Abstract
The host restriction factor Apobec3G is a cytidine deaminase that incorporates into HIV-1 virions and interferes with viral replication. The HIV-1 accessory protein Vif subverts Apobec3G by targeting it for proteasomal degradation. We propose a model in which Apobec3G N-terminal domains symmetrically interact via a head-to-head interface containing residues 122 RLYYFW 127. To validate this model and to characterize the Apobec3G–Apobec3G and the Apobec3G–Vif interactions, the mammalian protein–protein interaction trap two-hybrid technique was used. Mutations in the head-to-head interface abrogate the Apobec3G–Apobec3G interaction. All mutations that inhibit Apobec3G–Apobec3G binding also inhibit the Apobec3G–Vif interaction, indicating that the head-to head interface plays an important role in the interaction with Vif. Only the D128K, P129A and T32Q mutations specifically affect the Apobec3G–Vif association. In our model, D128, P129 and T32 cluster at the edge of the head-to-head interface, possibly forming a Vif binding site composed of two Apobec3G molecules. We propose that Vif either binds at the Apobec3G head-to-head interface or associates with an RNA-stabilized Apobec3G oligomer.
INTRODUCTION
Efficient replication of lentiviruses, such as HIV-1, requires the viral infectivity factor Vif. HIV-1 viruses deficient in Vif are unable to form infective particles in non-permissive cells due to the host restriction factor Apolipoprotein B messenger RNA (mRNA)-editing catalytic polypeptide-like 3G (Apobec3G) (1). Apobec3G and the closely related Apobec3F are incorporated into the virions of Vif-deficient HIV-1 and restrict the virus (1,2). Apobec3G contains two zinc-binding deaminase domains: the N-terminal domain binds to RNA, while the C-terminal domain is responsible for the DNA-binding and DNA-editing cytidine deaminase activity (3,4). Upon reverse transcription of the HIV-1 RNA, Apobec3G deaminates deoxycytidine to deoxyuridine in the nascent DNA minus strand (5). The resulting plus strand contains multiple dG to dA mutations, leading to premature stop codons and other aberrant viral transcripts (6,7). However, Apobec3G and Apobec3F do not strictly require cytidine deaminase activity for their antiviral effect (8,9). Apobec3G binding to the viral RNA partially inhibits the reverse transcription process, probably by inhibiting complementary DNA (cDNA) elongation (10). Apobec3G is packaged into viral particles via interactions with the nucleocapsid domain of the gag polyprotein and this interaction probably requires host or viral RNA (11–13). HIV Vif prevents the incorporation of Apobec3G and Apobec3F into the virions, thus restoring virus infectivity (14–16). Vif contains a C-terminal SOCS-box domain, and is able to recruit elongins B and C, cullin 5 and rbx1, leading to formation of an E3 ubiquitin ligase complex (17,18). Vif binding to Apobec3G results in ubiquitination and proteasomal degradation of Apobec3G. Therefore, inhibitors of the Apobec3G–Vif interaction may restore the antiviral effect of Apobec3G and could offer a novel mechanism for therapeutic intervention for HIV-1 (19,20).
Apobec3G and Apobec3F belong to the Apobec3 family which consists of 7 members (20,21). Apobec3A, C and H contain a single deaminase domain, where Apobec3B, DE, F and G contain tandem duplications of a deaminase domain with a similar sequence. The structure of the C-terminal domain of Apobec3G was determined by X-ray crystallography and nuclear magnetic resonance (NMR). This domain folds into a five-stranded central beta sheet, surrounded by six alpha helices (22–24). The X-ray crystallography structure of Apobec2, which resembles the structures of the Apobec3 C-terminal domain, revealed the formation of tetramers. Apobec2 dimers occur via pairing of the β strand 2 and subsequent head-to-head interaction of two dimers results in the formation of Apobec2 tetramers (25).
The N-terminal domain of Apobec3G has no cytidine deaminase activity but binds RNA and the HIV gag nucleocapsid and mediates packaging of Apobec3G in the virion (3,4,26,27). An 80-amino-acid sequence comprising the putative zinc-coordinating motif in this N-terminal domain was shown to be important for binding of Vif (28). Residues Y124 and W127 in the N-terminal domain are important for this packaging process (29). Different models have been proposed for the N-terminal domain of Apobec3G (30,31) or for complete Apobec3G (24,27). In these models, residues 124 and 127 are surface-exposed in the monomeric N-terminal domain. In a recent model (27), the N-and C-terminal domain of an Apobec3G monomer are proposed to interact with each other via their β2 strand as in the Apobec2 dimer interface. Two studies propose that the N-terminal domains of Apobec3G further interact with each other as in the Apobec2 head-to-head interface (27,31). Residues 122–127 maintain important interactions in this head-to-head interface, and the interaction is stabilized by RNA which binds to a cluster of positively charged residues at one edge of the head-to-head interface (27). The structure of Vif remains unsolved and it is thus far unknown how Vif interacts with Apobec3G. In this article, we study the Vif and Apobec3G interactions using the mammalian protein–protein interaction trap (MAPPIT) mammalian two-hybrid system (32,33), site-directed mutagenesis and homology modeling. Our study provides new information on detailed interfaces that govern the Apobec3G–Apobec3G and Apobec3G–Vif interactions.
MATERIALS AND METHODS
Cell culture, transfection and reporter assays
Culture conditions, transfection procedures and luciferase assays in Hek293T cells were as previously described (34). For a typical luciferase experiment, Hek293T cells were seeded in six-well plates 24 h before overnight transfection with the desired constructs together with the luciferase reporter gene. Twenty-hour hours after transfection, cells were washed, trypsinized and seeded into 96-well plates. The seeded cells were left untreated or were stimulated with 0.5 IU/ml hEpo overnight. The luciferase activity of the transfected cells was measured by chemiluminescence.
