Abstract
Tpa1 (for termination and polyadenylation) from Saccharomyces cerevisiae is a component of a messenger ribonucleoprotein (mRNP) complex at the 3′ untranslated region of mRNAs. It comprises an N-terminal Fe(II)- and 2-oxoglutarate (2OG) dependent dioxygenase domain and a C-terminal domain. The N-terminal dioxygenase domain of a homologous Ofd1 protein from Schizosaccharomyces pombe was proposed to serve as an oxygen sensor that regulates the activity of the C-terminal degradation domain. Members of the Tpa1 family are also present in higher eukaryotes including humans. Here we report the crystal structure of S. cerevisiae Tpa1 as a representative member of the Tpa1 family. Structures have been determined as a binary complex with Fe(III) and as a ternary complex with Fe(III) and 2OG. The structures reveal that both domains of Tpa1 have the double-stranded β-helix fold and are similar to prolyl 4-hydroxylases. However, the binding of Fe(III) and 2OG is observed in the N-terminal domain only. We also show that Tpa1 binds to poly(rA), suggesting its direct interaction with mRNA in the mRNP complex. The structural and functional data reported in this study support a role of the Tpa1 family as a hydroxylase in the mRNP complex and as an oxygen sensor.
INTRODUCTION
The central processes of gene expression, from mRNA synthesis to translation and degradation, are mediated by the messenger ribonucleoprotein (mRNP) complexes. These mRNP complexes consist of individual transcripts bound by a changing repertoire of proteins that are the actual target for regulation and dictate the fate of an mRNA in the gene expression pathway (1). The ‘mRNP code’ is unique for each transcript in a given cell type and changes as the primary nuclear transcript undergoes processing and export (2). The mRNA-binding proteins mediate co-transcriptional 5′-end capping, splicing, 3′ polyadenylation and quality control of mRNAs within nascent mRNP complexes (3–5). While passing through each stage of mRNA processing, an mRNP complex is successively remodeled through the loss or gain of RNPs and other proteins, and is implicated in mRNA localization, translation initiation/termination and mRNA turnover in the cytoplasm (2).
Eukaryotic mRNAs are synthesized with two integral stability determinants: a cap at their 5′-end and a poly(A) tail at their 3′-end, that are believed to provide a basal level of stability to all mRNAs by preventing their degradation by cytoplasmic exonucleases (6). The mRNA-stabilizing properties of the poly(A) tail have been linked both to its binding to the poly(A)-binding protein (PABP) (7) and to its capacity to stimulate active translation (8). The length of the poly(A) tail is regulated by polymerase-deadenylase (9) and plays an essential role in controlling the mRNA function, subcellular localization and half-life. The removal of the mRNA poly(A) tail (deadenylation process) is known to be the first step in normal mRNA decay and is the major factor that controls the rate of mRNA decay (10,11). The processes of mRNA deadenylation and degradation, and the protein complexes that are involved in these processes are evolutionarily well conserved from Saccharomyces cerevisiae to humans (12).
Tpa1 (termination and polyadenylation) of S. cerevisiae was shown to be part of an mRNP complex at the 3′ untranslated region of mRNAs (13). It associates specifically with components of the translation termination complex, and is involved in both translation termination and in the regulation of normal mRNA decay through translation termination-coupled poly(A) shortening (13). Genetic studies demonstrated that the tpa1Δ mutation led to a 1.5- to 2-fold increase in the half-lives of mRNAs degraded by the general 5′→3′ pathway or the 3′→5′ nonstop decay pathway (13). S. cerevisiae Tpa1 was found to interact with a variety of proteins involved in mRNA processing, the translation termination factors eRF1 and eRF3, and a tRNA methyltransferase TRM1 (13,14).
It was also shown that Tpa1 interacts with the PABP (13). PABP, part of the 3′-end RNA-processing complex, mediates interactions between the 5′ cap structure and the 3′ mRNA poly(A) tail, is involved in the control of poly(A) tail length, and interacts with the translation factor eIF-4G (15). PABP binds poly(A) using one or more RNA-recognition motifs (16). Nuclear PABPs are necessary for the synthesis of the poly(A) tail, regulating its ultimate length and stimulating maturation of mRNAs (17). In the cytoplasm, PABPs facilitate the formation of the ‘closed loop’ structure of the mRNP particles, which is crucial for additional PABP activities that promote translation initiation/termination, recycling of ribosomes and stability of mRNAs (17).
A comprehensive bioinformatics study revealed that S. cerevisiae Tpa1 contains an N-terminal domain (NTD) belonging to the non-heme Fe(II)- and 2-oxoglutarate (2OG) dependent dioxygenase family (Pfam03171), and a C-terminal domain (CTD) belonging to the ofd1_CTDD (2OG and iron-dependent oxygenase C-terminal degradation domain) family (Pfam10637) (18,19). Members of the Fe(II)-/2OG-dependent dioxygenase family catalyze a remarkable diversity of enzymatic reactions (20), including post-translational modification of protein side chains (as hydroxylases) and the repair of alkylated DNA/RNA (as demethylases). Some members of this family play a role as cellular sensors and regulators (21). Recently, the N-terminal dioxygenase domain of Schizosaccharomyces pombe Ofd1 protein, a homolog of S. cerevisiae Tpa1 showing sequence identity of 40% (55% similarity), was shown to be required for oxygen sensing (19,22). Members of the Fe(II)-/2OG-dependent dioxygenase family are characterized by a core consisting of eight antiparallel β-strands that adopt the double-stranded β-helix (DSBH) fold, also known as the β-strand jelly-roll fold or double Greek key motif (23,24). They bind Fe(II) through the conserved H1X(D/E)…H2 motif (20,25,26). For many members of this family, the oxidation of the primary protein substrate is coupled to the conversion of the co-substrate 2OG to succinate and CO2 (20).
Homologs of S. cerevisiae Tpa1 are present not only in other fungi but also in higher eukaryotes. Human Tpa1 shows 28% sequence identity (43% similarity) to S. cerevisiae Tpa1 (Figure 1). Despite the importance of the biological roles that have been suggested for the Tpa1 family members in yeast, further studies are required to clearly establish their molecular functions. To provide a structural basis for a better understanding of the function, we have determined the crystal structure of S. cerevisiae Tpa1 as a representative member of the Tpa1 protein family. The structure reveals that two domains of Tpa1 have the common DSBH fold. However, a dioxygenase-type active site for binding Fe(II) and 2OG is present only in the NTD of S. cerevisiae Tpa1, whereas the CTD lacks such a feature. Furthermore, our electrophoretic mobility shift assay demonstrates that Tpa1 is capable of binding poly(A), suggesting that S. cerevisiae Tpa1 directly interacts with the mRNA in the mRNP complex.
Figure 1.
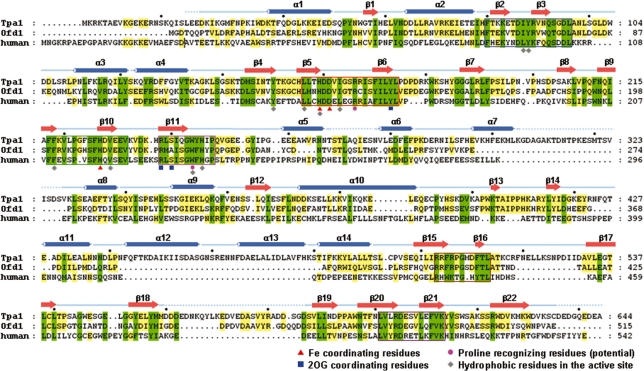
Sequence alignment of Tpa1 family. A sequence alignment of S. cerevisiae Tpa1 (Tpa1; SWISS-PROT accession code: P40032), S. pombe Ofd1 (Ofd1; Q11120) and Homo sapiens Tpa1 (human; Q8N543). Every 20th residue is marked with a black circle above the sequence of S. cerevisiae Tpa1. Highly conserved residues and partially conserved residues are shaded in green and yellow, respectively. Cylinders and arrows denote α-helices and β-strands, respectively. Disordered residues in the ternary structure are indicated by blue dotted lines. Six conserved sequence motifs of S. cerevisiae Tpa1 are enclosed by colored boxes: (1) motif I (Phe80–Leu96) in blue, (2) motif II (Leu156–Leu174) in red, (3) motif III (Phe217–Val232) in green, (4) motif IV (Arg238–Pro248) in pink, (5) motif V (Arg503–Leu513) in brown and (6) motif VI (Leu601–Lys613) in purple. The residues that coordinate the Fe(III) ion and 2OG are indicated by red triangles and blue squares below the sequences, respectively. This figure was drawn with ClustalX (53) and GeneDoc (http://www.nrbsc.org/downloads/).
