Abstract
DNA methylation and histone modifications play a central role in the epigenetic regulation of gene expression and cell differentiation. Recently, Np95 (also known as UHRF1 or ICBP90) has been found to interact with Dnmt1 and to bind hemimethylated DNA, indicating together with genetic studies a central role in the maintenance of DNA methylation. Using in vitro binding assays we observed a weak preference of Np95 and its SRA (SET- and Ring-associated) domain for hemimethylated CpG sites. However, the binding kinetics of Np95 in living cells was not affected by the complete loss of genomic methylation. Investigating further links with heterochromatin, we could show that Np95 preferentially binds histone H3 N-terminal tails with trimethylated (H3K9me3) but not acetylated lysine 9 via a tandem Tudor domain. This domain contains three highly conserved aromatic amino acids that form an aromatic cage similar to the one binding H3K9me3 in the chromodomain of HP1ß. Mutations targeting the aromatic cage of the Np95 tandem Tudor domain (Y188A and Y191A) abolished specific H3 histone tail binding. These multiple interactions of the multi-domain protein Np95 with hemimethylated DNA and repressive histone marks as well as with DNA and histone methyltransferases integrate the two major epigenetic silencing pathways.
INTRODUCTION
DNA methylation and histone modifications are crucially involved in the regulation of gene expression, inheritance of chromatin states, genome stability and differentiation (1–3). Although the biochemical networks controlling these epigenetic marks have been the subject of intensive investigation, their interconnection is still not well resolved in mammals. DNA methylation patterns are established by de novo DNA methyltransferases Dnmt3a and 3b, while Dnmt1 is largely responsible for maintaining genomic methylation after DNA replication (4,5). Dnmt1 possesses an intrinsic preference for hemimethylated DNA substrates (6,7) and associates with proliferating cell nuclear antigen (PCNA) at replication sites in vivo (8–10). The transient interaction of Dnmt1 with PCNA enhances methylation efficiency but is not strictly required to maintain genomic methylation in human and mouse cells (11,12).
Recently, Np95 has emerged as a central regulatory factor for DNA methylation and interacts with all three Dnmts (13). Np95 localizes at replication foci and its genetic ablation leads to genomic hypomethylation and developmental arrest (14–19). Np95 and its SET- and Ring- associated (SRA) domain were shown to bind hemimethylated DNA with higher affinity than corresponding symmetrically methylated or unmethylated sequences both in vitro and in vivo (17,18,20–22). In addition, crystal structures of the SRA domain complexed with hemimethylated oligonucleotides revealed flipping of the 5-methylcytosine out of the DNA double helix, a configuration that would stabilize the SRA–DNA interaction (20–22). Thus, recruitment of Dnmt1 to hemimethylated CpG sites by Np95 has been proposed as mechanism for the maintenance of genomic methylation.
In addition to its role in controlling DNA methylation, Np95 has been shown to take part in several other chromatin transactions. Np95 or its human homolog ICBP90/UHRF1 were reported to interact with the histone deacetylase HDAC1 and the histone methyltransferase G9a and to mediate silencing of a viral promoter, suggesting a role of Np95 in gene silencing through histone modification (13,23,24). Np95 binds histone H3 and displays a Ring domain-mediated E3 ubiquitin ligase activity for core histones in vitro and possibly histone H3 in vivo (25,26). The plant homeodomain (PHD) of Np95 has been linked to decondensation of replicating pericentric heterochromatin (PH), but it is still unclear which domains recognize specific histone modifications (16,26,27).
In this study we systematically analyzed the binding properties of Np95 and its individual domains to DNA and histone tails in vitro and their binding kinetics in living cells. Our data reveal a multi-functional modular structure of Np95 interconnecting DNA methylation and histone modification pathways.