Constructs
All constructs were obtained by standard polymerase chain reaction (PCR)- or restriction-based cloning procedures. The pXP2d2-rPAPI-luciferase reporter, originating from the rPAPI (rat pancreatitis associated protein I) promoter was described earlier (32). Generation of the standard chimeric bait receptor pCEL, containing the extracellular part of the EpoR and the intracellular part of the leptin receptor and standard chimeric prey receptor pMG2, containing part of the gp130 chain (amino acids 905–918) in duplicate, were previously reported (32,35,36). The pMet7Flagtag vector, the pMet7Etag vector and the pMet7EtagCIS construct were previously described (34,37).
Human peripheral blood leukocyte (PBL) cDNA was purchased from PrimGen. The pNL4-3 Plasmid which carries the full HIV-1 genome was a gift from Prof. C. Verhofstede. The pCMV4-HA-A3G-L123A and pCMV4-HA-A3G-Y124A mutants were kindly provided by Prof. M. Malim and Dr H. Huthoff. The VifSLQ144-146AAA prey (represented as VifSLQ prey) used in Figure 3C was created as stated in the table in the Supplementary Data. A human codon optimized VifSLQ prey was purchased from GENEART (represented in Supplementary Data) and was used in Figures 3D, 6 and 9 and most of the data represented in Table 1. All other constructs used in this paper are created as represented in the table in the Supplementary Data.
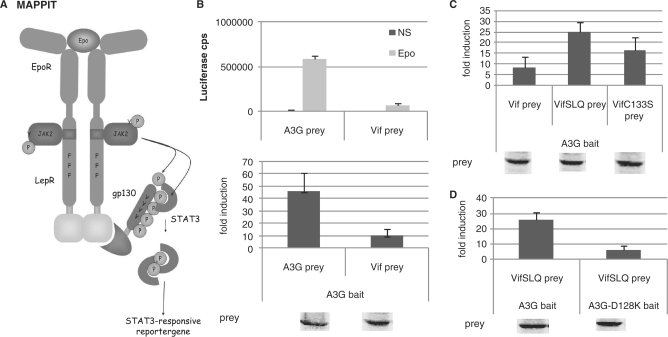
Figure 3.
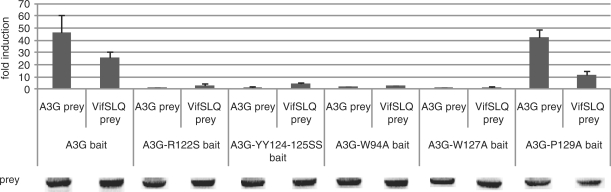
(A) The MAPPIT technique is based on signal transduction of the leptin receptor. The extracellular and transmembrane part of the Epo receptor is fused to the intracellular part of the leptin receptor. The membrane-distal tyrosine at position Y1138 in the cytoplasmic tail of the receptor is replaced by phenylalanine to prevent STAT3 recruitment. The two membrane-proximal tyrosines Y985 and Y1077 are mutated to overcome negative regulation and maximize the intensity of the signal. The bait protein is attached to this signaling-deficient leptin receptor. The prey protein is fused to several STAT3 recruitment sites of the gp130 chain. Interaction of bait and prey proteins results in STAT3 recruitment and activation. A STAT3-responsive luciferase reporter gene allows detection of the interaction. (B–D) Hek293T cells were transiently co-transfected with plasmids encoding the chimeric Apobec3G bait construct (A3G bait) and the prey constructs encoding for proteins Vif, VifSLQ144-146AAA (VifSLQprey), VifC133S, A3G or A3G-D128K fused to the gp130 chain, combined with the pXP2d2-rPAP1-luci reporter. The transfected cells were either stimulated for 24 h with Epo or were left untreated (NS, not stimulated). Luciferase measurements were performed in triplicate. Data are presented as either absolute luciferase counts per second (cps) or fold induction (Epo stimulated/NS) of luciferase activity. Western blot control of the different prey constructs in the experiment is shown.
Table 1.
