Abstract
Hypoxia-inducible factors (HIFs) are critical transcription factors that mediate cell survival during reduced oxygen conditions (hypoxia). At regular oxygen conditions (normoxia), HIF-1α and HIF-2α are continuously synthesized in cells and degraded via the ubiquitin–proteasome pathway. During hypoxia, these proteins are stabilized and translocate to the nucleus to activate transcription of target genes that enable cell survival at reduced oxygen levels. HIF proteins are tightly regulated via post-translational modifications including phosphorylation, acetylation, prolyl-hydroxylation and ubiquitination. Here we show for the first time that exogenous and endogenous HIF-2α are also regulated via the ubiquitin-like modifier small ubiquitin-like modifiers (SUMO). Using mutational analysis, we found that K394, which is situated in the sumoylation consensus site LKEE, is the major SUMO acceptor site in HIF-2α. Functionally, sumoylation reduced the transcriptional activity of HIF-2α. Similar to HIF-1α, HIF-2α is regulated by the SUMO protease SENP1. The proteasome inhibitor MG132 strongly stabilized SUMO-2-conjugated HIF-2α during hypoxia but did not affect the total level of HIF-2α. The ubiquitin E3 ligases von Hippel–Lindau and RNF4 control the levels of sumoylated HIF-2α, indicating that sumoylated HIF-2α is degraded via SUMO-targeted ubiquitin ligases.
INTRODUCTION
Many different post-translational modifications including phosphorylation, acetylation, methylation and glycosylation are involved in regulating the activity of proteins. Sumoylation, the process of attaching a small ubiquitin-like modifier (SUMO) protein to a target protein, is a more recently discovered reversible post-translational modification (1,2). SUMOs are covalently conjugated to acceptor lysines in sumoylation consensus motifs (ψKx(E/D), where ψ stands for V, L, I, M or F and x can be any amino acid) in target proteins. Sumoylation regulates many different cellular processes such as gene expression, signal transduction, chromatin structure and the maintenance of the genome by attachment to and modulation of target proteins.
Three SUMO family members have been described in vertebrates, SUMO-1, -2 and -3, encoded by three different genes (3). The mature forms of SUMO-2 and SUMO-3 are very similar (∼95% identical) but differ from SUMO-1 (∼50% identical) (4). SUMO-1 and SUMO-2 each have a unique subset of substrates as well as a shared subset to which both isoforms can be conjugated (5).
Low oxygen concentration leads to adaptive changes in the transcription of a range of genes. The Hypoxia-Inducible Factors (HIFs) mediate the transcriptional activation of genes that allow cells and tissues to cope with low oxygen conditions (6,7). HIFs are heterodimers, composed of one α and one β subunit. While the HIF-1β/ARNT (aryl hydrocarbon receptor nuclear translocator) subunit is stable, the HIF-α proteins (HIF-1α, HIF-2α and HIF-3α) are continuously synthesized and degraded under normoxic conditions. During normoxia, two conserved proline residues in the oxygen-dependent degradation domain (ODD) of HIF-α are hydroxylated by HIF-specific prolyl hydroxylase-domain proteins (PHD 1, 2 and 3). Hydroxylation of the prolyl residues mediates binding of the von Hippel–Lindau (VHL) ubiquitin ligase complex responsible for ubiquitination and degradation of HIF-1α in a proteasome-dependent manner (8,9). The PHD proteins require molecular oxygen for their enzymatic activity and no longer function during hypoxia. Consequently, degradation no longer occurs during hypoxia and the HIF-α subunits rapidly accumulate and translocate to the nucleus (10–12).
HIF-1α and HIF-2α are 48% identical and their stability and transcriptional activity are regulated via shared mechanisms (13,14). Both proteins play important and similar, but non-redundant roles in fetal development and tumor angiogenesis. Mouse embryos in which HIF-1α expression was disrupted exhibited multiple defects in cardiovascular development and died early during development (E11.5) (15). HIF-2α–/– mice generated by different groups differed somewhat in phenotype, possibly due to differences in genetic backgrounds. Phenotypes observed include defective vascular remodeling with local hemorrhage (16), defective fetal catecholamine production (17) or altered lung maturation secondary to impaired surfactant secretion by alveolar type 2 cells (18).
The activity of HIF-2α is tightly controlled by post-translational modifications including prolyl-hydroxylation, ubiquitination and phosphorylation. We have investigated the sumoylation of HIF-2α, which contains two consensus sumoylation sites, LK394EE and LK497IE. We have generated HIF-2α mutants in which these consensus sumoylation sites were disrupted and demonstrate that K394 is used for SUMO conjugation and plays a role in the regulation of HIF-2α activity. Interestingly, SUMO-2-conjugated HIF-2α is rapidly degraded during hypoxia via SUMO-targeted ubiquitin ligases.