MATERIALS AND METHODS
Expression constructs
Expression construct for GFP–Dnmt1 and RFP–PCNA were described previously (10,28,29). All Np95 constructs were derived by PCR from corresponding myc- and His6-tagged Np95 constructs (25). To obtain GFP– and Cherry-fusion constructs the Dnmt1 cDNA in the pCAG–GFP–Dnmt1-IRESblast construct (11) or the pCAG–Cherry–Dnmt1–IRESblast was replaced by Np95 encoding PCR fragments. The GFP–Np95ΔTudor expression construct was derived from the GFP–Np95 construct by overlap extension PCR (30). The GFP–Tudor mutant (Y188A, Y191A) was derived from the GFP–Np95 construct by PCR-based mutagenesis (31). All constructs were verified by DNA sequencing. Throughout this study enhanced GFP (eGFP) or monomeric Cherry (mCherry) constructs were used and for simplicity referred to as GFP– or Cherry-fusions.
Cell culture, transfection and immunofluorescence staining
HEK293T cells and embryonic stem cells (ESCs) were cultured and transfected as described (11), with the exception that FuGENE HD (Roche) was used for transfection of ESCs. The dnmt1−/− J1 ESCs used in this study are homozygous for the c allele (4). For immunofluorescence staining, TKO ESCs were grown on cover slips, fixed with 3.7% formaldehyde in PBS for 10 min and permeabilized with 0.5% Triton X-100 for 5 min. After blocking with 3% BSA in PBS for 1 h endogenous Np95 was detected with a polyclonal rabbit anti-Np95 serum (32). The secondary antibody was conjugated to Alexa Fluor 568 (Molecular Probes). Nuclear counterstaining was performed with DAPI and cells were mounted in Vectashield (Vector Laboratories). Images were obtained using a TCS SP5 AOBS confocal laser scanning microscope (Leica) using a 63x/1.4 NA Plan-Apochromat oil immersion objective. Fluorophores were excited with 405 and 561 nm lasers.
In vitro DNA binding assay
The in vitro DNA binding assay was performed as described previously (33) with the following modifications. Two different double-stranded DNA probes were labeled with distinct fluorophores and used in direct competition (see Supplementary Figures S3 and S6 for details). DNA oligos were controlled for CG methylation state by digestion with either a CG methylation-sensitive (HpaII) or -insensitive (MspI) enzyme (Supplementary Figure S4). For extract preparation 2 mM MgCl2 and 1 mg/ml DNaseI were included in the lysis buffer. Extracts from 1-3 transfected 10 cm plates were diluted to 500-1000 µl with immunoprecipitation (IP) buffer and 1 µg of GFP–Trap (34) (ChromoTek, Germany) per final assay condition was added. After washing and equilibration beads were resuspended in 500 µl of binding buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 100 ng/µl BSA). Two oligonucleotide substrates were added to a final concentration of 50 nM each and incubated at room temperature (RT) for 60 min with constant mixing. Fluorescence intensity measurements were performed with a Tecan Infinite M1000 plate reader using the following excitation/emission wavelengths: 490±10 nm/511±10 nm for GFP, 550 ± 15 nm/580±15 nm for ATTO550 and 650±10 nm/670±10 nm for ATTO647N. Values were adjusted using standard curves obtained with ATTO-dye coupled oligonucleotide primers and purified GFP. Binding activity was expressed as the ratio between the fluorescent signals of bound DNA probe and GFP fusion protein bound to the beads, so that the signals from bound probes are normalized to the amount of GFP fusion. Furthermore, values were normalized using a control set of DNA probes having identical sequences but distinct fluorescent labels (see Supplementary Figures S3 and S6 for details).
Peptide pull-down assay
Peptides were purchased as TAMRA conjugates (PSL, Germany) and are listed in Supplementary Figure S7. The peptide pull-down assay was performed analogously to the DNA binding assay described above. After one-step purification of GFP fusion proteins with the GFP–Trap (ChromoTek, Germany), the beads were equilibrated in 1 ml IP buffer and resuspended in 500 µl binding buffer supplemented with 100 ng/µl of BSA. Peptides were added to a final concentration of 0.74 µM and the binding reaction was performed at RT for 15 min to 60 min with constant mixing. The beads were washed twice with 1 ml of IP buffer and resuspended in 100 µl of the same. Wavelengths for excitation and measurement of TAMRA were 490±5 nm and 511±5 nm, respectively. Fluorescence intensity measurements were adjusted using standard curves from TAMRA coupled peptide and purified GFP.