Overview of the effect of mutations in the Apobec3G MAPPIT bait on the interaction with the Apobec3G prey (left) or VifSLQ prey (right)
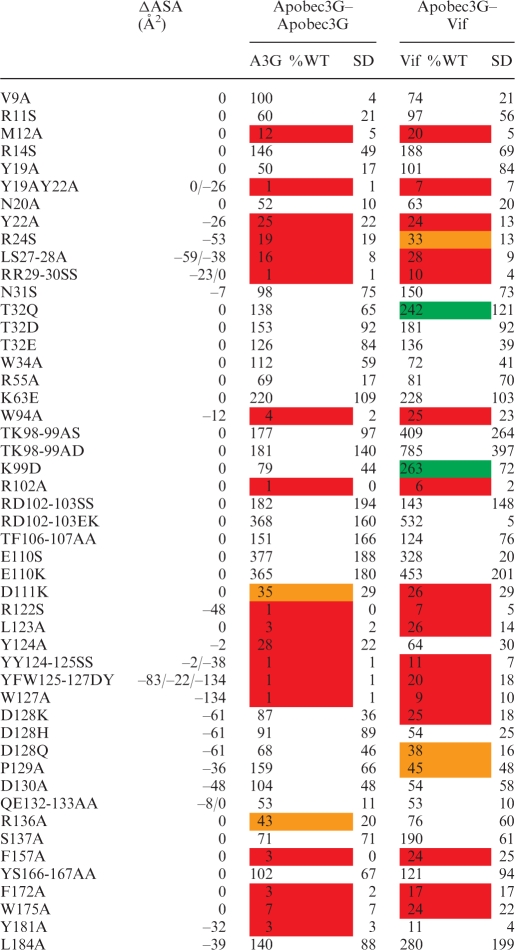 |
MAPPIT luciferase fold inductions for each mutant are expressed as percentage of Wild Type Apobec3G bait. The averages of these percent values of several independent MAPPIT experiments and standard deviation (SD) are shown. Strongly increased or decreased values that are significantly different from the WT in a paired t-test are indicated by a color code. Red and orange indicate a 70 and 50% reduction of the MAPPIT signal; green indicates a >200% increase of the MAPPIT signal. Hek293T cells were transiently co-transfected with plasmids encoding the chimeric Apobec3G WT or mutant Apobec3G bait constructs and Vif or Apobec3G prey constructs, combined with the pXP2d2-rPAP1-luci reporter. The transfected cells were either stimulated for 24 h with Epo or were left untreated (NS, not stimulated). Luciferase measurements were performed in triplicate. Data are expressed as a percentage of the WT fold inductions (stimulated/NS). ΔASA gives the decrease of the water accessibility (in square Angströms) of the mutated residue sidechains upon interaction of two monomers via the head-to-head interface. Residue sidechains that get buried in the interface have a negative ΔASA.
Western blot analysis and degradation assay
Protein expression levels in MAPPIT assays and degradation assays were confirmed by western blot analysis. Transfected Hek293T were lysed in RIPA buffer [200 mM NaCl, 50 mM Tris–HCl pH 8, 0.05% sodium dodecyl sulfate (SDS), 2 mM EDTA, 1% NP40, 0.5% DOC, Complete™ Protease Inhibitor Cocktail (Roche)]. Then 4X loading buffer (125 mM Tris–HCl pH 6.8, 6% SDS, 20% glycerol, 0.02% BFB, 10% β-mercapto-ethanol) was added to the lysates. Samples were heated and then loaded on a 7.5 or 10% polyacrylamide gel and blotted onto nitrocellulose membrane. Blotting efficiency was checked using PonceauS staining (Sigma). FLAG-tagged, E-tagged and V5-tagged proteins (data not shown) were revealed using monoclonal anti-Flag antibody M2 (Sigma), monoclonal anti-E-tag antibody (Amersham Biosciences) and monoclonal anti-V5-tag antibody (invitrogen). Secondary anti-mouse-DyLight800 or 680 antibodies (Thermo Scientific) were used and western blot analysis was performed using the Odyssey Infrared Imaging System (Li-Cor).
Alpha screen
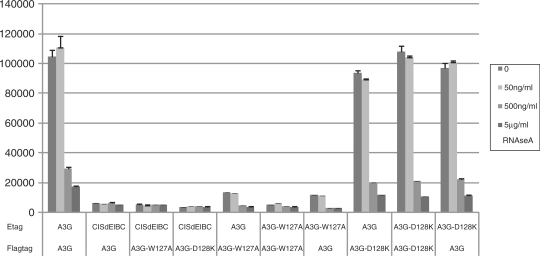
Alpha screen experiments were conducted according to the manufacturer’s protocol (PerkinElmer). Typically, cells were co-transfected overnight with the appropriate E- and Flag-tagged expression vectors, washed after 24 h and lysed 24 h later (lysis buffer: 50 mM Tris–HCl pH 7.5, 125 mM NaCl, 5% glycerol, 0.2% NP40, 1.5 mM MgCl2, 25 mM NaF, 1 mM Na3VO4, Complete™ Protease Inhibitor without EDTA Cocktail). Lysates were cleared by centrifugation. Streptavidin donorbeads and anti-Flag acceptorbeads were added together with biotinylated anti-Etag antibody and the appropriate amount of RNAse A (Roche) and incubated for at least 2 h under constant rotation. Samples were measured in triplicate using the EnVision platereader (PerkinElmer). The anti-Etag antibody (Amersham Biosciences) was biotinylated with sulfo-NHS-Biotin (PIERCE) according to the manufacturer’s guidelines.
Homology modeling
The Apobec3G N-and C-terminal domains were aligned to all human Apobec domains, using Multiple Alignment using Fast Fourier Transform (MAFFT) (38). The structures of Apobec2 and the C-terminal domain of Apobec3G were superposed and aligned to this MAFFT alignment in MOE (molecular operating environment, chemical computing group). Based on the superposition, the MAFFT alignment was edited. Using this edited alignment as input, homology models were built using MODELLER 9v7 (39) and alternative models were built in MOE. Both types of models were compared. A dimer model was built for the Apobec3G N-terminal domain using the structure of Apobec2 as template. For the dimer model, symmetry restraints were used for the backbone atoms of domains that interact via the head-to-head interface. The MODELLER models with the best DOPE score and the lowest energy MOE models were further subjected to conjugate gradients energy minimization using the OPLS/AA force field in MOE. The model protein geometry quality was assessed in MOE. Solvent accessibility of the models was determined using NACCESS (Hubbard,S.J. and Thornton, J.M. (1993), ‘NACCESS’, Computer Program, Department of Biochemistry and Molecular Biology, University College London.).
