbd is present in micrococcal nuclease-resistant insoluble chromatin. (A) Flowchart of the micrococcal nuclease digestion of mouse testes (left hand) (B) and of HeLa cells ectopically expressing MmH2A.Bbd-3 (C) and a representative PAGE of the DNA extracted from the different chromatin fractions generated in this way (right hand). (B) SDS–PAGE (left hand) and AU–PAGE (right hand) of the protein composition of the fractions indicated in (A). (C) Digestion at different times (in minutes) as indicated on top of the gel. (D) Flowchart of an extensive digestion of mouse testes to generate mononucleosome chromatin fragments. (E) PAGE of the DNA (left) and SDS–PAGE of the proteins (right) of the fractions thus obtained. Western blots in this figure were carried out with mouse polyclonal H2A.Bbd antibody. C and CM are CfoI pBR322 digested DNA and chicken erythrocyte histone markers, respectively.MR, mouse recombinant H2A.Bbd. The roman numerals (I–IV) in (A) and (E) denote DNA fragments corresponding to mono-, di-. tri- and tetra-nucleosmes, respectively.
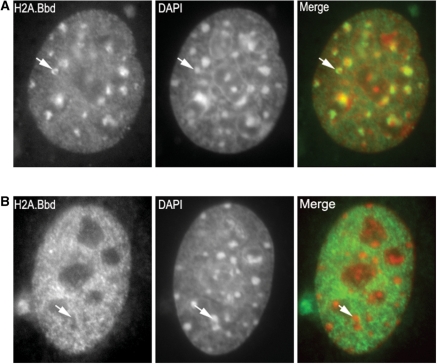
In order to try to ascertain the potential nature of the genomic regions associated with the micrococcal nuclease insoluble fraction, immunofluorescence studies using mouse and human cell lines transfected with Mm-H2A.Bbd-3-GFP were carried out (Figure 6). These experiments when carried in combination with FRAP (results not shown) indicated, that like in the case of the human counterpart (3) this histone exhibits an unusually high rate of exchange when compared to any other histone. Human (not shown) and mouse cell lines showed similar distributions. The results with mouse are shown because of the prominent chromocenters that facilitate the assessment of distribution. Two distributions were observed. A minority of the cells showed the distribution shown in Figure 6A, where there was an unusual enrichment at the periphery of the chromocenters (e.g. arrows in Figure 6A). This unusual organization has been previously reported for topoisomerase IIα during early G2 (38). The majority of cells showed a more broadly distributed pattern (Figure 6B). In both cases, the Mm-H2A.Bbd was largely excluded from heterochromatin. This is an unusual distribution for a histone. It is possible to speculate that the presence of the native form in the elongating spermatids of fraction 2 of mouse testis (Figure 3C) has a similar distribution. Interestingly, a murine homologue of HP1 (M31) localizes to the centromeric chromocenter of mice round spermatids. Like with H2A.Bbd, the protein is also present in mature sperm. It was proposed to be involved in higher order organization of sperm chromatin (39).
Figure 6.
Fluorescence microscopy of MmH2A.Bbd ectopically expressed in mouse C3H10T/2 embryonic fibroblast cells. Examples of the two distributions observed in mouse cell lines are shown. (A) A nucleus showing enrichment of MmH2A.Bbd on the periphery of chromocenters. The arrow illustrates an example of a chromocenter where the distinction between the H2A.Bbd staining and the DNA distribution can be clearly seen. The chromocenters, which are clearly visible in the mouse cell line, appear yellow in the portions of the chromocenter that overlap with H2A.Bbd. (B) A nucleus showing a broad distribution with exclusion from heterochromatin. In these images, the left panel shows H2A.Bbd-GFP, the middle panel is stained with DAPI and the panel on the right is the merged image. Sites that are enriched in H2A.Bbd are green and DNA stained with DAPI is red. The arrow indicates a region of the nucleus containing a chromocenter. The exclusion of H2A.Bbd from this chromocenter is evident as an area of exclusion in the H2A.Bbd image and an area of enrichment in the DAPI image. Reflecting the absence of H2A.Bbd in these heterochromatin domains, the chromocenter in the composite image are red.