Live cell microscopy and fluorescence recovery after photobleaching analysis
Live cell imaging and fluorescence recovery after photobleaching (FRAP) analysis were performed as described previously (11). For presentation, we used linear contrast enhancement on entire images.
Statistical analysis
Results were expressed as mean±SEM. The difference between two mean values was analyzed by Student's; t-test and was considered to be statistically significant in case of P < 0.05 and highly significant with P < 0.001.
Electrophoretic mobility shift and supershift assays
Un- and hemimethylated DNA substrates (1 pmol UMB550 and HMB647N, respectively) were incubated with 0.6 pmol purified GFP–Np95 and 0.4 pmol GFP–antibody (mouse monoclonal antibody, Roche). Samples were subjected to a 3.5% non-denaturing PAGE and analyzed with a fluorescence scanner (Typhoon Trio scanner; GE Healthcare) to detect ATTO550 (unmethylated substrate), ATTO647N (hemimethylated substrate) and green fluorescence (GFP–Np95).
RESULTS AND DISCUSSION
Np95 binding kinetics is largely independent of DNA methylation levels in vivo
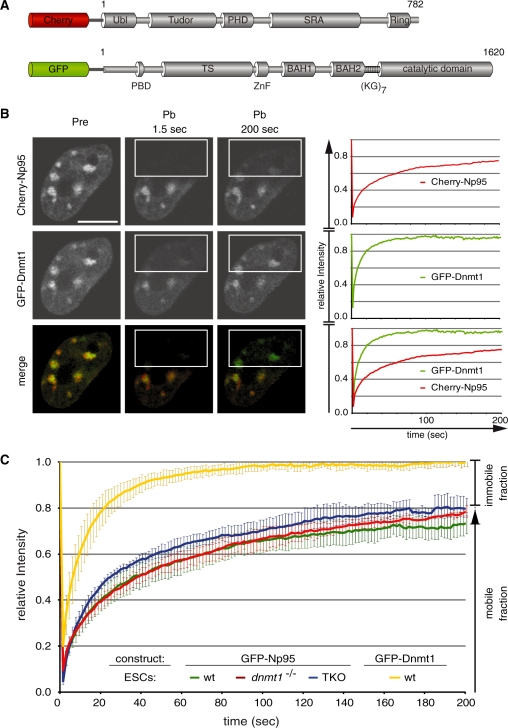
Recent studies showed Np95 bound to hemimethylated DNA, suggesting that the essential function of Np95 in the maintenance of DNA methylation consists of substrate recognition and recruitment of Dnmt1. To investigate the dynamics of these interactions in vivo we transiently transfected wild-type (wt) J1 ESCs with expression constructs for Cherry-Np95 and GFP–Dnmt1 and monitored their subcellular distribution using live-cell microscopy (Figure 1A and B). Np95 showed a nuclear distribution with a cell cycle-dependent enrichment at replicating PH, similar to Dnmt1. Consistent with earlier observations (8,12,14–16) we detected co-localization of Np95 and Dnmt1 at sites of DNA replication. We investigated the dynamics of Np95 binding by quantitative fluorescence recovery after photobleaching (FRAP) analysis (Figure 1B). As chromocenters (aggregates of PH) are not homogeneously distributed in the nucleus, we chose to bleach half nuclei to ensure that the bleached region contains a representative number of potential binding sites. We observed a relatively fast and full recovery of relative GFP–Dnmt1 fluorescence intensity (Figure 1B), reflecting a transient and dynamic interaction as described before (11). In contrast, Cherry-Np95 showed a considerably slower and only partial (∼80%) recovery within the same observation period. These results indicated a relatively stable binding of Np95 to chromatin and revealed an immobile protein fraction of about 20%. These in vivo binding properties would be consistent with tight binding of Np95 to hemimethylated CpG sites and flipping of the methylated cytosines out of the DNA double helix as shown in recent co-crystal structures of the SRA domain of Np95 (20–22).
Figure 1.