RESULTS
Homology model of the N-terminal domain of Apobec3G
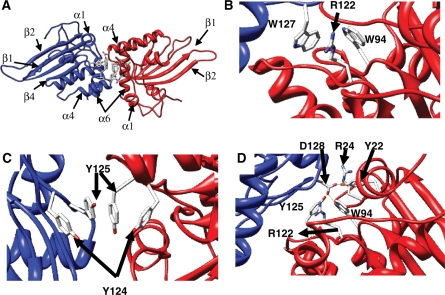
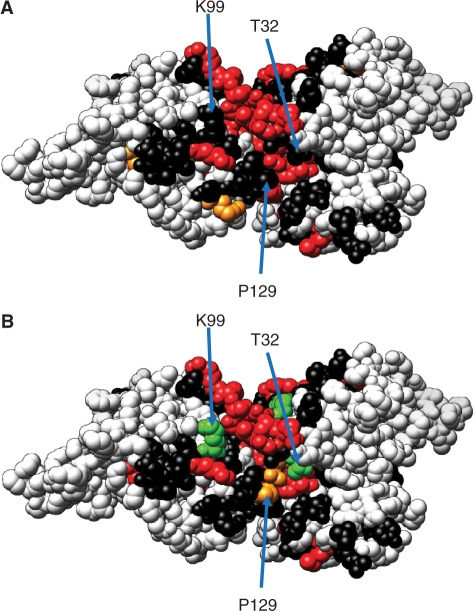
Homology models were built for the N-terminal domain of Apobec3G using the crystal structure of Apobec2 as template. In the crystal structure, Apobec2 forms tetramers. Two monomers dimerize via pairing of the β strand 2 with formation of an extended beta sheet. These dimers subsequently form a tetramer by head-to-head interactions of two dimers with symmetric interactions via helix 4 and 6 and via the loops between helix1 and β1 and between β4 and helix 4. Models were built for a head-to-head interaction of two Apobec3G N-terminal domains (Figure 1A). In the MODELLER models, the indole groups of W94 and W127 of both monomers stack against each other. During OPLSA/AA energy minimization, the guanidinium group of R122 intercalates between both indole groups (Figure 1B). Another interaction in the interface is formed by the aromatic rings of tyrosines 124 and 125 of both monomers, which stack on top of each other, and tyrosine 125 in both monomers interact symmetrically (Figure 1C). D128 is buried in our model and D128 of one monomer forms a hydrogen bond network with Y22, Y125 and W94 of the other monomer (Figure 1D). After OPLSA/AA energy minimization, a hydrogen bond is formed between D128 and R24.
Figure 1.
Homology model of the Apobec3G N-terminal deaminase domain. A dimer of two N-terminal domains (red and blue), interacting via the 122–127 motif is shown. (A) Overview with indication of secondary structure elements. (B–D) Detail of selected sidechain interactions between the different monomers.
Two conserved protein interfaces in Apobec3G and Apobec3F
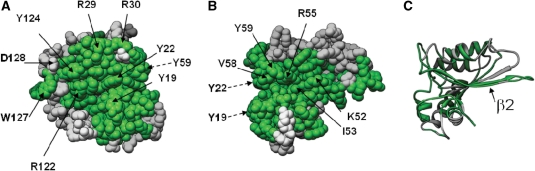
We compared the sequences of human Apobec3G and Apobec3F. The entire head-to-head interface area around W127 is nearly identical in Apobec3G and Apobec3F (Figure 2A). Apobec3G and Apobec3F may, therefore, homodimerize in a very similar way. This interface is possibly involved in RNA-dependent Apobec3G–3F heterodimerization (16,40). The conserved zone consists of two major subzones: a first subzone is centered around the 122 RFYYW 127 motif and forms the core of the head-to-head interface, a second subzone in the N-terminal domain contains residues such as Y19, Y22, R24 and T32, and extends beyond this modeled dimer interface. Another zone of identical surface residues is found around β strand 2 (Figure 2B and C). As β strand 2 forms a dimer interface in the Apobec2 crystal structure, it is possible that β strand 2 in the Apobec3G and Apobec3F N-terminal domain is similarly involved in dimerization.
Figure 2.
Model of the Apobec3G N-terminal domain. Residues which are identical in the N-terminal domain of Apobec3F are colored green. (A) A first identical area includes the 122–127 motif. (B) By turning the model 90° around the Y-axis, a second identical area appears. (C) Ribbon presentation with model orientation as in (B). The identical area in (B) coincides with the β-2 strand.
MAPPIT analysis of Apobec3G–Apobec3G and Apobec3G–Vif interactions
We tested the importance of the predicted head-to-head interface for the Apobec3G–Apobec3G and Apobec3G–Vif interactions using the MAPPIT configuration shown in Figure 3A (32). In MAPPIT, a bait protein is coupled to the C-terminus of a chimeric protein that consists of the extracellular and transmembrane domain of the erythropoietin receptor (EpoR) and the intracellular domain of the leptin receptor (LepR). The tyrosine residues in the leptin receptor domain are mutated to phenylalanine, thereby preventing the chimera from inducing JAK/STAT signaling. The prey is coupled to a duplication of a fragment of the gp130 receptor. When bait and prey interact, and the receptor is stimulated with Epo, the gp130 tyrosines are phosphorylated by the activated JAK2 in the bait construct. The phosphorylated tyrosines in the prey then recruit STAT3, which in turn is phosphorylated. Activated STAT3 is translocated to the nucleus where it induces expression from a luciferase reporter. We first tested whether we could detect an interaction using Apobec3G and Vif as bait or prey. Apobec3G cloned as bait shows a clear MAPPIT interaction with Vif or Apobec3G cloned as prey (Figure 3B). The loss-of function mutants Vif C133S and Vif SLQ144-146AAA were previously shown to disrupt Apobec3G ubiquitination and degradation, whereas these mutants retain their interaction with Apobec3G (17,41). Introduction of the SLQ144-146AAA and C133S mutations in the Vif prey resulted in increased MAPPIT signals (Figure 3C). In the following studies, we used the Vif SLQ144-146AAA mutant prey (VifSLQ prey) in our MAPPIT configuration to study the Apobec3G–Vif interaction.