MATERIALS AND METHODS
Expression vectors
Plasmids containing the human wild-type HIF-1α and -2α cDNA tagged at the N-terminus with three consecutive FLAG tags were a kind gift from Dr A. Groot (Utrecht University, Utrecht, The Netherlands) (19,20). The 5xHREpGL3-Luciferase reporter was a kind gift from Dr M. Duyndam (VUMC, Amsterdam, The Netherlands). The FLAG-SUMO-2 lentiviral construct was generated by inserting his6-SUMO-2 into the EcoRV site of plasmid pLV-CMV-IRES-eGFP (21). The FLAG epitope was subsequently introduced in this plasmid via oligonucleotide insertion into the PstI site. Plasmids encoding shRNAs were obtained from the MISSION shRNA Library (Sigma Aldrich): control shRNA, SHC-002; RNF4 shRNA, TRCN0000017053; RSUME shRNA, TRCN0000004095; SENP1 shRNA, TRCN0000004399 and VHL shRNA, TRCN0000010460.
Mutagenesis
The K394R, K497R, E396A and E499A mutations in HIF-2α were generated by site-directed mutagenesis using the Quickchange II kit according to the instructions of the manufacturer (Stratagene). The following oligonucleotides were used to generate these mutations: K394R, forward 5′-ctattcaccaagctaagggaggagcccgag-3′ and reverse 5′-ctcgggctcctcccttagcttggtgaatag-3′; K497R, forward 5′-gataacgacctgaggattgaagtgattg-3′ and reverse 5′-caatcacttcaatcctcaggtcgttatc-3′; E396A, forward 5′-caagctaaaggaggcgcccgaggagctg-3′ and reverse 5′-cagctcctcgggcgcctcctttagcttg-3′; E499A, forward 5′-gacctgaagattgcagtgattgagaag-3′ and reverse 5′-cttctcaatcactgcaatcttcaggtc-3′. All mutants were sequence verified.
Cell culture, transfection and infection
HeLa cells stably expressing his6-SUMO-1 or his6-SUMO-2 were previously described (5). The HeLa cell line stably expressing FLAG-SUMO-2 was generated by infection with the lentivirus encoding FLAG-SUMO-2. HeLa cells were grown in Dulbecco’s modified Eagle medium (DMEM), supplemented with 10% FCS and 100 U/ml penicillin and streptomycin (Invitrogen). Hypoxic stabilization of HIF-α was performed at 1% O2 for 24 h. Transient transfections were performed using 2.5 µl Polyethylenimine (PEI, 1 mg/ml, Alpha Aesar) per µg DNA. Trichostatin A (TSA, Sigma Aldrich) was added to the culture medium at 300 nM for 24 h where indicated. MG132 (Sigma Aldrich) was added to the culture medium at 10 µM for 4 or 7 h where indicated. For both drugs, control cells were treated with dimethyl sulphoxide (DMSO). Cycloheximide was added to the culture medium at 10 µg/ml where indicated. Lentiviral infections were performed at an MOI of 1 for 24 h. Cells were subsequently incubated for another 48 h prior to lysis.
Luciferase assays
Cells were cultured in 24-well plates and cotransfected with 0.2 µg luciferase reporter plasmid and 0.5 µg expression plasmid as indicated. Experiments were carried out in triplicate and additional wells were prepared for control immunoblotting experiments. Cells were lysed in Reporter Lysis Buffer (Promega) for luciferase activity measurements or in NuPage LDS protein sample buffer (Invitrogen) for immunoblotting.
Cell fixation and immunostaining
HeLa cells were cultured on glass cover slips in 24-well plates and transfected with 0.5 µg expression plasmid. The cells were fixed for 20 min in 3.7% paraformaldehyde at room temperature and permeabilized in phosphate-buffered saline (PBS) containing 0.5% Triton X-100 for 10 min. Cells were blocked with 10% goat serum for 30 min. The cells were incubated with primary antibody (mouse anti FLAG, 1:1000 in PBS/T, 10% NGS) for 1 h, washed and incubated with secondary antibody (goat anti mouse-Alexa 488, 1:1000 in PBS/T, 10% NGS) for 1 h. The cells were washed 5× with PBS/T, mounted in Vectashield, sealed with nail varnish and stored at −20°C.
Microscopy
Microscopy experiments were carried out using a confocal microscope system (model TCS/SP2, Leica). Images were acquired with a 100× NA 1.4 plan Apo objective and were analysed with Leica confocal software.