Binding kinetics of Dnmt1 and Np95 in living cells. (A) Schematic representation of Np95 and Dnmt1 fluorescent fusions. Ubl, ubiquitin-like domain; Tudor, tandem Tudor domain; PHD, plant homeodomain; SRA, SET- and Ring-associated domain; Ring domain; PBD, PCNA-binding domain; TS, targeting sequence; ZnF, zinc-finger; BAH, bromo adjacent homology domain; (KG)7, lysine-glycine repeat. (B) Dnmt1 and Np95 display different kinetics. Representative images from FRAP experiments on wt J1 ESCs transiently co-transfected with Cherry-Np95 and GFP–Dnmt1 constructs. Images show co-localization at chromocenters before (Pre) and at the indicated time points after (Pb) bleaching half of the nucleus. Bleached areas are outlined. Corresponding FRAP curves are shown on the right. Bars, 5 µm. (C) FRAP kinetics of GFP–Np95 in J1 ESCs with different genetic backgrounds [wt, dnmt1–/– and dnmt1–/–, 3a–/–, 3b–/– (TKO)]. Kinetics of GFP–Dnmt1 is shown for comparison. Mobile and immobile fractions are indicated on the right. Values represent mean ± SEM.
To directly test the contribution of DNA methylation and the interaction with Dnmt1 to protein mobility, we compared the binding kinetics of GFP–Np95 in wt ESCs and ESCs lacking either Dnmt1 or all three major DNA methyltransferases Dnmt1, 3a and 3b (triple knockout, TKO). Surprisingly, Np95 binding to chromatin was not affected by either drastic reduction (dnmt1−/−) or even complete loss (TKO) of genomic methylation and showed in both cases remarkably similar FRAP kinetics compared to wt J1 ESCs (Figure 1C). Similar results were obtained with a C-terminal GFP fusion (Np95-GFP; Supplementary Figure S1), arguing against conformational or sterical impairments of the N-terminal fusion protein that could affect the binding kinetics. Also, both, the levels of endogenous Np95 and its degree of accumulation at chromocenters were highly variable in TKO cells, with chromocenter accumulation clearly visible in some cells (Supplementary Figure S2). These results show that DNA methylation and the three DNA methyltransferases do not have a major effect on the overall binding kinetics of Np95 in living cells.
The SRA domain of Np95 is necessary and sufficient for DNA binding in vitro
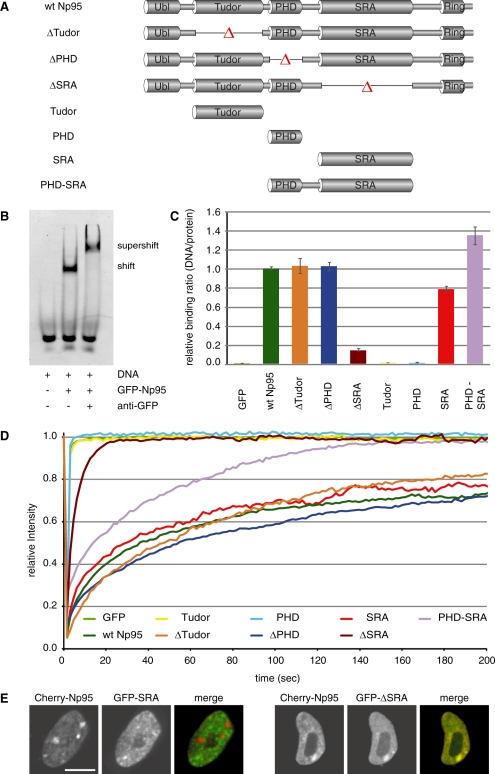
Next, we investigated the DNA binding activity of Np95 and the contribution of distinct Np95 domains by generating a systematic set of individual domains and deletion constructs fused to GFP (Figure 2A). To directly compare the in vitro binding affinity of Np95 regarding different methylation states, we synthesized double-stranded DNA-binding substrates with either one or three un- or hemimethylated CpG sites and labeled them with two distinct fluorophores (Supplementary Figure S3). DNA probes were controlled for CG methylation state by digestion with either a CG methylation-sensitive (HpaII) or -insensitive (MspI) enzyme (Supplementary Figure S4). Performing conventional electrophoretic mobility shift and supershift assays we confirmed the DNA binding activity of Np95 and detected a preference for hemimethylated DNA substrates (Figure 2B and Supplementary Figure S5).