Next, we validated our method for studying mutations that disrupt protein interactions. D128 of Apobec3G was previously reported to be involved in Vif association (29). We introduced the D128K mutation in our Apobec3G bait construct and verified the effect on VifSLQ prey association. The Vif–Apobec3G interaction was clearly abrogated by the D128K mutation in Apobec3G (Figure 3D).
Mutations in the predicted head-to-head interface affect the Apobec3G homodimerization interaction
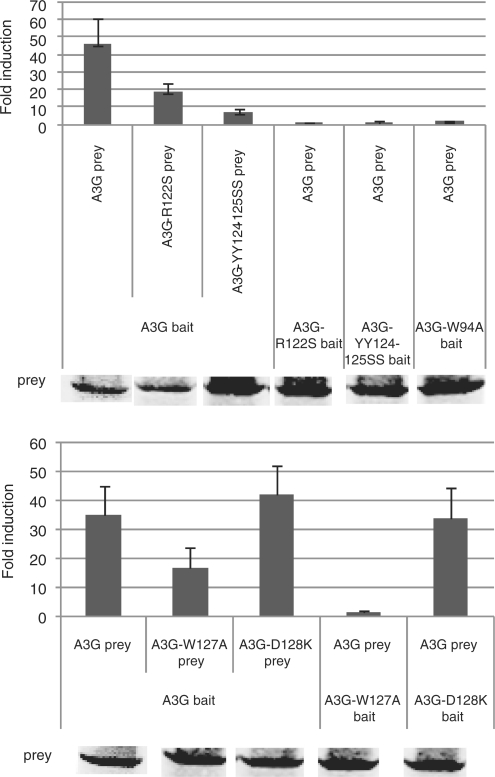
In the Apobec3G–Apobec3G interaction model, W94, R122, Y124, Y125 and W127 play an important role in maintaining the interaction at the dimer interface. We mutated these residues in the Apobec3G bait and/or prey and tested the effect of the mutations on the Apobec3G–Apobec3G interaction. All mutations clearly disrupt the interaction, supporting the hypothesis that the mutated residues are involved in mediating the Apobec3G–Apobec3G interaction (Figure 4). In addition, we used alphascreenTM technology (PerkinElmer) to test the effect of the W127A and D128K mutations. A strong alphascreen signal was detected between E-tagged and Flag-tagged Apobec3G, which was abrogated by the addition of RNAse A indicating that the Apobec3G-Apobec3G interaction is RNA mediated (Figure 5). The W127A mutation strongly decreased the alphascreenTM signal, supporting an important role of W127 in homodimerization, as predicted from the analysis of the modeled interface. The D128K mutation did not inhibit Apobec3G–Apobec3G interaction in a MAPPIT experiment or alphascreenTM setup (Figures 4 and 5).
Figure 4.
Hek293T cells were transiently co-transfected with plasmids encoding the chimeric Apobec3G WT or mutant constructs and the Apobec3G WT or mutant prey constructs, combined with the pXP2d2-rPAP1-luci reporter. The transfected cells were either stimulated for 24 h with Epo or were left untreated (NS, not stimulated). Luciferase measurements were performed in triplicate. Data are expressed as fold induction (Epo stimulated/NS) of luciferase activity. Western blot control of the prey constructs is shown.
Figure 5.
Hek293T cells were transiently co-transfected with plasmids encoding the E- and Flag tagged Apobec3G WT or mutants, a BC-box deficient mutant of CIS was included as a negative control. The transfected cells were lysed after 48 h and protein interactions were detected with the alphascreen FLAGTM (M2) detection kit (Perkinelmer) with or without RNAse treatment.
Mutations in the predicted Apobec3G–Apobec3G head-to-head interface affect the interaction with Vif
We next determined the effect of mutations in the predicted interface on the Apobec3G–Vif interaction using MAPPIT with the Apobec3G mutants as bait and VifSLQ as prey. Mutations of R122, Y124 and Y125, W94 or W127 in the predicted Apobec3G interface completely abolished the interaction with the VifSLQ prey (Figure 6). These data suggest that the residues in the predicted interface are important for both the interaction with Apobec3G and interaction with Vif.
Figure 6.
Hek293T cells were transiently co-transfected with plasmids encoding the chimeric Apobec3G WT or mutant constructs and the Apobec3G or VifSLQ prey constructs, combined with the pXP2d2-rPAP1-luci reporter. The transfected cells were either stimulated for 24 h with Epo or were left untreated (NS, not stimulated). Luciferase measurements were performed in triplicate. Data are expressed as fold induction (Epo stimulated/NS) of luciferase activity. Western blot control of the different prey constructs in the experiment is shown.
For eight different Apobec3G mutants, we compared the interaction with Vif and VifSLQ preys. Similar results were obtained with both Vif and VifSLQ preys, justifying the use of VifSLQ preys in this study (data not shown).