MATERIALS AND METHODS
Protein expression and purification
The S. cerevisiae tpa1 gene (YER049W) encoding the N-terminal truncated form (residues 21–644) of Tpa1, which is called Tpa1Δ20, was PCR-amplified and cloned into the expression vector pET-21a(+) (Novagen). The recombinant protein fused with a hexahistidine-containing tag at its C-terminus was overexpressed in Escherichia coli Rosseta2(DE3)pLysS cells using Terrific Broth culture medium. Protein expression was induced by 0.5 mM isopropyl β-d-thiogalactopyranoside and the cells were incubated for additional 18 h at 30°C following growth to mid-log phase at 37°C. The cells were lysed by sonication in buffer A—50 mM Tris–HCl at pH 7.9, 0.50 M NaCl and 10% (v/v) glycerol—containing 5.0 mM imidazole. The crude lysate was centrifuged at ∼36 000g for 60 min. The supernatant was applied to an affinity chromatography column of HiTrap Chelating HP (GE Healthcare), which was previously equilibrated with buffer A. Upon eluting with a gradient of imidazole in the same buffer, the protein was eluted at 120–150 mM imidazole concentration. The eluted Tpa1 was applied to a HiLoad XK-16 Superdex 200 prep-grade column (Amersham-Pharmacia), which was previously equilibrated with 20 mM Tris–HCl (pH 7.5) and 200 mM NaCl. Fractions containing the yeast Tpa1 were concentrated to 5 mg ml–1 for crystallization using an Amicon Ultra-15 centrifugal filter unit (Millipore). The procedure for preparing the SeMet-substituted protein was the same except for the presence of 1.0 mM Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) in all buffers used during purification steps besides buffer A. When overexpressing the SeMet-substituted protein in E. coli Rosseta2(DE3)pLysS cells, we used the M9 cell culture medium that contained extra amino acids including SeMet. The CTD (residues 273–644) fused with a hexahistidine-containing tag at its C-terminus was overexpressed and purified essentially as described earlier. We tried to overexpress several constructs of the NTD (residues 21–246, 21–250, 21–253 and 21–277) but all of them were expressed as insoluble.
Crystallization
Crystals were grown by the sitting-drop vapor diffusion method at 14°C by mixing equal volumes (2 μl each) of the protein solution (5 mg ml–1 concentration in 20 mM Tris–HCl, pH 7.5 and 200 mM NaCl) and the reservoir solution. Crystals of the native Tpa1–Fe(III) binary complex were grown using a reservoir solution consisting of 200 mM lithium sulfate, 100 mM Tris–HCl (pH 8.5) and 25% (w/v) PEG 3350 and 0.14 mM of ferrous ascorbate. They grew to approximate dimensions of 0.2 mm × 0.2 mm × 0.1 mm within a few days. Crystals of the binary complex of the SeMet-substituted Tpa1 were obtained as above for the native crystals except for the presence of 0.10 mM TCEP and 10 mM strontium chloride in the protein solution. To elucidate how Tpa1 binds its co-substrate 2OG, crystals of the native Tpa1–Fe(III)–2OG ternary complex were obtained by co-crystallization in the presence of 0.14 mM ferrous ascorbate and 7.0 mM sodium 2OG.
X-ray data collection and structure determination
A crystal of the native Tpa1–Fe(III) binary complex was dipped for a few seconds into 10 µl of a cryoprotectant solution, which consisted of 20% (v/v) glycerol added to the reservoir solution. We found that an addition of ∼2 µl of 50 mM ferrous ascorbate solution to the cryoprotectant solution was necessary to improve the resolution limit of the crystals. The soaked crystal was frozen in the cold nitrogen gas stream at 100 K. X-ray diffraction data were collected at 100 K using an ADSC Quantum 210 CCD detector system (Area Detector Systems Corporation, Poway, CA, USA) at the beamline NW12A of Photon Factory (PF), Japan. The raw data were processed and scaled using the program suit HKL2000 (27). The native Tpa1–Fe(III) binary complex crystal belongs to the space group P321, with unit cell parameters of a = b = 136.3 Å, c = 79.92 Å, α = β = 90° and γ = 120°. One Tpa1 monomer is present in an asymmetric unit, giving a solvent fraction of 58.2%. X-ray diffraction data from a crystal of the native Tpa1–Fe(III)–2OG ternary complex were collected essentially as described above at the BL-4A experimental station of Pohang Light Source (PLS), Pohang, Korea. The addition of 50 mM ferrous ascorbate solution to the cryoprotectant solution was unnecessary to improve the resolution limit of the ternary complex crystals. The native Tpa1–Fe(III)–2OG ternary complex crystal belongs to the space group P321, with unit cell parameters of a = b = 136.3 Å, c = 79.83 Å, α = β = 90° and γ = 120°. One Tpa1 monomer is present in an asymmetric unit, giving a solvent fraction of 58.2%.
Single-wavelength anomalous diffraction (SAD) data using a binary complex crystal of the SeMet-substituted Tpa1 were collected at 100 K on a Quantum 210 CCD area detector (Area Detector Systems Corporation) at the BL-4A experimental station of PLS, Pohang, Korea, at the absorption peak for selenium. The addition of 50 mM ferrous ascorbate solution to the cryoprotectant solution was not helpful in improving the resolution limit of the binary complex crystals of the SeMet-labeled Tpa1. Instead, the crystal was dehydrated by incubating for ∼10 min in 10–20 µl of a cryoprotectant solution containing 40% (v/v) glycerol. The SeMet-substituted crystal belongs to the space group P321, with unit cell parameters of a = b = 136.3 Å, c = 83.28 Å, α = β = 90° and γ = 120°. One Tpa1 monomer is present in an asymmetric unit, giving a solvent content of 59.9%. A summary of the data collection statistics is given in Table 1. Selenium atoms were located with the program SOLVE (28). The phases were further improved by density modification using the program RESOLVE (29), yielding an interpretable electron density map. Phasing statistics are summarized in Table 1.
Table 1.