Figure 2.
In vitro DNA binding and in vivo mobility of Np95 domains. (A) Schematic representation of the analyzed GFP–Np95 fusion constructs. (B) Electrophoretic mobility shift and supershift assay. GFP–Np95 binding to hemimethylated DNA substrates is shown by the shifted GFP–Np95:DNA complex. The addition of a GFP–antibody supershifted the GFP–Np95:DNA complex (supershift assay with unmethylated DNA substrates in direct competition with hemimethylated DNA substrates is shown in Supplementary Figure S6). (C) In vitro DNA-binding properties of Np95 constructs. Binding assays were performed using fluorescently labeled double stranded oligonucleotide probes containing one central hemimethylated CpG site. Shown are fluorescence intensity ratios of bound probe/bound GFP fusion. Values represent means and SD of three to six independent experiments. GFP was used as control. Further control experiments with either one or three central CpG sites and alternating fluorescent labels are shown in Supplementary Figure S3. (D) Kinetics of Np95 constructs in living np95–/– ESCs determined by half nucleus FRAP analysis. GFP is shown as reference. Curves represent mean values from 6 to 15 nuclei. SEM (0.001–0.005) is not shown for clarity of illustration. (E) Confocal mid-sections of living np95–/– ESCs transiently expressing the indicated Np95 fusion constructs (left and mid-panels). Merged images are displayed on the right. Bar, 5 µm.
As a second line of evidence and to quantify binding preferences, we applied our recently developed non-radioactive DNA binding assay (33) and tested GFP–fused wt Np95 as well as a systematic set of individual domains and deletion constructs for their DNA-binding properties in vitro (Figure 2C and Supplementary Figure S6). This assay allows fast comparison of different potential binding substrates in direct competition as well as the simultaneous quantification of GFP–labeled protein to calculate relative binding activity. The different GFP–Np95 fusion constructs were expressed in HEK293T cells, purified with the GFP–Trap (34) and incubated with the fluorescently labeled DNA substrates. GFP–fusion protein and bound DNA substrates were quantified with a fluorescence plate reader (Figure 2C and Supplementary Figure S6). Furthermore, results were corrected for any bias due to incorporation of different fluorescent labels (Supplementary Figures. S3 and S6). Under these assay conditions we observed an up to 2-fold preference (factor 1.6–1.9) of Np95 for DNA substrates containing one or three hemimethylated CpG sites (Supplementary Figure S6). Deletion of the SRA domain completely abolished the DNA-binding activity of Np95, whereas deletion of either the PHD or the Tudor domain had no effect (Figure 2C). Consistently, the isolated PHD and Tudor domains did not bind to DNA, while the SRA domain alone showed similar binding strength and sequence preference as full-length Np95. Together, these results clearly demonstrate that the SRA domain of Np95 preferentially binds to hemimethylated CpG sites, although this preference is only about 2-fold with purified proteins and substrates in vitro.
The SRA domain dominates binding kinetics but not localization of Np95
Next, we investigated the role of distinct Np95 domains in nuclear interactions in vivo. To this aim, we expressed the same GFP–Np95 constructs in np95−/− ESCs and tested their binding kinetics with FRAP experiments (Figure 2D). Importantly, GFP–Np95 showed similar FRAP kinetics in Np95 deficient, wt or Dnmt-deficient ESCs (Figures 1C and 2D). Among all domains tested, only the SRA domain showed similar kinetics as full-length Np95, including the relatively slow recovery and an immobile fraction of about 20%, while the Tudor and PHD domain displayed the same high mobility as GFP. Also, FRAP curves of the corresponding deletion constructs indicated that the Tudor and the PHD domains have only a minor contribution to in vivo binding kinetics, while deletion of the SRA domain drastically increased the mobility of Np95. These data indicate that the SRA domain dominates the binding kinetics of Np95 in vivo. Curiously, the addition of the PHD to the SRA domain (GFP–PHD-SRA) resulted in intermediate kinetics and loss of the immobile fraction. This effect was, however, not observed in the context of the full-length protein, suggesting that nuclear interactions of Np95 are controlled by a complex interplay among its domains. To directly study the role of the SRA domain in controlling the subcellular localization of Np95 we co-transfected np95−/− ESCs with expression constructs for Cherry-Np95 and either GFP–SRA or GFP–Np95ΔSRA (Figure 2E). This direct comparison showed that the isolated SRA domain does not co-localize with full-length Np95 at PH. Together, these results indicate that the SRA domain of Np95 is necessary and sufficient for DNA binding in vitro and also dominates the binding kinetics in vivo, but is per se not sufficient for proper subnuclear localization. The fact that the Np95ΔSRA construct co-localized with Np95 suggests that other domains than the SRA control the subcellular targeting of Np95.