Antibodies
Peptide antibody AV-SM23-0100 (Eurogentec) against SUMO-2/3 was described previously (22). Monoclonal antibody T5326 against γ-tubulin, polyclonal antibody HPA011765 against SENP1 and monoclonal antibody M2 against FLAG were obtained from Sigma Aldrich. Polyclonal antibody NB100-122 against HIF-2α was obtained from Novus Biologicals. Polyclonal antibody against RNF4 was a kind gift of Dr J. Palvimo. Secondary antibodies used were anti-rabbit HRP and anti-mouse HRP (Pierce Chemical Co.). The secondary antibody used for immunofluorescence was goat anti mouse Alexa 488 (Invitrogen).
Purification of his6-SUMO conjugates
Cells were cultured in 14 cm diameter culture dishes and transfected with 10 µg FLAG-HIF-1α or -2α expression plasmids as indicated. His6-SUMO conjugates were purified essentially as previously described (23). Cells were scraped in ice-cold PBS. Two small aliquots of each sample were lysed in LDS protein sample buffer (Invitrogen) as input control, or in 8 M Urea, 100 mM Na2HPO4/NaH2PO4, 10 mM Tris–HCl, pH 7.0 to determine the protein concentration. The remaining cells were solubilized in lysis buffer (6 M Guanidinium-HCl, 100 mM Na2HPO4/NaH2PO4, 10 mM Tris–HCl, pH 8.0, 20 mM Imidazole, 10 mM β-mercaptoethanol) and sonicated to reduce the viscosity. His6-SUMO conjugates were enriched on Ni-NTA Agarose beads (Qiagen) and washed using wash buffers A–D. (Buffer A: 6 M Guanidinium–HCl, 100 mM NaH2PO4/Na2HPO4, 10 mM Tris–HCl pH 8.0 and 0.2% Triton-X-100. Buffer B: 8 M Urea, 100 mM NaH2PO4/Na2HPO4, 10 mM Tris–HCl pH 8.0 and 0.2% Triton-X-100. Buffer C: 8 M Urea, 100 mM NaH2PO4/Na2HPO4, 10 mM Tris–HCl pH 6.3 and 0.2% Triton-X-100. Buffer D: 8 M Urea, 100 mM NaH2PO4/Na2HPO4, 10 mM Tris–HCl pH 6.3 and 0.1% Triton-X-100). These wash buffers also contained 10 mM β-mercaptoethanol. Samples were eluted in 6.4 M Urea, 80 mM NaH2PO4/Na2HPO4, 8 mM Tris–HCl pH 7.0, 200 mM imidazole.
Purification of FLAG-SUMO conjugates
Cells were grown in 14 cm diameter dishes and cultured for 7 h at 1% O2 before lysis. Cell extracts were prepared in SDS buffer [2% SDS, 50 mM Tris–HCl, pH 7.5 and 10 mM iodoacetamide (23)] and incubated at room temperature for 15 min. The viscosity of the samples was reduced by sonication. SDS concentration of the samples was reduced by dilution in buffer B (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP40 and 0.5 mM β-mercaptoethanol). Samples were centrifuged at 9000g for 30 min at 4°C and incubated for 3 h at 4°C with Anti-FLAG M2 beads (M8823, Sigma Aldrich). The beads were collected, washed 5 times with wash buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl and 0.5% NP40) and eluted with 15 µl wash buffer containing 100 µg/ml FLAG peptide for 15 min at 4°C. Supernatant was collected for immunoblotting.
Immunoblotting
Protein samples were size fractionated on Novex 4–12% Bis-TRIS gradient gels using 4-morpholineprop-anesulfonic acid buffer (Invitrogen) or on regular SDS-PAGE gels with a tris-glycine buffer. Note that these different methods cause slight differences in the running behavior of proteins (Invitrogen). Size fractionated proteins were subsequently transferred onto Hybond-C extra membranes (Amersham Biosciences) using a submarine system (Invitrogen). The membranes were incubated with specific antibodies as indicated. Bound antibodies were detected via chemiluminescence with ECL Plus (Amersham Biosciences).
RESULTS
HIF-2α contains two conserved consensus sumoylation sites
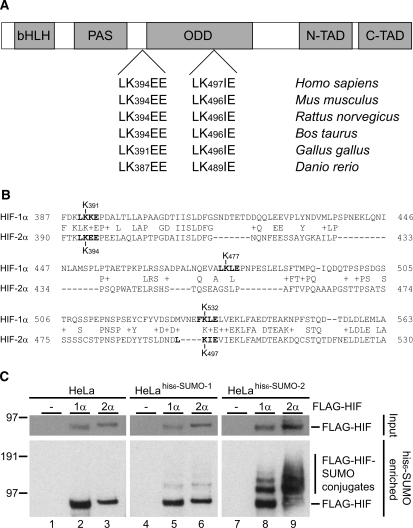
The transcription factor HIF-1α is regulated by SUMO conjugation. As a homologue of HIF-1α, HIF-2α shows close similarity in sequence, function and regulation. HIF-1α contains three consensus sumoylation sites, two of which are involved in the regulation of its transcriptional activity (6,24–26). Two potential SUMO acceptor sites matching the consensus sequence are present in HIF-2α (Figure 1). These sites are conserved in a wide range of species including Mus musculus, Rattus norvegicus, Bos taurus, Gallus gallus and Danio rerio.