Effect of extensive mutagenesis of the Apobec3G–Apobec3G head-to-head interface
Mutations of W94, R122, Y124 and Y125 or W127 disrupt both the Apobec3G–Apobec3G interaction and the Apobec3G–Vif interaction, suggesting that both interactions are functionally coupled. We created point mutations in the entire area around the model interface and compared the effect of the mutations on both interactions. Table 1 summarizes the results of these experiments. The effect of the mutations was mapped in color code on the Apobec3G model (Figures 7 and 8). The data show a patch of residues Y22, R24, R29, R30, W94, R122, Y125, W127 and Y181 that strongly affect the Apobec3G–Apobec3G interaction when mutated. Comparison of Figure 8A and B shows that all mutations that affect the Apobec3G– Apobec3G interaction also affect the Apobec3G–VifSLQ interaction. These data confirm that the predicted interface is important for mediating both protein–protein interactions.
Figure 7.
Effect of mutations on A3G–A3G (A) and A3G–vif (B) interactions. (A) Residues are colored according to the effect of mutation of the residue on the Apobec3G–Apobec3G MAPPIT interaction. (B) Residues are colored according to the effect of mutation on the Apobec3G–Vif MAPPIT interaction. The effect on the MAPPIT signal in(A) and (B) is indicated by color as in Table 1. red is < 30% of WT; orange is < 50% of WT; green is > 200% of WT; black: the other mutants.
Figure 8.

View of a dimer interface in a monomer model of the Apobec3G N-terminal domain. (A) Residues are colored according to the effect of mutation of the residue on the Apobec3G–Apobec3G MAPPIT interaction. (B) Residues are colored according to the effect of mutation on the Apobec3G–Vif MAPPIT interaction. The effect on the MAPPIT signal in (A) and (B) is indicated by color as in Table 1. red is < 30% of WT; orange is < 50% of WT; green is > 200% of WT; black: the other mutants.
P129 and T32 are possibly part of a Vif-binding site at the edge of the Apobec3G–Apobec3G interface
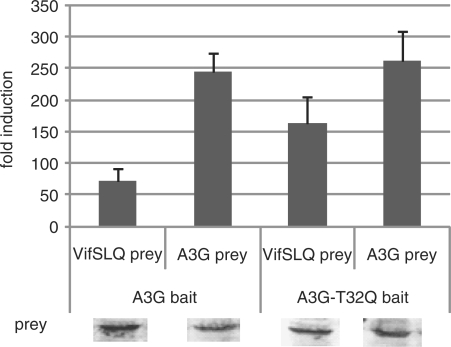
HIV Vif cannot bind Apobec3G of some nonhuman primate species. A single D128K mutation found in Apobec3G of African green monkeys abrogates the binding of Vif and subsequent degradation (42–45). D128 is proposed to be part of a Vif-binding site and is surface exposed in different models of the Apobec3G N-terminal domain (27,30). Mutation of residues D128, P129 or D130 protects Apobec3G against Vif-mediated degradation, and these residues have been proposed as a possible Vif interaction site (29,42–45). In our MAPPIT analysis, the D128K and P129A mutations only disrupt the interaction with Vif (Figure 3, 4 and 6). In Apobec3G, T32 is phosphorylated by protein kinase A, reducing the interaction with Vif (46). A T32Q mutation increases the MAPPIT interaction with the VifSLQ prey and has no effect on the Apobec3G homodimerization interaction (Figure 9 and Table 1). A K99D mutation similarly affects the MAPPIT A3G-Vif interaction, without affecting the homodimerization of Apobec3G (Table 1).
Figure 9.
Hek293T cells were transiently co-transfected with plasmids encoding the chimeric Apobec3G WT or mutant constructs and the Apobec3G or VifSLQ prey constructs, combined with the pXP2d2-rPAP1-luci reporter. The transfected cells were either stimulated for 24 h with Epo or were left untreated (NS, not stimulated). Luciferase measurements were performed in triplicate. Data are expressed as fold induction (stimulated/NS). Western blot control of the different prey constructs in the experiment is shown.
We mapped the position of D128, P129, T32 and K99 on our dimer model (Figures 7 and 8). D128 is located at the edge of the conserved Apobec3G–Apobec3G interface. P129, T32 and K99 are all found at the edge of the interface and are surface exposed. P129 of one monomer comes very close to T32 of another monomer (7.5 Å). These residues may thus be part of a composite Vif-binding site formed by interaction of two Apobec3G N-terminal domains.
DISCUSSION
Vif binds to Apobec3G and Apobec3G homodimerize via an RNA-dependent mechanism (27,40,47). The N-terminal deaminase domain of Apobec3G was reported to play an important role in both interactions (4). Huthoff et al. (27) recently proposed a homology model for an Apobec3G dimer and demonstrated the importance of the area around W127 for RNA-mediated dimerization of Apobec3G. Mutations of Y124 and W127 inhibit incorporation of Apobec3G into virion particles and oligomerization (29). Arginine 24, 30 and 136 are important for interaction with RNA and for RNA-mediated oligomerization. Mutation of these arginines reduces packing of Apobec3G into the virion (27).
We built a dimer model for the N-terminal domain of Apobec3G where W127 is involved in head-to-head interactions, consistent with the model proposed by Huthoff et al. The residues in the predicted interface are well conserved between Apobec3G and Apobec3F, suggesting a similar mode of homodimerization for Apobec3F. The zone of sequence identity between Apobec3G and Apobec3F extends beyond the model interface and includes a subzone that centers around W127 and a subzone formed by more N-terminal residues including Y19, Y22 and T32. Interestingly, these two subzones coincide with two regions that determine subcellular localization of Apobec3G. Residues 113–128 were shown to be important for nuclear exclusion and cytoplasmic retention of Apobec3G, while mutations of Y19, Y22 and T32, F126 and W127 demonstrated the importance of these two regions for maintaining the cytoplasmic localization (48,49). Probably, the oligomerization status of Apobec3G and Apobec3F plays a determining role in their localization.