Statistics on data collection, phasing and refinement
| A. Data collection | |||
| Data set | Binary complex | Ternary complex | Se-Met (peak) |
| X-ray source | PF NW12A | PLS BL-4A | PLS BL-4A |
| Wavelength (Å) | 1.00000 | 1.23990 | 0.97954 |
| Space group | P321 | P321 | P321 |
| a, b, c (Å) | 136.3, 136.3, 79.92 | 136.3, 136.3, 79.83 | 136.3, 136.3, 83.28 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution range (Å) | 50–2.50 (2.59–2.50)a | 50–1.77 (1.80–1.77)a | 50–2.73 (2.80–2.73)a |
| Total/unique reflections | 489 231/29 916 | 1 660 636/82 927 | 731 176/23 991 |
| Completeness (%) | 100.0 (100.0)a | 99.8 (96.5)a | 100.0 (100.0)a |
| I/σI | 39.5 (8.5)a | 80.7 (8.3)a | 45.5 (7.7)a |
| Rmerge b (%) | 12.5 (41.7)a | 6.1 (41.9)a | 16.5 (65.5)a |
| B. Phasing | |||
| Figure of merit for 20.0–3.10 Å range (before/after density modification) | 0.30/0.72 | ||
| C. Model refinement | |||
| Resolution range (Å) | 20.0–2.50 | 20.0–1.77 | 20.0–2.73 |
| Rwork/Rfree c (%) | 18.1/23.1 | 18.0/20.5 | 19.0/24.7 |
| Number of atoms/average B-factor (Å2) | |||
| Protein nonhydrogen atoms | 4556/27.7 | 4563/22.5 | 4603/36.7 |
| Water oxygen atoms | 324/28.0 | 791/37.1 | 199/36.8 |
| Ligand nonhydrogen atoms | |||
| Fe(III) ion | 1/43.4 | 1/17.5 | 1/45.8 |
| 2-Oxoglutarate | 10/21.8 | ||
| Sulfate | 20/51.5 | 20/46.4 | |
| Glycerol | 6/58.8 | 6/44.0 | |
| R.M.S. deviations from ideal geometry | |||
| Bond lengths (Å)/angles (°) | 0.008/1.21 | 0.007/1.07 | 0.010/1.25 |
| Ramachandran plot | |||
| Most favorable (%) | 89.5 | 90.3 | 89.3 |
| Allowed (%) | 9.9 | 9.7 | 10.5 |
| Generously allowed (%) | 0.6 | 0.0 | 0.2 |
| Disallowed (%) | 0.0 | 0.0 | 0.0 |
aValues in parentheses refer to the highest resolution shell.
bRmerge = ∑h∑i | I(h)i – <I(h)> | /∑h∑i I(h)i, where I(h) is the intensity of reflection h, ∑h is the sum over all reflections, and ∑i is the sum over i measurements of reflection h.
cRwork = ∑ | |Fobs| – |Fcalc| | /∑ |Fobs|, where Rfree is calculated for a randomly chosen 5% of reflections, which were not used for structure refinement and Rwork is calculated for the remaining reflections.
Model building and refinement
The SAD-phased electron density map was interpreted by the automatic model building program RESOLVE to build an initial polyalanine model, which accounted for ∼30% of the residues. Subsequently, the missing residues were built and the chains were traced manually using the program COOT (30). The model of the SeMet-labeled Tpa1 was refined with the program REFMAC (31), including the bulk solvent correction. The refined model of the Fe(III)-bound Tpa1, accounting for 563 residues in one monomer of Tpa1, 199 water molecules and a Fe(III) ion in the asymmetric unit, gave Rwork and Rfree values of 19.0 and 24.7% for the 20.0–2.73 Å data, respectively (Table 1). Of the total 5% of the data were randomly set aside as the test data for the calculation of Rfree (32). The model of the SeMet-labeled Tpa1 was used to refine the structures of native Tpa1. The refined model of the Fe(III)-bound Tpa1, accounting for 557 residues in one monomer of Tpa1, 324 water molecules, a Fe(III) ion and four sulfate ions in the asymmetric unit, gave Rwork and Rfree values of 18.1 and 23.1% for the 20.0–2.50 Å data, respectively (Table 1). One of the sulfate ions is located at the crystallographic 2-fold symmetry axis. The model of the ternary complex was refined against 20.0–1.77 Å data to Rwork and Rfree values of 18.0 and 20.5%, respectively. It accounts for 558 residues in one monomer of Tpa1, 791 water molecules, a Fe(III) ion, one molecule of 2OG, four sulfate ions and one glycerol molecule in the asymmetric unit. The electron density map indicated that the side chains of 11 residues (His59, Ile87, Val90, Ser169, Ser183, Asn266, Ser339, Gln340, Ser343, Glu345, Cys494 and Asp592) have dual conformations. The refined models of both binary and ternary complexes have excellent stereochemistry (Table 1), as evaluated by the program PROCHECK (33).
Equilibrium sedimentation
Equilibrium sedimentation studies were performed using a Beckman ProteomeLab XL-A analytical ultracentrifuge in 20 mM Tris–HCl buffer (pH 7.5) containing 4.0 μM ferrous ascorbate, 4.0 μM sodium 2OG and 150 mM NaCl at 20°C. Tpa1Δ20 samples containing ferrous ascorbate and 2OG were measured at 235, 237 and 280 nm using a six-sector cell at three speeds (8000, 10 000 and 12 000 r.p.m.) and two different protein monomer concentrations (1.50 and 2.02 μM) with the loading volume of 130 μl. The Tpa1 CTD (residues 273–644) samples in 20 mM Tris–HCl buffer (pH 7.5) containing 200 mM NaCl were measured at 240 and 280 nm using a two-sector cell at 30 000 r.p.m. and two different monomer concentrations of 1.97 and 3.93 μM with the loading volume of 180 μl at 20°C. The concentrations of the Tpa1Δ20 and CTD proteins were calculated using ε280 nm = 98 905 M–1cm–1 and ε280 nm = 50 880 M–1cm–1, respectively, obtained from their amino acid compositions.
Electrophoretic mobility shift assay
To assess the mRNA-binding ability of the purified Tpa1, a 20-mer poly(rA) was synthesized (Bioneer, Korea) and was 5′-labeled with [γ-32P]ATP by T4 polynucleotide kinase. The reaction buffer contained 15 mM sodium HEPES at pH 7.5, 100 mM potassium acetate, 0.2% bovine serum albumin, 20 unit of RNase inhibitor and 5.0 mM DTT. The Tpa1Δ20 proteins (final concentration of 5.0 μM) were added to the reaction mixture prior to the labeled poly(rA). Ferrous ascorbate (2.0 mM), 2OG (2.0 mM) or EDTA (10 mM) was also added to the reaction mixture. As a control, 15 μM bovine serum albumin was added instead of Tpa1Δ20. The mixture was incubated for 1 h at 37°C and the incubated mixture was resolved on a 5% (w/v) polyacrylamide gel. After electrophoresis at 4°C, the gel was exposed to an X-ray film.
RESULTS AND DISCUSSION
Structure determination, model quality and comparison of monomer structures
Because overexpression of the full-length yeast Tpa1 (residues 1–644) was not successful, several truncated constructs were made. Although we could express the C-terminal truncated form of Tpa1 (residues 1–635) and both N- and C-terminal truncated form of Tpa1 (residues 21–635) in E. coli cells and crystallize them, the quality of these crystals was poor. Only the N-terminal truncated form of Tpa1 (residues 21–644) yielded well-diffracting crystals in the presence of ferrous ascorbate, a cofactor of Tpa1. The structure of Tpa1 was solved by SAD phasing (Table 1).
We have determined three crystal structures of S. cerevisiae Tpa1: (i) the binary complex of the native protein with Fe(III), (ii) the ternary complex of the native protein with both Fe(III) and 2OG and (iii) the binary complex of the SeMet-labeled protein. Refinement statistics are summarized in Table 1. An example of the electron density for the refined model of the ternary complex is shown in Figure 2A. Since the crystals were grown under aerobic conditions, the Tpa1-bound iron in the crystal likely exists in the inactive Fe(III) state, which can be produced by either direct attacking of molecular oxygen or an uncoupled reaction pathway (i.e. conversion of 2OG to CO2 and succinate with no corresponding substrate transformation) (20). In the ternary complex model, three internal regions of the polypeptide chain (Thr270–Ser276, Gly306–Ser327 and Asp561–Gly585) and terminal residues (Leu21–Glu23, Asp636–Ala644 and a C-terminal affinity tag LEHHHHHH) are disordered. In the binary complex model of the native Tpa1, Ser269 is also disordered. In the binary complex structure of the SeMet-labeled Tpa1, three internal regions of the polypeptide chain (Asp95–Leu99, Gly306–Ser327 and Asp561–Gly585) and terminal residues (Leu21, Glu637–Ala644 and a C-terminal affinity tag LEHHHHHH) are disordered. In this model, the linker (Ser269–Ser276) between NTD and CTD is ordered and forms an α-helix. The disordered internal regions are not well conserved among the Tpa1 family members (Figure 1). They correspond to or occur adjacent to long insertions in S. cerevisiae Tpa1. The root-mean-square (R.M.S.) deviation between the binary and ternary complexes of the native protein is 0.23 Å for 557 Cα atom pairs. The R.M.S. deviations >1.0 Å occur at Pro210, Thr268 and Asn520, with a maximum deviation of 1.37 Å for Thr268. This result indicates that the binding of 2OG does not accompany a large structural change. The R.M.S. deviations between the models of the SeMet-labeled Tpa1 and the binary (or ternary) complex of the native Tpa1 is 0.58 Å for 552 (or 553) Cα atom pairs.