Figure 1.
HIF-2α contains two conserved consensus sumoylation sites. (A) Structural domains of HIF-2α. Two consensus sumoylation sites are present in human HIF-2α, LK394EE and LK497IE, which are well conserved in other species. (B) Sequence alignment between HIF-1α and HIF-2α. Both potential HIF-2α sumoylation sites have equivalent sites in HIF-1α, a known SUMO substrate. (C) FLAG-HIF-1α and -2α are conjugated to SUMO-1 and to SUMO-2. HeLa, HeLahis6-SUMO-1 and HeLahis6-SUMO-2 cells were transfected with plasmids encoding FLAG-HIF-1α, -2α or an empty vector control. The cells were grown at 1% O2 for 24 h, lysed and his6-SUMO conjugates were purified. Purified fractions were separated by SDS–PAGE, transferred to membranes and probed with an anti-FLAG antibody. Total cell lysates were included as input controls. Several SUMO-modified forms of FLAG-HIF-1α and -2α could be detected in the his6-SUMO-2 enriched samples whereas only mono-sumoylated forms could be detected in the his6-SUMO-1 enriched samples. bHLH, basic helix-loop-helix; PAS, Per/Arnt/Sim domain; ODD, oxygen dependent degradation domain; N-TAD, N-terminal transactivation domain; C-TAD, C-terminal transactivation domain.
To compare SUMO conjugation between HIF-1α and -2α in cells, we made use of published stable cell lines expressing his6-SUMO-1 and his6-SUMO-2 (5). HeLa, HeLahis6-SUMO-1 and HeLahis6-SUMO-2 cells were transfected with plasmids encoding FLAG-HIF-1α, -2α or an empty vector. The cells were placed at 1% O2 24 h post-transfection and cultured for another 24 h before lysis and purification of his6-SUMO conjugates. Expression levels of HIF-1α and -2α were checked in total cell lysate samples. SUMO-modified forms of FLAG-HIF-1α and -2α could be detected in both the his6-SUMO-1 and his6-SUMO-2 purified samples (Figure 1C). Note that unmodified, but not SUMO-modified, FLAG-HIF could be detected in the HeLa negative control. These results indicate that analogous to HIF-1α, HIF-2α is a substrate for sumoylation. HIF-2α was found to be conjugated to both SUMO proteins at least as efficiently as HIF-1α.
HIF-2α is sumoylated on K394
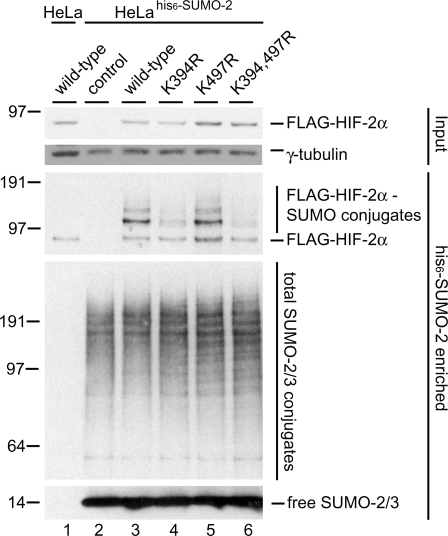
To investigate whether the consensus sumoylation sites present in HIF-2α are used for SUMO conjugation in cells, we created plasmids encoding mutant forms of FLAG-HIF-2α lacking one or both SUMO acceptor lysines. HeLahis6-SUMO-2 cells were transfected with plasmids encoding FLAG-HIF-2α wild-type and mutants and cultured at 1% O2 for 24 hours. His6-SUMO-2 conjugates were purified from these cells and analysed by immunoblotting. At least 2 SUMO-modified forms of FLAG-HIF-2α wild-type could be detected (Figure 2, lane 3) that were absent in the control samples (lanes 1 and 2). Disrupting the first consensus site by mutating K394 (lane 4) strongly reduced sumoylation of FLAG-HIF-2α compared to wild-type. In contrast, disrupting the second consensus site had no effect on sumoylation levels (lane 5). A mutant in which both sites were disrupted did not exhibit a further reduction in sumoylation compared to the K394R single mutant (lane 6 compared to 4). These results indicate that lysine 394 is the major SUMO acceptor site of HIF-2α.
Figure 2.