During mammalian spermiogenesis, histones at the onset of the differentiation process are gradually replaced by transition proteins and protamines (14,22) and although the replacement is almost complete in most species, in humans ∼15% of the mature sperm chromatin remains associated with histone variants (33). In vitro studies of H2A.Bbd have shown that it participates in chromatin destabilization and unfolding (2,3,21). Therefore, it is possible to envisage how this variant may play a critical role in assisting the displacement of histones by protamines and in facilitating the transition from a nucleohistone to a nucleoprotamine chromatin organization during the late stages of mammalian spermiogenesis. The presence of H2A.Bbd within the histone complement that remains bound to DNA in mature human sperm is very intriguing (40). The role of these remaining histones is currently the focus of intense investigation. Evidence exists for both structural (41) and functional (37) involvement and a recent study indicates that these histones could have potential implications for fertility (27).
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC) (OGP-0046399-02 grant to J.A.), by a contract within the Ramon y Cajal Program of the Spanish Ministry of Science and Innovation-MICINN (to J.M.E.-L.); National Institutes of Health (grant HD-16843 to M.L.M.); Cancer Center Support Grant (CA-16672 to M.D. Anderson Cancer Center); Canadian Institutes of Health Research grant (to M.H.). T.I. is the recipient of a Michael Smith Health Research Foundation post-doctoral fellowship and A.L. is the recipient of a Natural Sciences and Engineering Research Council of Canada (NSERC) CGSD doctoral fellowship. M.H. is an Alberta Heritage Foundation for Medical Research Senior Scholar. Funding for open access charge: NSERC.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are very thankful to Steven Henikoff, Victor Vacquier and Deanna Dryhurst for carefully reading the manuscript and for their insightful criticism and comments. We also would like to thank Igor Nazarov and Andrei Zalensky at the Jones Institute for Reproductive Medicine in Eastern Virginia Medical School (Norfolk, Virginia) for providing us with the mature human sperm nucleosomal histone fractions used for the western shown in Figure 4B and C.
REFERENCES
- 1.Chadwick BP, Willard HF. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J. Cell Biol. 2001;152:375–384. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao Y, Konesky K, Park YJ, Rosu S, Dyer PN, Rangasamy D, Tremethick DJ, Laybourn PJ, Luger K. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 2004;23:3314–3324. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gautier T, Abbott DW, Molla A, Verdel A, Ausio J, Dimitrov S. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 2004;5:715–720. doi: 10.1038/sj.embor.7400182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montel F, Fontaine E, St-Jean P, Castelnovo M, Faivre-Moskalenko C. Atomic force microscopy imaging of SWI/SNF action: mapping the nucleosome remodeling and sliding. Biophys J. 2007;93:566–578. doi: 10.1529/biophysj.107.105569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyen CM, Montel F, Gautier T, Menoni H, Claudet C, Delacour-Larose M, Angelov D, Hamiche A, Bednar J, Faivre-Moskalenko C, et al. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 2006;25:4234–4244. doi: 10.1038/sj.emboj.7601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelov D, Verdel A, An W, Bondarenko V, Hans F, Doyen CM, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, et al. SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J. 2004;23:3815–3824. doi: 10.1038/sj.emboj.7600400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongelard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okuwaki M, Kato K, Shimahara H, Tate SI, Nagata K. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell. Biol. 2005;25:10639–10651. doi: 10.1128/MCB.25.23.10639-10651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marushige K. Activation of chromatin by acetylation of histone side chains. Proc. Natl Acad. Sci. USA. 1976;73:3937–3941. doi: 10.1073/pnas.73.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calestagne-Morelli A, Ausio J. Long-range histone acetylation: biological significance, structural implications, and mechanisms. Biochem. Cell Biol. 2006;84:518–527. doi: 10.1139/o06-067. [DOI] [PubMed] [Google Scholar]