Np95 binds to histone H3 via a tandem Tudor domain
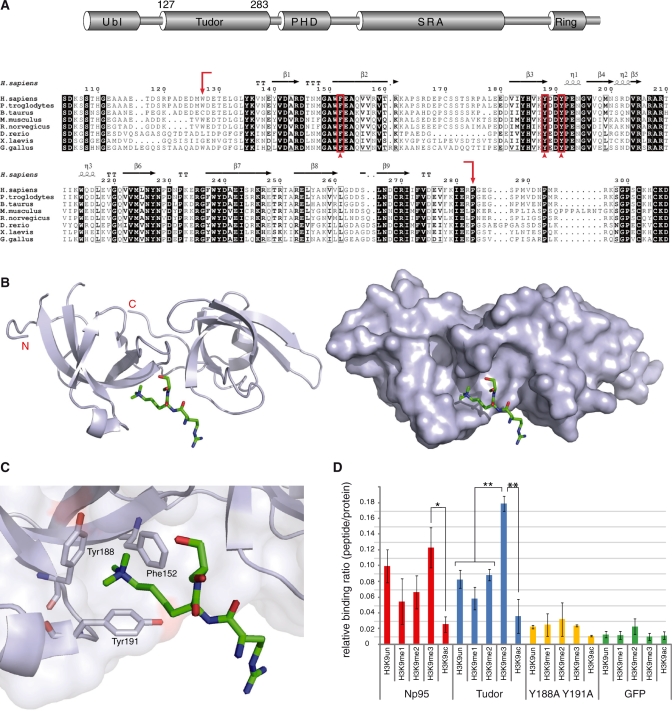
Database searches showed that the sequence between the Ubl and PHD domains of Np95 is highly conserved in vertebrates and displays structural similarity to the family of Tudor domains [(35); PDB 3db4; Figure 3A and B]. The crystal structure revealed that the Tudor domain is composed of two subdomains (tandem Tudor) forming a hydrophobic pocket that accommodates a histone H3 N-terminal tail trimethylated at K9 (H3K9me3) (PDB 3db3; Figure 3C). This hydrophobic-binding pocket is created by three highly conserved amino acids (Phe152, Tyr188, Tyr191) forming an aromatic cage (Figure 3A and C). Interestingly, a very similar hydrophobic cage structure has been described for the chromodomain of the heterochromatin protein 1ß (HP1ß) (Supplementary Figure S7) that is known to bind trimethylated lysine 9 of histone H3 and associates with PH (36).
Figure 3.
Structure and H3 N-terminal tail binding of the tandem Tudor domain. (A) Schematic drawing of the multi-domain architecture of Np95 (top) and alignment of tandem Tudor domains from vertebrate Np95 homologs (bottom). Arrows show the end and start positions of the crystallized tandem Tudor domain shown in (B). Residues forming the aromatic cage shown in (C) are indicated by arrowheads. Absolutely conserved residues of the tandem Tudor domain are black shaded, while positions showing conservative substitutions are boxed with residues in bold face. Secondary-structure elements were generated with EsPript (37) using the crystal structure of human UHRF1 (PDB 3db3 and 3db4) and are shown above the amino acid sequence: α-helices (η), β-strands, strict alpha turns (TT) and strict beta turns (TTT). Accession numbers: Homo sapiens Q96T88.1; Pan troglodytes XP_001139916.1; Bos Taurus AAI51672.1; Mus musculus Q8VDF2.2; Rattus norvergicus Q7TPK1.2; Dario rerio NP_998242.1; Xenopus laevis ABY28114.1, Gallus gallus XP_418269.2. (B) Side view of the tandem Tudor domain as a cartoon model (left) and as surface representation (right) in complex with a histone H3 N-terminal tail peptide trimethylated at lysine 9 (green stick model; only Arg8-Lys9-Ser10 of the H3 peptide are resolved). The image was generated with PyMOL (38). (C) An aromatic cage is formed by Phe152, Tyr188 and Tyr191 and accommodates the trimethylated lysine 9 of H3 (H3K9me3). (D) Histone H3 N-terminal tail binding specificity of GFP–Np95, GFP–Tudor and GFP–Tudor (Y188A Y191A) in vitro. Shown are fluorescence intensity ratios of bound probe/bound GFP fusion. GFP was used as negative control. Shown are means ± SEM from four to ten independent experiments and two-sample t-tests were performed that do or do not assume equal variances, respectively. Statistical significance compared to the binding ratio of H3K9me3 is indicated: *P < 0.05, **P < 0.001.