K394 in HIF-2α is a SUMO acceptor site. HeLa and HeLahis6-SUMO-2 cells were transfected with plasmids encoding FLAG-HIF-2α wild-type and the indicated mutant proteins. The cells were cultured at 1% O2 for 24 h and lysed 48 h post-transfection. His6-SUMO-2 conjugates were enriched using metal affinity chromatography. Enriched fractions were separated by SDS-PAGE, transferred to membranes and probed with anti-FLAG or anti-SUMO-2/3 antibody. Total cell lysates were included as input controls. K394 was found to be the major SUMO acceptor lysine, whereas K497 appeared not to be used for SUMO conjugation. Total cell lysates were probed with an antibody specific for γ-tubulin as a loading control.
To rule out the possibility that our results could be attributed to potential modifications of these lysine residues other than sumoylation, a second set of mutants was prepared. In these mutants, the consensus sumoylation sequence was disrupted by mutating the glutamic acid residues, while leaving the lysines intact. This second set of mutants was used to repeat this experiment and confirmed our findings with the lysine to arginine mutants (Supplementary Figure S1). Mutating E396 resulted in a similar reduction of sumoylation compared to the K394R mutant. Disrupting the second consensus site by mutating E499 had no effect on sumoylation.
Sumoylation of HIF-2α leads to a reduction in transcriptional activity
Since sumoylation of transcription factors often leads to a reduction in their activity (27), we compared the transcriptional activity of wild-type and mutant FLAG-HIF-2α using an HRE-luciferase reporter plasmid (Figure 3). Mutating either K394 or E396 increased the transcriptional activity of HIF-2α while the K497R and E499A mutants showed a level of activity similar to that of the wild-type (Figure 3A). Neither double mutant was transcriptionally more active than HIF-2α proteins carrying only the single K394R or E396A mutation. Increased transcriptional activity of the sumoylation-impaired HIF-2α mutants was not due to an increase in expression of these mutant proteins (Figure 3B).
Figure 3.
SUMO modification of HIF-2α on K394 reduces its transcriptional activity. HeLa cells were cotransfected with wild-type or mutant forms of FLAG-HIF-2α and an HRE-luciferase reporter vector (cartoon). Cells were kept normoxic (21% O2) or cultured hypoxic (1% O2) for 24 h before being lysed in Reporter Lysis Buffer (Promega) for luciferase activity measurements. Control cell lysates were prepared in LDS sample buffer and analysed by immunoblotting to determine the FLAG-HIF-2α expression levels. (A) The transcriptional activity of wild-type FLAG-HIF-2α was compared to the mutants. Mutating either the K394 or E396 in the first consensus sumoylation site resulted in an increase in transcriptional activity over the wild-type both in hypoxic and normoxic samples. Mutations made in the second sumoylation consensus site had no effect on the transcriptional activity of FLAG-HIF-2α. The activity of wild-type HIF-2α under normoxic and under hypoxic conditions was set to 1. A single representative experiment is shown that was carried out in triplicate. The experiment was independently repeated twice with similar results. (B) The expression levels of wild-type and mutant FLAG-HIF-2α proteins were determined by immunoblotting using an anti-FLAG antibody, loading was verified using an anti-γ-tubulin antibody. The increase in transcriptional activity of the sumoylation-impaired HIF-2α mutants was not due to increased expression.
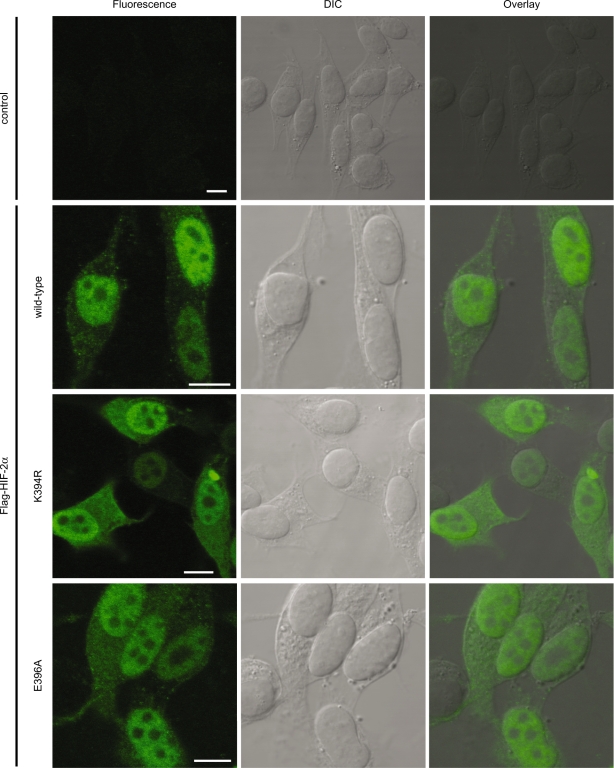
Sumoylation does not affect the subcellular localization of HIF-2α
Sumoylation has previously been shown to alter the subcellular localization of a subset of target proteins including RanGAP1 (28,29). To investigate whether sumoylation has an effect on the subcellular localization of HIF-2α, cells were transfected with plasmids encoding wild-type or mutant FLAG-HIF-2α and cultured for 24 h at 1% O2 before fixation. The localization of the FLAG-HIF-2α proteins was determined using immunofluorescence (Figure 4). FLAG-HIF-2α wild-type was found to be predominantly localized to the nucleoplasm. Disrupting the major sumoylation consensus site by mutating K394 or E396 did not influence the localization of FLAG-HIF-2α.