Figure 2.
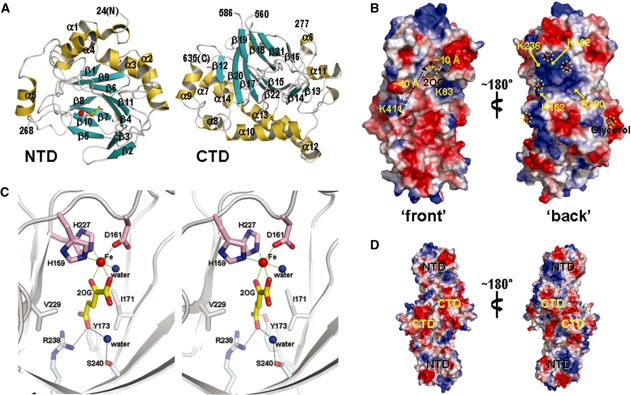
Overall structure of S. cerevisiae Tpa1 and electron density of the bound Fe(III) and 2OG. (A) 2Fo − Fc electron density map of Fe(III)–2OG and Fe(III)-coordinating residues in the ternary complex of S. cerevisiae Tpa1 (contoured at 1.5 σ). All the figures except Figure 1 were prepared using PyMol (DeLano, W. L., 2002, The PyMol Molecular Graphics System. DeLano Scientific, Palo Alto, CA, USA). (B) Topology diagram of Tpa1. α-Helices are shown as circles and β-strands as triangles. DSBH folds of NTD and CTD of S. cerevisiae Tpa1 are enclosed in gray boxes. (C) Stereo ribbon diagram of the S. cerevisiae Tpa1. α-Helices, β-strands and loops are colored in yellow, cyan and gray, respectively. Iron and 2OG bound near the center of Tpa1 NTD are shown in a sphere and sticks, respectively.
Two-domain structure of Tpa1
The structure of Tpa1 bound to both Fe(III) and 2OG is shown in Figure 2C. S. cerevisiae Tpa1 is elongated with approximate dimensions of 90 Å × 55 Å × 45 Å. It comprises two domains of unequal size: the smaller NTD (residues 24–268) and the larger CTD (residues 277–635) (Figure 2C). Unexpectedly, both NTD and CTD adopt the same DSBH fold (24) in their cores (Figure 2B), although no sequence similarity is detected between them. The R.M.S. difference between NTD and CTD is 2.8 Å for 180 Cα atom pairs.
NTD contains 8 helices (composed of five α-helices and three 310-helices) and 11 β-strands (Figure 3A). Five antiparallel β-strands β1↑-β9↓-β6↑-β11↓-β4↑ form the larger sheet of the β-sandwich, while four antiparallel β-strands β8↓-β7↑-β10↓-β5↑ form the smaller sheet. The remaining two antiparallel β-strands β2↑-β3↓ close the opening of the two sheets, resembling a lid. A cluster of four helices (α1, α2, α3 and α4) cover the larger β-sheet of the β-sandwich. Eight β-strands—β9, β6, β11, β4, β8, β7, β10 and β5—constitute the DSBH fold of Tpa1 NTD. The DSBH topology may be considered as a special case of β-sandwiches and is built of eight β-strands that form the two four-stranded antiparallel β-sheets of the β-sandwich (34,35).
Figure 3.
Surface diagram and the active site of S. cerevisiae Tpa1. (A) Ribbon diagram of the Tpa1 NTD (left panel) and CTD (right panel), colored as in Figure 2C and labeled as in Figure 2B. (B) Electrostatic potential at the surface of Tpa1 monomer. Blue and red colors correspond to positive and negative potentials, respectively. The bound 2OG, glycerol and sulfate ions are shown in stick models and are circled. The binding groove is indicated with two orthogonal dotted lines. (C) Stereo view of the active site around the bound iron and 2OG. Red and blue balls represent the iron ion and a water molecule, respectively. Green dotted lines denote the iron coordination. Black dotted lines denote hydrogen bonds. (D) Two different views of the electrostatic potential at the molecular surface of Tpa1 dimer, colored as in Figure 3B.
CTD contains 11 helices (composed of nine α-helices and two 310-helices) and 11 β-strands (Figure 3A). Five antiparallel β-strands β15↑-β22↓-β17↑-β20↓-β12↑ form the larger sheet of the β-sandwich, while four antiparallel β-strands β16↓-β21↑-β18↓-β19↑ form the smaller sheet. The remaining two antiparallel β-strands β13↑-β14↓ close the opening of the two sheets. Eight β-strands—β15, β22, β17, β20, β16, β21, β18 and β19—constitute the DSBH fold of Tpa1 CTD. A cluster of seven helices (α7, α8, α9, α10, α12, α13 and α14) cover the larger β-sheet of the β-sandwich and another helix α11 covers the smaller β-sheet. Two cysteine residues (Cys494 and Cys635) in CTD are close to each other. In the native Tpa1–Fe(III) binary complex, a disulfide bond is formed between Cys494 and Cys635, with a distance of 2.04 Å between the sulfur atoms. In the native Tpa1–Fe(III)–2OG ternary complex, the disulfide bond appears to be only partially formed, with a distance of either 2.03 or 3.75 Å between the sulfur atoms for the two conformations. In the SeMet-labeled Tpa1–Fe(III) binary complex, no disulfide bond exists between Cys494 and Cys635, with a distance of 4.65 Å between the sulfur atoms. This may be due to the presence of a reducing agent (TCEP) in the crystallization condition.
NTD and CTD of S. cerevisiae Tpa1 are arranged in a face-to-face fashion, with their β-sandwiches being nearly orthogonal to each other (Figure 2C). The smaller sheets of the NTD and CTD β-sandwiches face toward the domain interface. The solvent accessible surface area buried at the interface between the two domains in the ternary complex is ∼1170 Å2 (∼4.7% of the monomer surface area), with 42.6% of the atoms in this interface being polar (Protein–Protein Interaction Server at http://www.biochem.ucl.ac.uk/bsm/PP/server/). The surface representation of S. cerevisiae Tpa1 reveals a deep cleft (∼40 Å long and ∼20 Å wide), which runs roughly from the 2OG-binding site of NTD to the corresponding, unoccupied site of CTD (Figure 3B). The deep cleft is contributed by β2 and the following loop (Lys83, Asp86, Ile87 and Tyr88), β3 (Val90 and Gln92), β5 and the following loop (Leu156, His159, Ile163 and Arg166), the loop between α6 and α7 (Glu283 and Asp287), the loop between β13 and β14 (His410 and Lys411) and the loop between β16 and β17 (Leu513 and Thr515). This cleft is open at both ends. Each of the positively charged residues Lys83 and Lys411 is located near the end of this cleft (Figure 3B). Lys411 is conserved in both S. pombe Ofd1 and human Tpa1, while Lys83 is conserved in S. pombe Ofd1 only (Figure 1). This cleft may provide a binding site for an unknown main substrate of Tpa1. In both the ternary and binary complexes, four sulfate ions are bound to the positively charged surface on the ‘back’ side of the S. cerevisiae Tpa1 NTD (Figure 3B). Lys152, Lys182, Arg190 and Lys236 on this surface (Figure 3B) are conserved in S. pombe Ofd1 (Figure 1).