MAPPIT can be used as a valid tool to study protein–protein interactions of potential therapeutic targets. Previously we employed MAPPIT in a detailed examination of the dimerization of HIV-1 reverse transcriptase in intact human cells (50). We here used MAPPIT to analyze the Apobec3G-Apobec3G and the Apobec3G–Vif interactions. The D128K and P129A mutations specifically disrupt the Apobec3G–Vif interaction. Mutations of R122, Y124 and Y125 or W127 inhibit the Apobec3G–Apobec3G interaction. These data are in accordance with previous reports (26,27,29,42–45), validating MAPPIT as a reliable tool to study the Apobec3G–Apobec3G and Apobec3G–Vif interaction. In line with this, alphascreenTM experiments confirmed the role of W127 in the Apobec3G homodimerization, as the W127A mutation strongly decreases the interaction. The A3G homodimerization is abrogated by RNAseA treatment, confirming that the interaction via W127 is dependent on RNA.
We tested the importance of the predicted Apobec3G homodimerization interface for the interaction with Vif. We found that all mutations that strongly reduce the Apobec3G–Apobec3G interaction also strongly reduce the interaction with Vif. These data seem to contradict the three previous reports that show that degradation of Apobec3G by Vif is not affected by mutations in the proposed Apobec3G homodimerization interface: (i) Huthoff et al. (27) found that point mutations in the 122 RLYYFW 127 motif have no effect on Apobec3G degradation by Vif; (ii) Bulliard et al. (31) reported that a W94L and W127A mutation in the head-to-head interface do not interfere with Apobec3G degradation by Vif; and (iii) Shirakawa et al. (31,46) demonstrated that an R24A mutant shows normal Vif binding and normal degradation by Vif. In our MAPPIT experiments, an R24S mutation severely reduces both Apobec3G dimerization and Vif binding.
However, an important role for the homodimerization interface for Vif binding is in line with a recent study by Gooch and Cullen (26). The RLYYFW motif in Apobec3G is replaced by RIYDY in Apobec3A, which is not a target of Vif. Gooch and Cullen (26) reported that mutation of 124 YYFW 127 in Apobec3G to the corresponding Apobec3A sequence YDY decreased Vif binding. This Apobec3G mutant is not targeted for degradation by Vif. The mutation drastically disrupts the Apobec3G–Apobec3G and Apobec3G–Vif interactions in our MAPPIT assay (Table 1).
Conversely, mutation of the RIYDY sequence in Apobec3A to RIYYFW turns Apobec3A into a target for Vif binding (26). This mutation introduces a RIYYFWDP sequence into Apobec3A, which is similar to the RLYYFWDP sequence in Apobec3G. The RI/LYYFWDP sequence may be central for recognition by Vif. Unfortunately, it is at present unclear whether the RIYYFW mutation in Apobec3A leads to dimerization of Apobec3A.
Different regions in Vif, 40 YRHHY 44 and 14 DRMR 17 were reported to be essential for binding to Apobec3G and 3F, respectively (51). While Vif associates with the N-terminal domain of Apobec3G, Vif binds to the C-terminal domain of Apobec3F (52). Interestingly, the C-terminal domain of Apobec3F contains a RLYYFWDTD sequence, which is almost identical to the RLYYFWDPD sequence of Apobec3G.
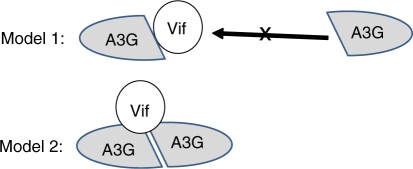
Two models explain why mutations that affect Apobec3G dimerization also affect Vif binding (Figure 10).
Figure 10.
Models for the Apobec3G–Vif interaction. In model 1, Vif binds to an Apobec3G monomer at the binding site used for Apobec3G–Apobec3G interaction. In this model, Vif binding may interfere with Apobec3G dimerization. In model 2, Vif binds to an Apobec3G dimer. In this model, Vif binding requires Apobec3G dimerization.
In a first model, Vif binds an Apobec3G monomer. The Vif interaction site overlaps with the Apobec3G head-to-head interface. This Vif-binding site would then include residues R122, Y125, W127, D128 and P129. In this model, Vif binding may compete with Apobec3G dimerization.
In a second model, Vif binds to an Apobec3G dimer. The Vif-binding site is composed of two interacting Apobec3G monomers. Four residues affect the Apobec3G–Vif interaction without apparent effects on the Apobec3G dimerization. Mutation of D128 or P129 disrupts the interaction with Vif. Mutation of T32 or K99 increases interaction with Vif. In the Apobec3G dimer model, T32 of one Apobec3G monomer comes close to K99, D128 and P129 of the other Apobec3G monomer. These four residues may be part of a composed Vif-binding site.