Figure 4.
Sumoylation does not affect the subcellular localization of FLAG-HIF-2α. HeLa cells were grown on coverslips and transfected with FLAG-HIF-2α expression constructs or empty vector as a control. The cells were incubated at 1% O2 for 24 h before paraformaldehyde fixation. The subcellular localization of wild-type and mutant FLAG-HIF-2α was determined by immunostaining using an anti-FLAG antibody. FLAG-HIF-2α shows a predominant nuclear staining. No difference in localization could be detected between wild-type and mutant proteins. Scale bars are 10 µm.
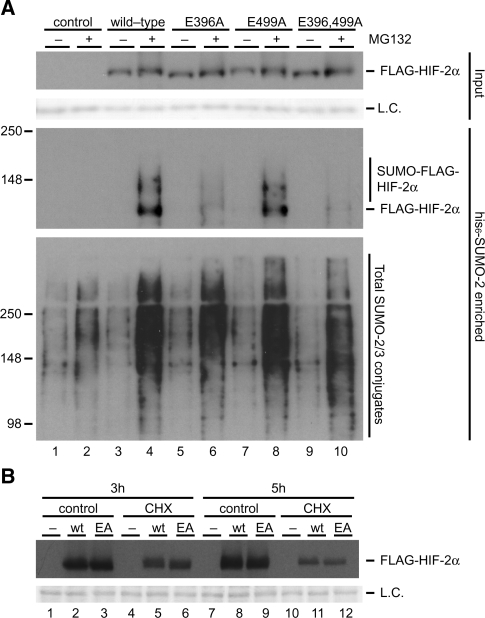
SUMO-modified HIF-2α is degraded by the ubiquitin–proteasome system
We have previously reported that proteasome inhibition strongly increases the total pool of SUMO-2/3 conjugates (30). Moreover, we have identified a subset of SUMO-2 conjugates that are stabilized by proteasome-inhibition and verified that the ubiquitin–proteasome system specifically degraded the SUMO-2-conjugated forms of hnRNP M, MCM-7 and PIAS-1 without affecting the total pools of these proteins. To investigate whether sumoylation regulates HIF-2α stability, we purified his6-SUMO-2 conjugates from HeLa cells that were transfected with plasmids encoding FLAG-HIF-2α wild-type or mutant proteins. The cells were cultured at 1% O2 in the presence or absence of the proteasome inhibitor MG132 for 4 h before being lysed. His6-SUMO-2 conjugates were analysed by immunoblotting. As expected, proteasome inhibition strongly stabilized SUMO-2 conjugates (Figure 5A). SUMO-modified FLAG-HIF-2α exhibited a very strong accumulation upon proteasome inhibition by MG132, whereas the stability of unmodified HIF-2α was not significantly affected. Accumulation of SUMO-modified HIF-2α could be strongly reduced by disrupting the LK394EE sumoylation site but not by disruption of the LK497IE site. The E to A mutants were used for this experiment to interfere as little as possible with potential direct ubiquitination of lysines.
Figure 5.
Proteasome inhibition results in an accumulation of SUMO-modified HIF-2α. (A) HeLa cells were cotransfected with plasmids encoding his6-SUMO-2 and FLAG-HIF-2α wild-type and the indicated mutant proteins. The cells were cultured at 1% O2 for 4 h in the presence of 10 µM MG132 or DMSO control and lysed 48 h post-transfection. His6-SUMO-2 conjugates were enriched using metal affinity chromatography. Enriched fractions were separated by SDS-PAGE, transferred to membranes and probed with anti-FLAG or anti-SUMO-2/3 antibody. Total cell lysates were included as input controls. Ponceau S staining was performed to verify equal loading. SUMO-2 conjugates and SUMO-2-modified forms of FLAG-HIF-2α were found to be strongly increased upon proteasome inhibition. Mutation of the major SUMO acceptor site LK394EE reduced HIF-2α-SUMO accumulation whereas mutating the LK497IE consensus site had no effect. Proteasome inhibition had only marginal effects on total FLAG-HIF-2α levels. (B) HeLa cells were transfected with FLAG-HIF-2α wild-type (wt) or the E396A mutant (EA). The cells were cultured at 21% O2 and treated with cycloheximide (CHX) for 3 or 5 h to inhibit protein synthesis. Equal loading was verified by Ponceau S staining. L.C., loading control.