To further investigate the histone tail-binding properties of Np95, we mutated two amino acids of the aromatic cage (Y188A, Y191A) and tested the isolated tandem Tudor domain and corresponding mutant in comparison with Np95 using a peptide binding assay. GFP–Np95, GFP–Tudor and GFP–Tudor (Y188A, Y191A) were expressed in HEK293T cells, purified with the GFP–Trap and incubated with TAMRA-labeled histone tail peptides. The fluorescence intensity of GFP fusion proteins and bound peptides was quantified and the relative binding activity calculated (Figure 3D and Supplementary Figure S7). The tandem Tudor domain showed a highly significant preference for the trimethylated (H3K9me3) peptide, while this effect was less pronounced in the full-length Np95. Interestingly, acetylation of K9 (H3K9ac), a modification largely underrepresented in silent chromatin, prevented binding of the tandem Tudor domain. Remarkably, point mutations targeting aromatic cage residues within the tandem Tudor domain completely abolished specific binding to N-terminal histone H3 peptides.
Consistent with these binding data the tandem Tudor domain also showed a weak enrichment at PH, while the PHD domain, previously proposed as potential histone H3-binding motif (26), did neither bind to H3K9 peptides in vitro nor to PH in vivo (Supplementary Figure S8). These results indicate that the tandem Tudor domain of Np95 features a peptide binding pocket with structural and functional striking similarity to HP1ß and confers selective binding to histone modification states associated with silent chromatin.
These multiple interactions of Np95 with heterochromatin components correlate well with functional data. The depletion of Np95 in mouse cells resulted in increased transcription of major satellite repeats (16). Also, an interaction of Np95 with G9a was described and both were found to be essential for transcriptional regulation (24) and epigenetic silencing of transgenes (13).
In summary, we showed that the SRA domain is necessary and sufficient for DNA binding of Np95 in vitro. Photobleaching experiments further indicated that the SRA domain also dominates the binding kinetics of Np95 in living cells which was however largely independent of the DNA methylation level. These results suggest that the SRA domain may also bind to unmethylated DNA or undergo additional, still unidentified interactions in vivo. While the essential role of Np95 in the maintenance of DNA methylation is well established, it is still unclear how a relatively weak preference for hemimethylated DNA can be sufficient to maintain DNA methylation patterns over many cell division cycles for an entire life time. We suggest that the multiple interactions of the multi-domain protein Np95 with hemimethylated DNA and H3K9 methylated histone tails as well as with histone (G9a) and DNA (Dnmt1, 3a and 3b) methyltransferases may add up to the necessary specificity in vivo. Clearly, these multiple interactions place Np95 at the center of various epigenetic silencing mechanisms and likely mediate epigenetic crosstalk.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Nanosystems Initiative Munich (NIM), the BioImaging Network Munich (BIN) and by grants from the Deutsche Forschungsgemeinschaft (DFG) to H.L. IMB was supported by the Italian Association for Cancer Research (AIRC), the Fondazione CARIPLO Progetto NOBEL. C.F. and G.P. were supported by the International Doctorate Program NanoBioTechnology (IDK-NBT) and the International Max Planck Research School for Molecular and Cellular Life Sciences (IMPRS-LS). Funding for open access charges: DFG.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Masahiro Muto and Haruhiko Koseki (RIKEN Research Center for Allergy and Immunology, Yokohama, Japan) for providing wild-type and np95−/− E14 ESCs, to En Li (Novartis Institutes for Biomedical Research, Boston, MA) for dnmt1−/− and J1 ESCs and to Masaki Okano (RIKEN Center for Developmental Biology, Kobe, Japan) for the TKO ESCs.