Oligomeric state of Tpa1 in solution
Although a monomer of S. cerevisiae Tpa1 exists in each asymmetric unit of the crystal, it forms a dimeric unit, with approximate dimensions of 140 Å × 70 Å × 55 Å, by association of two monomers related by a crystallographic 2-fold symmetry through their CTDs (Figure 3D). The solvent accessible surface area buried at the interface between the two monomers in this dimeric unit is ∼2030 Å2 (∼8.1% of the monomer surface area), with 31.5% of the atoms in this interface being polar (Protein–Protein Interaction Server at http://www.biochem.ucl.ac.uk/bsm/PP/server/). This finding raises the possibility that S. cerevisiae Tpa1 may exist as dimers in solution.
To assess the oligomeric state of Tpa1 in solution, we carried out sedimentation equilibrium ultracentrifugation analyses. Typical data obtained at 2.02 μM of Tpa1Δ20 at 280 nm and at two speeds of 10 000 and 12 000 r.p.m. are shown in Supplementary Figure S1. Supplementary Figure S1A also shows fits for different models of the Tpa1Δ20 oligomer; monomer (1x), dimer (2x) and monomer–dimer equilibrium (1x–2x). Ultracentrifugal data at two speeds (10 000 and 12 000 r.p.m.) were jointly fitted to the given models and analyzed. The R.M.S. errors for the 1x and 2x models are 1.38×10–2 and 1.23×10–2, respectively, which indicate the poor quality of the fits. In contrast to the homogeneous models, the R.M.S. error for the reversible 1x–2x model gave a much improved value of 5.88×10–3 indicating the superior quality of the fit with a dissociation constant (Kd) of 3.5×10–6 M. The reversibility of the monomer–dimer equilibrium was confirmed by multiple speed experiments (8000, 10 000 and 12 000 r.p.m.). These data clearly indicate that Tpa1 exists as a reversible monomer–dimer mixture in solution. Other data on Tpa1Δ20 lead to the same conclusion.
For Tpa1 CTD (residues 273–644), typical data obtained at two concentrations of 1.97 and 3.93 μM of Tpa1 CTD at 280 nm and at the speed of 30 000 r.p.m. are presented in Supplementary Figure S1B. The R.M.S. errors for the 1x and 2x models are 3.63×10–2 and 2.99×10–2, respectively, which indicate the poor quality of the fits. In contrast to the homogeneous models, the R.M.S. error for the reversible 1x–2x model gave a much improved value of 1.05×10–2 indicating the superior quality of the fit with a dissociation constant (Kd) of 1.3×10–5 M. Based on these results, we suggest that the dimerization of Tpa1, which is observed to be mediated through CTD in the crystal likely represents the mode of Tpa1 dimerization in solution.
Structurally similar proteins
To identify structurally similar proteins we carried out a search using the DALI server (36). The three best matches belong to the Fe(II)-/2OG-dependent dioxygenase family. They are (i) the human PHD2, the hypoxia-inducible factor (HIF) prolyl 4-hydroxylase (37) (PDB code 2G19; an R.M.S. deviation of 3.0 Å for 194 equivalent Cα positions in residues 32–266 of Tpa1, a Z-score of 18.0 and a sequence identity of 18%), (ii) the Chlamydomonas reinhardtii prolyl 4-hydroxylase (P4H) (38) (PDB code 2JIG; an R.M.S. deviation of 2.4 Å for 162 equivalent Cα positions in residues 28–249 of Tpa1, a Z-score of 14.2 and a sequence identity of 15%) and (iii) the human ABH3 protein, an oxidative DNA/RNA repair enzyme (demethylase) (39) (PDB code 2IUW; an R.M.S. deviation of 3.0 Å for 161 equivalent Cα positions in residues 37–248 of Tpa1, a Z-score of 12.8 and a sequence identity of 10%).
We further elaborated the structural similarity search with individual domains of S. cerevisiae Tpa1. Using the NTD (residues 24–269) alone, the result is similar to that obtained using the whole structure of Tpa1. The highest structural similarity is obtained with the human PHD2 (an R.M.S. deviation of 2.9 Å for 193 equivalent Cα positions in residues 32–268 of Tpa1, a Z-score of 18.0 and a sequence identity of 17%). The second highest similarity is found with the C. reinhardtii P4H (an R.M.S. deviation of 2.4 Å for 162 equivalent Cα positions in residues 28–268 of Tpa1, a Z-score of 14.0 and a sequence identity of 15%). Using CTD (residues 277–635) alone, the highest Z-score is obtained with the human PHD2 (an R.M.S. deviation of 3.1 Å for 183 equivalent Cα positions in residues 344–635 of Tpa1, a Z-score of 13.1 and a sequence identity of 6%) and the next highest similarity is found with the human ABH3 protein (an R.M.S. deviation of 3.3 Å for 157 equivalent Cα positions in residues 349–635 of Tpa1, a Z-score of 10.0 and a sequence identity of 11%).
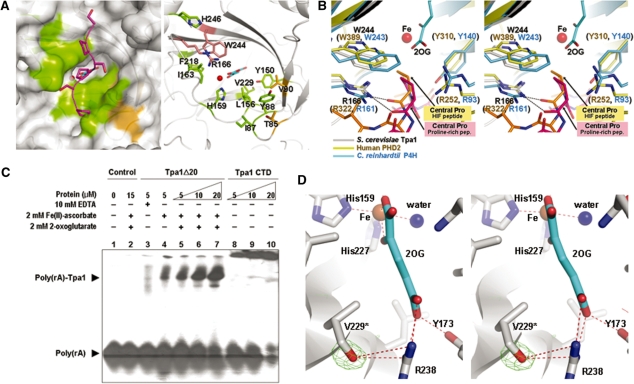
The observed structural similarity of Tpa1 with human and algal P4Hs implies functional relatedness. Indeed, the Tpa1 NTD has a deep cleft that resembles the active sites of the human PHD2 and C. reinhardtii P4H (37,38; Figure 4A, the left panel). The putative active site cleft of S. cerevisiae Tpa1 NTD displays a high structural similarity with these P4Hs. It is lined with Ile87, Tyr88, Tyr150, Leu156, His159, Ile163, Arg166, Phe218, Val229, Trp244 and His246 (Figure 4A), many of which are hydrophobic. These residues are well conserved in other Tpa1 family members (marked using gray diamonds in Figure 1). Unlike the NTD of S. cerevisiae Tpa1, the human PHD2 and C. reinhardtii P4H, the substrate-binding pocket of the human ABH3 is considerably more polar (39). These findings suggest that S. cerevisiae Tpa1 could possibly function as a hydroxylase targeting a proline residue. When the Tpa1 NTD structure is superimposed with the (Ser-Pro)5 peptide-bound structure of the C. reinhardtii P4H (PDB code: 3GZE) (40), the (Ser-Pro)5 peptide is well fitted into the active site cleft of Tpa1 (Figure 4A, left panel). In the human PHD2 complexed with the C-terminal oxygen-dependent degradation domain of HIF-1α (41), Arg252, Tyr310, Arg322 and Trp389 surround the central proline (a hydroxylated target) of the peptide substrate (Figure 4B). In the peptide complex of C. reinhardtii P4H, Arg93, Tyr140, Arg161 and Trp243 surround the central proline of the peptide substrate (Figure 4B). Among them, Arg322/Trp389 of the human PHD2 and Arg161/Trp243 of C. reinhardtii P4H are strictly conserved as Arg166/Trp244 in S. cerevisiae Tpa1 (Figures 1 and 4B). Arg252/Tyr310 of the human PHD2 and Arg93/Tyr140 of C. reinhardtii P4H are not conserved in S. cerevisiae Tpa1 and are replaced as Tyr88/Leu156.