In order to further investigate whether sumoylation has an effect on the stability of the total pool of HIF-2α, we compared the stability of FLAG-HIF-2α wild-type versus the E396A mutant employing cycloheximide to block protein synthesis. HeLa cells were transfected with plasmids encoding wild-type FLAG-HIF-2α or the E396A mutant. The cells were kept at 21% oxygen and 24 h post-transfection cells were cultured for 3 or 5 h in the presence or absence of cycloheximide. Total cell lysates were prepared and analysed by immunoblotting to compare total FLAG-HIF-2α levels. Cells treated with cycloheximide showed greatly reduced levels of FLAG-HIF-2α compared to control samples (Figure 5B). No differences in stability were observed for the total pools of wild-type and E396A mutant FLAG-HIF-2α.
Endogenous HIF-2α is sumoylated in cells
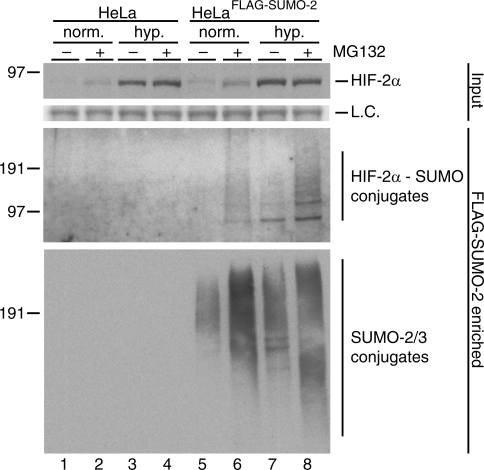
Subsequently, we investigated the conjugation of endogenous HIF-2α to SUMOs by employing a HeLa cell line stably expressing FLAG-SUMO-2. HeLaFLAG-SUMO-2 and control HeLa cells were cultured in the presence or absence of MG132 at 1 or 21% O2 for 7 h before lysis and purification of FLAG-SUMO-2 conjugates. Several SUMO-modified forms of endogenous HIF-2α could specifically be detected in FLAG-SUMO-2 purified samples from hypoxic or MG132-treated HeLaFLAG-SUMO-2 cells (Figure 6). A substantial increase in the amount of SUMO-modified HIF-2α could be observed after treating the cells with MG132. We conclude that endogenous HIF-2α is a target for SUMO-2 during hypoxia and normoxia.
Figure 6.
Endogenous HIF-2α is conjugated to SUMO-2. HeLaFLAG-SUMO-2 and HeLa cells were cultured for 7 h at 21% (norm.) or 1% O2 (hyp.) in the presence or absence of MG132. FLAG-SUMO-2 conjugates were purified from cell lysates and purified samples were analysed by immunoblotting using an antibody against HIF-2α. Total cell lysates were included as input controls. Sumoylated forms of HIF-2α were specifically detected in the FLAG-SUMO-2 purified fractions from hypoxic cells and accumulated upon proteasome inhibition also in the purified fractions from the normoxic cells. The membrane was reprobed to verify SUMO-2/3 levels in the purified samples. Ponceau S stained protein bands were used as loading controls (L.C.).
Sumoylated HIF-2α is regulated by SENP1, RNF4 and VHL
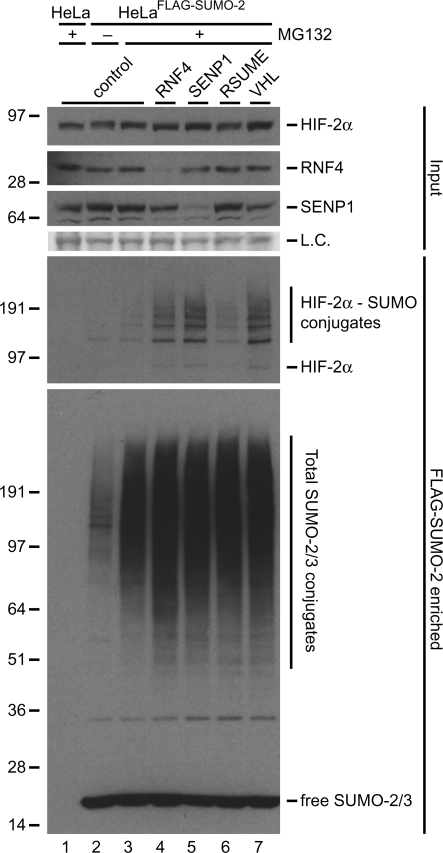
Proteasome inhibition strongly stabilized SUMO-2-conjugated HIF-2α without affecting the total levels of HIF-2α (Figures 5 and 6), indicating that HIF-2α might be controlled by SUMO-targeted ubiquitin ligases. RNF4 has previously been shown to act as a SUMO-targeted ubiquitin ligase (31), therefore we have performed RNF4 knockdown experiments (Figure 7). RNF4 depletion resulted in a significant accumulation of SUMO-modified HIF-2α, consistent with a role for RNF4 as a SUMO-targeted ubiquitin ligase for HIF-2α.