REFERENCES
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 4.Lei H, Oh S, Okano M, Juttermann R, Goss K, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 5.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 6.Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl Acad. Sci. USA. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pradhan S, Talbot D, Sha M, Benner J, Hornstra L, Li E, Jaenisch R, Roberts R. Baculovirus-mediated expression and characterization of the full-length murine DNA methyltransferase. Nucleic Acids Res. 1997;25:4666–4673. doi: 10.1093/nar/25.22.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 9.Chuang L.S.-H, Ian H.-I, Koh T.-W, Ng H.-H, Xu G, Li BFL. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 10.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schermelleh L, Haemmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cristina Cardoso M, Leonhardt H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–43012. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Langst G, Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. EMBO Rep. 2009;10:1259–1264. doi: 10.1038/embor.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uemura T, Kubo E, Kanari Y, Ikemura T, Tatsumi K, Muto M. Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct. Funct. 2000;25:149–159. doi: 10.1247/csf.25.149. [DOI] [PubMed] [Google Scholar]
- 15.Miura M, Watanabe H, Sasaki T, Tatsumi K, Muto M. Dynamic changes in subnuclear NP95 location during the cell cycle and its spatial relationship with DNA replication foci. Exp. Cell Res. 2001;263:202–208. doi: 10.1006/excr.2000.5115. [DOI] [PubMed] [Google Scholar]
- 16.Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol. Biol. Cell. 2007;18:1098–1106. doi: 10.1091/mbc.E06-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bostick M, Kim JK, Esteve P.-O, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining dna methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 18.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 19.Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. doi: 10.1038/sj.onc.1210855. [DOI] [PubMed] [Google Scholar]
- 20.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 21.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 22.Qian C, Li S, Jakoncic J, Zeng L, Walsh MJ, Zhou MM. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J. Biol. Chem. 2008;283:34490–34494. doi: 10.1074/jbc.C800169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 24.Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papait R, Pistore C, Grazini U, Babbio F, Cogliati S, Pecoraro D, Brino L, Morand AL, Dechampesme AM, Spada F, et al. The PHD domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol. Biol. Cell. 2008;19:3554–3563. doi: 10.1091/mbc.E07-10-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schermelleh L, Spada F, Easwaran HP, Zolghadr K, Margot JB, Cardoso MC, Leonhardt H. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Methods. 2005;2:751–756. doi: 10.1038/nmeth794. [DOI] [PubMed] [Google Scholar]
- 29.Sporbert A, Domaing P, Leonhardt H, Cardoso MC. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 2005;33:3521–3528. doi: 10.1093/nar/gki665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 31.Ko JK, Ma J. A rapid and efficient PCR-based mutagenesis method applicable to cell physiology study. Am. J. Physiol. Cell Physiol. 2005;288:C1273–1278. doi: 10.1152/ajpcell.00517.2004. [DOI] [PubMed] [Google Scholar]
- 32.Bonapace IM, Latella L, Papait R, Nicassio F, Sacco A, Muto M, Crescenzi M, Di Fiore PP. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J. Cell Biol. 2002;157:909–914. doi: 10.1083/jcb.200201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frauer C, Leonhardt H. A versatile non-radioactive assay for DNA methyltransferase activity and DNA binding. Nucleic Acids Res. 2009;37:e22. doi: 10.1093/nar/gkn1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Adams-Cioaba MA, Min J. Structure and function of histone methylation binding proteins. Biochem. Cell Biol. 2009;87:93–105. doi: 10.1139/O08-129. [DOI] [PubMed] [Google Scholar]
- 36.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 37.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. DeLano Scientific LLC. CA, USA: Palo Alto; 2008. The PyMOL Molecular Graphics System. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.