Figure 4.
Structural comparison of Tpa1 and other P4Hs and RNA binding assay. (A) Surface representation (left panel) and ribbon diagram (right panel) of the conserved residues around the active site of Tpa1. The left and right panels are the same view. The left panel shows the (Ser–Pro)5 peptide in a purple stick model after structural superimposition of the algal P4H–peptide complex (PDB code: 3GZE) and S. cerevisiae Tpa1. On the surface representation of S. cerevisiae Tpa1 (left panel), residues conserved among yeast Tpa1, Ofd1 and human Tpa1 are colored in green (Ile87, Tyr88, Tyr150, Leu156, His159, Ile163, Arg166, Phe218, Val229, Trp244, His246); residues conserved between yeast Tpa1 and Ofd1 as brown (Thr85, Val90). In the right panel, potential proline-recognizing residues of S. cerevisiae Tpa1 (Arg166, Trp244) are shown in stick models and colored in salmon. (B) Stereo view of the superposition of S. cerevisiae Tpa1 (gray), human PHD2 (PDB code: 3HQR) (yellow) and algal P4H (PDB code: 3GZE) (blue) around the active site. The HIF-1α peptide from human PHD2 complex and the (Ser–Pro)5 peptide from the algal P4H complex are shown in orange and pink stick models, respectively. (C) Electrophoretic mobility shift assay of S. cerevisiae Tpa1. Poly(rA) is a radiolabeled 20-mer RNA. Poly(rA)-Tpa1 indicates the shifted band. The protein concentration is for bovine serum albumin in lane 2, for Tpa1Δ20 in lanes 3−7 and for Tpa1 CTD in lanes 5−10. (D) Stereo view of the extra electron density around Val229. The averaged kick omit Fo – Fc map was calculated using PHENIX (54) after omitting the hydroxyl group that modifies the side chain of Val229 (contoured at 3.0 σ).
Conserved sequence motifs of Tpa1 and ligand binding
When the sequences of the Tpa1 family members are aligned, we can recognize six sequence segments that are highly conserved (motifs I–VI in Supplementary Figure S2). Our structures show that motifs II, III and IV of the NTD of S. cerevisiae Tpa1 are involved in the binding of Fe(III) and 2OG. Motif II [L(L/M)xHDDx(I/L)xxRxIx(F/Y)ILYL] encompasses Leu156–Leu174 of S. cerevisiae Tpa1 (boxed in red in Supplementary Figure S2). Motif III [FFxVxPx1∼2SFHxVxEV] covers Phe217–Val232 (boxed in green in Supplementary Figure S2), while motif IV [R(L/M)(S/A)xIxGW(Y/F)xxP] covers Arg238–Pro248 (boxed in pink in Supplementary Figure S2). ‘h’ is a hydrophobic residue, ‘x’ stands for any amino acid and the strictly conserved residues are in boldface. Despite the common DSBH fold for both NTD and CTD of S. cerevisiae Tpa1, Fe(III) and 2OG are bound to NTD only (Figure 2C). The NTD of S. cerevisiae Tpa1 possesses the highly conserved H1x(D/E)…H2 iron-binding motif (His159–Xxx–Asp161…His227) contributed by sequence motifs II and III (Supplementary Figure S2), and the 2OG-binding motif (Arg238–Xxx–Ser240) as part of motif IV (Figures 1 and 3C). ‘Xxx’ stands for any amino acid. His159 and His227 of S. cerevisiae Tpa1 are located at the C-terminal end of the strand β5 and the N-terminal end of β10, respectively (Figure 1). The two ligand-binding motifs are also present in other Fe(II)-/2OG-dependent dioxygenases (20). Their absence in the CTD of Tpa1 suggests a role of CTD other than the dioxygenase-like function.
In the binary structure of the native Tpa1–Fe(III) complex, the active site metal ion is coordinated by the side chains of His159, Asp161 and His227 as well as two tightly bound and one loosely bound water molecules. In the ternary structure of the Tpa1–Fe(III)–2OG complex, the C-1 carboxylate and C-2 keto group of 2OG replace two water molecules that coordinate the Fe(III) ion (Figure 3C). The third water molecule, trans to His159, remains attached to the Fe(III) ion. The C-5 carboxylate of 2OG makes a salt bridge with Arg238 of motif IV (Figure 3C). In the current EXPASY database, Lys236 is suggested to be a potential binding residue of 2OG. This assignment needs to be revised in view of our structural information. The closest distance between the side chain NZ atom of Lys236 to 2OG is 18.7 Å. Hydrogen bonds are present between the hydroxyl group of Tyr173 (on β6) and the C-5 carboxylate of 2OG, as well as between the hydroxyl group of Ser240 in motif IV and the C-5 carboxylate of 2OG via a water molecule. Furthermore, the aliphatic portion of 2OG is sandwiched between the side chains of Ile171 (on β6) and Val229 (on β10).
On the basis of the 2OG binding mode, the iron-binding H1x(D/E)…H2 motif of the Fe(II)-/2OG-dependent dioxygenase family members has been grouped into two categories (20). The first category (‘in line’ binding mode) is represented by clavaminic acid synthase (CAS) (42), taurine dioxygenase (TauD) (43), factor inhibiting HIF (FIH) (44,45) and alkylsulfatase (AtsK) (46). In these enzymes, the side chain of His1 and the C-1 carboxylate of 2OG are approximately co-planar, with the C-1 carboxylate lying on the opposite side of His1 and the C-2 keto group lying on the opposite side of the acidic residue. In the second category (‘off line’ binding mode), represented by carbapenem synthase (CarC) (47), anthocyanidin synthase (ANS) (48) and AlkB (49), the C-1 carboxylate of 2OG is positioned on the opposite side of His2, and the C-2 keto group is located on the opposite side of the acidic residue. The active site of Tpa1 bears resemblance to the ‘off line’ binding mode, with the C-1 carboxylate of 2OG being positioned on the opposite side of His227 (His2) (Figure 3C).
Other conserved sequence motifs I, V and VI (Supplementary Figure S2) are not directly involved in the binding of iron and 2OG. Motif I [FxxKxxD(I/L)Y(R/K)hxQ(S/T)xDL] encompasses Phe80–Leu96 in the NTD of S. cerevisiae Tpa1 (boxed in blue in Figure 1 and Supplementary Figure S2). It covers the strands β2 and β3, and a loop C-terminal to β3 (Supplementary Figure S2). It is located near the active site in the NTD of S. cerevisiae Tpa1. This motif contributes to one side of the active site cleft (Supplementary Figure S3). The side chains of Ile87 and Tyr88 of this motif line the deep hydrophobic cleft in the active site. The hydroxyl group of Tyr88 points toward the bulk solvent, making hydrogen bonds with the conserved Lys83 and Gln92. Asp86 and Arg89 interact with the backbone of CTD residues Leu513 and Glu550, respectively. Except for Lys83 (on β2), well-conserved residues from motif I reside on the loop regions (α2–β2, β2–β3, β3–α3). Motif I is neither related to the core of the DSBH fold nor involved in the binding of iron and 2OG. In other P4Hs, the loop region corresponding to the sequence motif I of Tpa1 displays a large conformational change upon binding of the peptide substrate (40,41). If this analogy can be extended, motif I of Tpa1 may play a role in recognizing the protein substrate.