- 11.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 12.Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat. Struct. Mol. Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 14.Oliva R, Dixon GH. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog. Nucleic Acid Res. Mol. Biol. 1991;40:25–94. doi: 10.1016/s0079-6603(08)60839-9. [DOI] [PubMed] [Google Scholar]
- 15.Lewis JD, Song Y, de Jong ME, Bagha SM, Ausio J. A walk though vertebrate and invertebrate protamines. Chromosoma. 2003;111:473–482. doi: 10.1007/s00412-002-0226-0. [DOI] [PubMed] [Google Scholar]
- 16.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 17.Sitnikova T, Rzhetsky A, Nei M. Interior-branch and bootstrap tests of phylogenetic trees. Mol. Biol. Evol. 1995;12:319–333. doi: 10.1093/oxfordjournals.molbev.a040205. [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 21.Eirin-Lopez JM, Ishibashi T, Ausio J. H2A.Bbd: a quickly evolving hypervariable mammalian histone that destabilizes nucleosomes in an acetylation-independent way. FASEB J. 2008;22:316–326. doi: 10.1096/fj.07-9255com. [DOI] [PubMed] [Google Scholar]
- 22.Unni E, Meistrich ML. Purification and characterization of the rat spermatid basic nuclear protein TP4. J. Biol. Chem. 1992;267:25359–25363. [PubMed] [Google Scholar]
- 23.Zhao M, Rohozinski J, Sharma M, Ju J, Braun RE, Bishop CE, Meistrich ML. Utp14b: a unique retrogene within a gene that has acquired multiple promoters and a specific function in spermatogenesis. Dev. Biol. 2007;304:848–859. doi: 10.1016/j.ydbio.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, He C, Moore SC, Ausió J. Effects of histone acetylation on the solubility and folding of the chromatin fiber. J. Biol. Chem. 2001;276:12764–12768. doi: 10.1074/jbc.M100501200. [DOI] [PubMed] [Google Scholar]
- 25.Sarcinella E, Zuzarte PC, Lau PN, Draker RR, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell Biol. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose KL, Li A, Zalenskaya I, Zhang Y, Unni E, Hodgson KC, Yu Y, Shabanowitz J, Meistrich ML, Hunt DF, et al. C-terminal phosphorylation of murine testis-specific histone H1t in elongating spermatids. J. Proteome Res. 2008;7:4070–4078. doi: 10.1021/pr8003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singleton S, Zalensky A, Doncel GF, Morshedi M, Zalenskaya IA. Testis/sperm-specific histone 2B in the sperm of donors and subfertile patients: variability and relation to chromatin packaging. Hum. Reprod. 2007;22:743–750. doi: 10.1093/humrep/del439. [DOI] [PubMed] [Google Scholar]
- 28.Nazarov IB, Shlyachtenko LS, Lyubchenko YL, Zalenskaya IA, Zalensky AO. Sperm chromatin released by nucleases. Syst. Biol. Reprod. Med. 2008;54:37–46. doi: 10.1080/19396360701876849. [DOI] [PubMed] [Google Scholar]
- 29.Abbott DW, Laszczak M, Lewis JD, Su H, Moore SC, Hills M, Dimitrov S, Ausio J. Structural characterization of macroH2A containing chromatin. Biochemistry. 2004;43:1352–1359. doi: 10.1021/bi035859i. [DOI] [PubMed] [Google Scholar]
- 30.Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 31.Retief JD, Winkfein RJ, Dixon GH, Adroer R, Queralt R, Ballabriga J, Oliva R. Evolution of protamine P1 genes in primates. J. Mol. Evol. 1993;37:426–434. doi: 10.1007/BF00178872. [DOI] [PubMed] [Google Scholar]
- 32.Torgerson DG, Singh RS. Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol. Biol. Evol. 2003;20:1705–1709. doi: 10.1093/molbev/msg193. [DOI] [PubMed] [Google Scholar]
- 33.Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 34.Churikov D, Zalenskaya IA, Zalensky AO. Male germline-specific histones in mouse and man. Cytogenet. Genome Res. 2004;5:203–214. doi: 10.1159/000078190. [DOI] [PubMed] [Google Scholar]
- 35.Zalenskaya IA, Bradbury EM, Zalensky AO. Chromatin structure of telomere domain in human sperm. Biochem. Biophys. Res. Commun. 2000;279:213–218. doi: 10.1006/bbrc.2000.3917. [DOI] [PubMed] [Google Scholar]
- 36.Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rattner JB, Hendzel MJ, Furbee CS, Muller MT, Bazett-Jones DP. Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol. 1996;134:1097–1107. doi: 10.1083/jcb.134.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoyer-Fender S, Singh PB, Motzkus D. The murine heterochromatin protein M31 is associated with the chromocenter in round spermatids and Is a component of mature spermatozoa. Exp. Cell Res. 2000;254:72–79. doi: 10.1006/excr.1999.4729. [DOI] [PubMed] [Google Scholar]
- 40.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J. Biol. Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 41.Zalenskaya IA, Zalensky AO. Telomeres in mammalian male germline cells. Int. Rev. Cytol. 2002;218:37–67. doi: 10.1016/s0074-7696(02)18011-9. [DOI] [PubMed] [Google Scholar]
- 42.Isenberg I. Histones. New York: Academic Press; 1978. [Google Scholar]