In summary, we present a model where the N-terminal domains of Apobec3G interact in a similar way as Apobec2. Using MAPPIT and alphascreenTM, we demonstrate that the interaction is mediated by RNA and that mutations in the head-to-head interface inhibit Vif binding. The dimer interface of Apobec3G is almost identical in the N-terminal domain of Apobec3F, suggesting that Apobec3G and 3F can form similar complexes. Mutations of T32, K99, D128 or P129 at the brim of the head-to-head interface in Apobec3G specifically affect Vif binding, and these residues may be part of a Vif-binding site that covers parts of two interacting N-terminal domains. Mutations in the head-to-head interface interfere with interaction of Apobec3G with gag and with incorporation of Apobec3G into the virion (26,27,29). Vif may select Apobec targets that incorporate into virions via binding to the dimer interface area.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT), Ghent University and the Interuniversity Attraction Poles (grant number P6:28). D.L. and I.U. are postdoctoral fellows of the Fund for Scientific Research-Flanders (FWO). Funding for open access charge: Methusalem grant from Ghent University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Prof. M. Malim and Dr H. Huthoff for the generous gift of the pCMV4-HA-A3G-L123A and pCMV4-HA-A3G-Y124A constructs and Prof. C. Verhofstede for kindly providing us the pNL4-3 vector. We thank Dr E. Pattyn for the construction of the pMG2-A3G vector.
REFERENCES
- 1.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 2.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 6.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 9.Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 10.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 15.Stopak K, de NC, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 16.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 18.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 19.Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos. Trans. Roy. Soc. Lond B Biol. Sci. 2009;364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 21.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 22.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 23.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J. Mol. Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 26.Gooch BD, Cullen BR. Functional domain organization of human APOBEC3G. Virology. 2008;379:118–124. doi: 10.1016/j.virol.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 29.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang KL, Mangeat B, Ortiz M, Zoete V, Trono D, Telenti A, Michielin O. Model structure of human APOBEC3G. PLoS ONE. 2007;2:e378. doi: 10.1371/journal.pone.0000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulliard Y, Turelli P, Rohrig UF, Zoete V, Mangeat B, Michielin O, Trono D. Functional analysis and structural modeling of human APOBEC3G reveals the role of evolutionarily conserved elements in HIV-1 and Alu inhibition. J. Virol. 2009;83:12611–12621. doi: 10.1128/JVI.01491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eyckerman S, Verhee A, Van der Heyden J, Lemmens I, Ostade XV, Vandekerckhove J, Tavernier J. Design and application of a cytokine-receptor-based interaction trap. Nat. Cell Biol. 2001;3:1114–1119. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- 33.Eyckerman S, Lemmens I, Lievens S, Van der Heyden J, Verhee A, Vandekerckhove J, Tavernier J. Design and use of a mammalian protein–protein interaction trap (MAPPIT) Sci. STKE. 2002;2002:L18. doi: 10.1126/stke.2002.162.pl18. [DOI] [PubMed] [Google Scholar]
- 34.Eyckerman S, Broekaert D, Verhee A, Vandekerckhove J, Tavernier J. Identification of the Y985 and Y1077 motifs as SOCS3 recruitment sites in the murine leptin receptor. FEBS Lett. 2000;486:33–37. doi: 10.1016/s0014-5793(00)02205-5. [DOI] [PubMed] [Google Scholar]
- 35.Lemmens I, Eyckerman S, Zabeau L, Catteeuw D, Vertenten E, Verschueren K, Huylebroeck D, Vandekerckhove J, Tavernier J. Heteromeric MAPPIT: a novel strategy to study modification-dependent protein-protein interactions in mammalian cells. Nucleic Acids Res. 2003;31:e75. doi: 10.1093/nar/gng075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavernier J, Eyckerman S, Lemmens I, Van der Heyden J, Vandekerckhove J, Van OX. MAPPIT: a cytokine receptor-based two-hybrid method in mammalian cells. Clin. Exp. Allergy. 2002;32:1397–1404. doi: 10.1046/j.1365-2745.2002.01520.x. [DOI] [PubMed] [Google Scholar]
- 37.Lavens D, Montoye T, Piessevaux J, Zabeau L, Vandekerckhove J, Gevaert K, Becker W, Eyckerman S, Tavernier J. A complex interaction pattern of CIS and SOCS2 with the leptin receptor. J. Cell Sci. 2006;119:2214–2224. doi: 10.1242/jcs.02947. [DOI] [PubMed] [Google Scholar]
- 38.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 40.Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- 41.Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl Acad. Sci. USA. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc. Natl Acad. Sci. USA. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, Strebel K, Pathak VK. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl Acad. Sci. USA. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 46.Shirakawa K, Takaori-Kondo A, Yokoyama M, Izumi T, Matsui M, Io K, Sato T, Sato H, Uchiyama T. Phosphorylation of APOBEC3G by protein kinase A regulates its interaction with HIV-1 Vif. Nat. Struct. Mol. Biol. 2008;15:1184–1191. doi: 10.1038/nsmb.1497. [DOI] [PubMed] [Google Scholar]
- 47.Wedekind JE, Gillilan R, Janda A, Krucinska J, Salter JD, Bennett RP, Raina J, Smith HC. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J. Biol. Chem. 2006;281:38122–38126. doi: 10.1074/jbc.C600253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett RP, Presnyak V, Wedekind JE, Smith HC. Nuclear exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J. Biol. Chem. 2008;283:7320–7327. doi: 10.1074/jbc.M708567200. [DOI] [PubMed] [Google Scholar]
- 49.Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 2008;82:9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pattyn E, Lavens D, Van der Heyden J, Verhee A, Lievens S, Lemmens I, Hallenberger S, Jochmans D, Tavernier J. MAPPIT (MAmmalian Protein–Protein Interaction Trap) as a tool to study HIV reverse transcriptase dimerization in intact human cells. J. Virol. Methods. 2008;153:7–15. doi: 10.1016/j.jviromet.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 51.Schrofelbauer B, Senger T, Manning G, Landau NR. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell RA, Smith J, Barr R, Bhattacharyya D, Pathak VK. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J. Virol. 2009;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.