Motif V [R(R/H)(F/W)(R/K)xGx1∼2(F/Y)TL] covers Arg503–Leu513 in the CTD of S. cerevisiae Tpa1 (boxed in brown in Figure 1 and Supplementary Figure S2), which resides on the strands β15 and β16, and the connecting loop between them. It is part of the DSBH fold of CTD. The tripeptide sequence Phe511–Thr512–Leu513 is located near β5 and the β2–β3 loop of NTD, providing an additional hydrophobic surface patch into the substrate-binding pocket of NTD (Supplementary Figure S3). Motif VI [LVhRDxxxLxFVK] covers Leu601–Lys613 in the CTD of S. cerevisiae Tpa1 (boxed in purple in Figure 1 and Supplementary Figure S2). It is also part of the DSBH fold of CTD and the conserved sequence Phe611–Val612–Lys613 is located adjacent to the Phe511–Thr512–Leu513 sequence of Motif V and the Asp86–Ile87–Tyr88–Arg89 sequence of Motif I. Lys613 of motif VI makes hydrogen bonds with the carbonyl groups of Thr85, Asp86 and Tyr88 of motif I in the NTD as well as the side chain of His155 of NTD.
Val229 is hydroxylated in the ternary complex
Interestingly, the side chain of Val229 in the ternary complex structure of Tpa1 showed an extra electron density, which was interpreted as a hydroxylated valine (Figure 4D). In contrast, no such extra electron density was present in the binary complex structures of either native or the SeMet-labeled Tpa1. In view of the weak electron density, we assumed half occupancy of the hydroxyl group, resulting in a B-factor of 19.5 Å2 for the oxygen atom. Val229 is C-terminal to His2 (His227) of the iron-binding H1x(D/E)…H2 motif. We confirmed by DNA sequencing that the codon corresponding to Val229 was not mutated. In the ternary complex structure, the oxygen atom of the hydroxylated Val229 is located 9.5 Å away from the iron center (Figure 4D). The modification of Tpa1 Val229 may have resulted from an attack by a hydroxyl radical formed by the uncoupled reaction. The uncoupled reaction is often stimulated by inhibitors or poor substrates, may also occur in the absence of a primary substrate and can result in aberrant self-hydroxylation reactions involving the protein side chains (20). Such irreversible protein modifications have been observed in other 2OG-dependent hydroxylases such as TfdA, AlkB, TauD and ABH3 (39,50−52). The hydroxylated residues in TfdA, AlkB, TauD and ABH3 are Trp113, Trp178, Trp128/Trp240/Trp248 and Leu177, respectively, and the distance between the oxygen atom of the hydroxyl group and the iron center is 10.5, 4.7, 6.0/7.5/10.5 and 6.8 Å, respectively. Similarly, hydroxylated Val229 of Tpa1 is not directly bound to the iron ion. Instead, its hydroxyl group forms a hydrogen bond with Arg238 that stabilizes the C-5 carboxylate of 2OG. Observation of self-hydroxylation in S. cerevisiae Tpa1 suggests that it could catalyze post-translational modification of other components in the mRNP complex.
Tpa1 binds poly(rA)
The report that Tpa1 plays a role as a component of the mRNP complex (13) raises the possibility that Tpa1 interacts directly with mRNA. To determine whether Tpa1 binds to mRNA, we performed gel-mobility shift assays using a radiolabeled 20-base poly(rA) and Tpa1Δ20. We found that the RNA band was shifted in the presence of Tpa1Δ20 complexed with Fe(II) and 2OG in a protein dose-dependent manner (Figure 4C, lanes 5−7). As a control, bovine serum albumin did not bind the same RNA in the presence of Fe(II) and 2OG (Figure 4C, lane 2). This result clearly indicates that the shift of the RNA band is due to the binding of poly(rA) by Tpa1. When residual metal ions bound to Tpa1Δ20 were removed by excess EDTA, the binding ability of Tpa1Δ20 was much reduced (Figure 4C, lane 3). The maximum poly(rA) binding by Tpa1Δ20 was observed when 2OG was added in addition to Fe(II) (Figure 4C, lanes 4 and 5). To assess the role of NTD and CTD of Tpa1 in RNA binding, we attempted to express each of them but we could express only CTD (residues 273−644). Tpa1 CTD also retarded the electrophoretic mobility of poly(rA) in a dose-dependent manner (Figure 4C, lanes 8−10). However, CTD showed a tendency to form high molecular weight aggregates upon binding poly(rA). This may have been caused by the lower solubility of the recombinant CTD compared to Tpa1Δ20. The structure of S. cerevisiae Tpa1 suggests that the binding of poly(rA) might occur on the positively charged surface patch on the ‘back’ side of Tpa1 opposite from the catalytic site (Figure 3B). This surface is lined with many positively charged residues (Lys152, Arg179, Lys180, Lys182, Arg190, Lys207, Lys233, Lys236, Lys354, Lys380, Lys384, Arg413 and Lys620). Four sulfate ions are bound to this surface in both the binary and ternary structures of the native S. cerevisiae Tpa1, possibly mimicking the phosphate backbone of poly(rA) (Figure 3B).
Biological implications
Our present study provides the structural details of S. cerevisiae Tpa1 and it supports the previous assignment of Tpa1 as a member of the Fe(II)-/2OG-dependent dioxygenase family on the basis of the primary sequence. The structural information on S. cerevisiae Tpa1 has significant biological implications. This is because the Tpa1 family members were shown to be required for the control of translation termination, mRNA poly(A) tail length and mRNA stability as an essential component of the mRNP complex in S. cerevisiae and for oxygen sensing in the transcriptional regulation of gene expressions in S. pombe. Members of the Tpa1 family are also present in higher eukaryotes including humans and they are likely to play important biological roles. Our crystal structure of S. cerevisiae Tpa1 is the first structure of the Tpa1 family members, representing an important step toward understanding the biological functions of this protein family. It shows that two domains (NTD and CTD) of Tpa1 have the common DSBH fold (Figure 2B). However, the binding of Fe(III) and 2OG via the iron-binding motif—H1X(D/E)…H2—and the Arg–Xxx–Ser sequence motif is observed only in the NTD of S. cerevisiae Tpa1 (Figure 3C). It suggests that the NTD of Tpa1 family members could function as a P4H-like enzyme or as a sensor of molecular oxygen.
Ofd1 of S. pombe, a member of the Tpa1 family, is an uncharacterized P4H-like Fe(II)/2OG-dependent dioxygenase. It was reported that its C-terminal degradation domain accelerates the degradation of Sre1N in the presence of oxygen (19). Under low oxygen conditions, the sterol regulatory element binding protein (Sre1), an endoplasmic reticulum membrane-bound transcription factor, is proteolytically cleaved and the released N-terminal transcription factor (Sre1N) activates gene expression essential for hypoxic growth (19). It was shown that the Ofd1 N-terminal dioxygenase domain is required for oxygen sensing and the Ofd1 CTD accelerates Sre1N degradation (19). These data support a model whereby the Ofd1 N-terminal dioxygenase domain is an oxygen sensor that regulates the activity of the C-terminal degradation domain (19). Our structure of S. cerevisiae Tpa1, together with the conserved sequence features, indicates that the NTD of S. pombe Ofd1 has the potential to function as an oxygen sensor. Unlike the fission yeast, however, S. cerevisiae does not contain an Sre1 pathway (19). The role of S. cerevisiae Tpa1 as part of the mRNP complex likely depends on the Fe(II)-/2OG-dependent dioxygenase-type hydroxylase domain in its N-terminal half. It may control mRNA stability indirectly via post-translational modification of other components of the mRNP complex. We also demonstrated through an RNA binding assay (Figure 4C) that the S. cerevisiae Tpa1 can bind to poly(rA), suggesting that it could interact directly with mRNA in the mRNP complex. In S. pombe, negative regulator of Ofd1 (Nro1) was found to bind to the Ofd1 CTD and to inhibit Sre1N degradation under low oxygen (22). The counterpart of Nro1 in S. cerevisiae has been detected by global affinity purification studies (14,22). The role of the CTD of S. cerevisiae Tpa1 has yet to be established; this study, together with the lack of the ligand-binding motifs, supports a non-enzymatic role.
ACCESSION NUMBER
The coordinates and structure factors for yeast Tpa1 have been deposited in the Protein Data Bank under accession numbers 3KT1, 3KT7 and 3KT4 for the binary and ternary complexes of the native Tpa1 and the binary complex of SeMet-substituted Tpa1, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Basic Science Outstanding Scholars Program, World-Class University Program (grant no. 305-20080089) and Basic Research Program Grant (R01-2006-000-10311-0) of the National Research Foundation of Korea, Korea Ministry of Education, Science and Technology. Funding for open access charge: Basic Science Outstanding Scholars Program and World-Class University Program (grant no. 305-20080089) of the National Research Foundation of Korea, Korea Ministry of Education, Science and Technology.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank beamline staffs for assistance during X-ray data collection at Photon Factory (beamlines BL-5A, BL-17A and NW12A) and Pohang Light Source (beamlines BL-4A and BL-6C). We also thank Dr V. Narry Kim for providing laboratory space and equipment for electrophoretic mobility shift assay.
REFERENCES
- 1.Hieronymus H, Silver PA. Systems view of mRNP biology. Genes Develop. 2004;18:2845–2860. doi: 10.1101/gad.1256904. [DOI] [PubMed] [Google Scholar]
- 2.Singh R, Valcárcel J. Building specificity with nonspecific RNA binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- 3.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 4.Neugebauer KM. On the importance of being co-transcriptional. J. Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 5.Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 6.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein P, Peltz SW, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translation initiation. Mol. Cell. Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 11.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 12.Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol. Cell. Biol. 2007;27:6243–6253. doi: 10.1128/MCB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM. Tpa1 is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover. Mol. Cell. Biol. 2006;26:5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 15.Prévôt D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell. 2003;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 16.Derry MC, Yanagiya A, Martineau Y, Sonenberg N. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb. Symp. Quant. Biol. 2006;71:537–543. doi: 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 17.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:research0007.1–research0007.8. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27:1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hausinger RP. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 21.Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 22.Lee CY, Stewart EV, Hughes BT, Espenshade PJ. Oxygen-dependent binding of Nro1 to the prolyl hydroxylase Ofd1 regulates SREBP degradation in yeast. EMBO J. 2009;28:135–143. doi: 10.1038/emboj.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roach PL, Clifton IJ, Fülöp V, Harlos K, Barton GJ, Hajdu J, Andersson I, Schofield CJ, Baldwin JE. Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature. 1995;375:700–704. doi: 10.1038/375700a0. [DOI] [PubMed] [Google Scholar]
- 24.Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins. J. Inorg. Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 25.Hegg EL, Que L., Jr The 2-His-1-carboxylate facial triad–an emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur. J. Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 26.Koehntop KD, Emerson JP, Que L., Jr The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J. Biol. Inorg. Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Brünger AT. The free R-value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 34.Zhang C, Kim SH. A comprehensive analysis of the Greek key motifs in protein β-barrels and β-sandwiches. Proteins. 2000;40:409–419. doi: 10.1002/1097-0134(20000815)40:3<409::aid-prot60>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 37.McDonough MA, Li V, Flashman E, Chowdhury R, Mohr C, Liénard BM, Zondlo J, Oldham NJ, Clifton IJ, Lewis J, et al. Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2) Proc. Natl Acad. Sci. USA. 2006;103:9814–9819. doi: 10.1073/pnas.0601283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koski MK, Hieta R, Böllner C, Kivirikko KI, Myllyharju J, Wierenga RK. The active site of algal prolyl 4-hydroxylase has a large structural plasticity. J. Biol. Chem. 2007;282:37112–37123. doi: 10.1074/jbc.M706554200. [DOI] [PubMed] [Google Scholar]
- 39.Sundheim O, Vågbø CB, Bjørås M, Sousa MM, Talstad V, Aas PA, Drabløs F, Krokan HE, Tainer JA, Slupphaug G. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koski MK, Hieta R, Hirsilä M, Rönkä A, Myllyharju J, Wierenga RK. The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J. Biol. Chem. 2009;284:25290–25301. doi: 10.1074/jbc.M109.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chowdhury R, McDonough MA, Mecinović J, Loenarz C, Flashman E, Hewitson KS, Domene C, Schofield CJ. Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure. 2009;17:981–989. doi: 10.1016/j.str.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Ren J, Stammers DK, Baldwin JE, Harlos K, Schofield CJ. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat. Struct. Biol. 2000;7:127–133. doi: 10.1038/72398. [DOI] [PubMed] [Google Scholar]
- 43.Elkins JM, Ryle MJ, Clifton IJ, Dunning Hotopp JC, Lloyd JS, Burzlaff NI, Baldwin JE, Hausinger RP, Roach PL. X-ray crystal structure of Escherichia coli taurine/α-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry. 2002;41:5185–5192. doi: 10.1021/bi016014e. [DOI] [PubMed] [Google Scholar]
- 44.Dann CE, III, Bruick RK, Deisenhofer J. Structure of a factor-inhibiting hypoxia-inducible factor 1: an essential asparaginyl hydroxylase involved in the hypoxic response pathway. Proc. Natl Acad. Sci. USA. 2002;99:15351–15356. doi: 10.1073/pnas.202614999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elkins JM, Hewitson KS, McNeill LA, Seibel JF, Schlemminger I, Pugh CW, Ratcliffe PJ, Schofield CJ. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J. Biol. Chem. 2003;278:1802–1806. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- 46.Müller I, Kahnert A, Pape T, Sheldrick GM, Meyer-Klaucke W, Dierks T, Kertesz M, Usón I. Crystal structure of the alkylsulfatase AtsK: insights into the catalytic mechanism of the Fe(II), α-ketoglutarate dependent dioxygenase superfamily. Biochemistry. 2004;43:3075–3088. doi: 10.1021/bi035752v. [DOI] [PubMed] [Google Scholar]
- 47.Clifton IJ, Doan LX, Sleeman MC, Topf M, Suzuki H, Wilmouth RC, Schofield CJ. Crystal structure of carbapenem synthase (CarC) J. Biol. Chem. 2003;278:20843–20850. doi: 10.1074/jbc.M213054200. [DOI] [PubMed] [Google Scholar]
- 48.Wilmouth RC, Turnbull JJ, Welford RW, Clifton IJ, Prescott AG, Schofield CJ. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure. 2002;10:93–103. doi: 10.1016/s0969-2126(01)00695-5. [DOI] [PubMed] [Google Scholar]
- 49.Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 50.Liu A, Ho RY, Que L., Jr, Ryle MJ, Phinney BS, Hausinger RP. Alternative reactivity of an α-ketoglutarate-dependent iron(II) oxygenase: enzyme self-hydroxylation. J. Am. Chem. Soc. 2001;123:5126–5127. doi: 10.1021/ja005879x. [DOI] [PubMed] [Google Scholar]
- 51.Ryle MJ, Liu A, Muthukumaran RB, Ho RY, Koehntop KD, McCracken J, Que L., Jr, Hausinger RP. O2- and α-ketoglutarate-dependent tyrosyl radical formation in TauD, an α-keto acid-dependent non-heme iron dioxygenase. Biochemistry. 2003;42:1854–1862. doi: 10.1021/bi026832m. [DOI] [PubMed] [Google Scholar]
- 52.Henshaw TF, Feig M, Hausinger RP. Aberrant activity of the DNA repair enzyme AlkB. J. Inorg. Biochem. 2004;98:856–861. doi: 10.1016/j.jinorgbio.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 53.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams PD, Grosse-Kunstleve RW, Hung L.-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.