Figure 7.
Sumoylated HIF-2α is regulated by RNF4, SENP1 and VHL. HeLaFLAG-SUMO-2 cells were infected with lentiviruses encoding shRNAs against RNF4, SENP1, RSUME or VHL. The cells were cultured at 1% O2 in the presence of MG132 or DMSO for 7 h and lysed 96 h after infection. FLAG-SUMO-2 conjugates were enriched. Enriched fractions were separated by SDS–PAGE, transferred to membranes and probed with anti-HIF-2α or anti-SUMO-2/3 antibody. Total cell lysates were included as input controls and used to verify the knockdown efficiencies of RNF4 and SENP1. Ponceau S stained proteins were included to show equal loading (L.C.). An increase in the amount of HIF-2α-SUMO-2 conjugates was observed upon knockdown of RNF4, SENP1 and VHL compared to the control sample. RSUME knockdown did not affect the levels of HIF-2α-SUMO-2 conjugates.
Previously, the sumoylation of HIF-1α was shown to be regulated via the SUMO protease SENP1 (25) and via the SUMO E3 ligase RSUME (32). Cheng et al. (25) also proposed a role for VHL as a SUMO-targeted ubiquitin ligase in SUMO-dependent HIF-1α degradation during hypoxia. We used shRNA interference to reduce the expression levels of endogenous SENP1, RSUME, and VHL (Figure 7). Downregulating the expression of VHL resulted in a significant accumulation of SUMO-modified HIF-2α, consistent with a role for VHL as a second SUMO-targeted ubiquitin ligase for HIF-2α. Furthermore, we observed an increase in the levels of sumoylated HIF-2α upon SENP1 knockdown, consistent with a role for this SUMO protease in HIF-2α desumoylation. SENP1 knockdown did not affect the non-sumoylated pool of HIF-2α but did decrease the total pool of HIF-1α, consistent with the results published by Cheng et al. (25) (Figure 7 and Supplementary Figure S2). The shRNA against RSUME did not lead to a change in the level of SUMO-modified HIF-2α. However, we cannot rule out a role for RSUME in HIF-2α regulation as we were unable to verify the efficiency of the downregulation of this protein in our experiment.
DISCUSSION
We have shown here that HIF-2α is a target protein for SUMOs; both endogenous and exogenous HIF-2α were found to be sumoylated in cells. HIF-2α contains two consensus sumoylation sites, LK394EE and LK497IE, and mutational analysis showed that K394 is the major SUMO acceptor site in HIF-2α. The FLAG-HIF-2α K394R and E396A mutants showed a clear reduction in sumoylation; however, sumoylation was not totally absent, indicating the presence of at least one non-consensus site for SUMO modification. Functionally, sumoylation inhibited the transcriptional activity of HIF-2α.
Recently, we have shown that SUMO-2 conjugation affects the stability of a subset of conjugates (30). HIFs are primarily controlled via stability; they are degraded during normoxia and stabilized during hypoxia (33). Interestingly, SUMO-2 conjugation enabled the degradation of HIF-2α during hypoxia. Since the total pool of HIF-2α was not affected, SUMO-2 conjugation appeared to be a requirement for HIF-2α degradation during hypoxia. This indicated that HIF-2α can be controlled by SUMO-targeted ubiquitin ligases (25,31,34). RNF4 is an important mammalian SUMO-targeted ubiquitin ligase that controls the stability of sumoylated PML, PML-RARα and PEA3 (35–37). Consistently, SUMO-2-conjugated HIF-2α accumulated upon RNF4 knockdown. Similarly, VHL is a key regulator of HIF stability during normoxia and part of an ubiquitin E3 ligase complex (8,9). In addition, Cheng et al. (25) have previously reported the involvement of VHL in the degradation of sumoylated HIF-1α under hypoxic conditions. Our results suggest that VHL also controls the stability of sumoylated HIF-2α during hypoxia since VHL knockdown strongly increased the amount of SUMO-2-modified HIF-2α.
Different studies have reported conflicting roles for HIF-1α sumoylation (6,24–26,32). Carbia-Nagashima et al. (32) and Bae et al. (24) reported that sumoylation stabilizes and activates HIF-1α whereas Cheng et al. (25) and Berta et al. (6) reported that sumoylation inhibits HIF-1α. Cheng et al. (25) showed that in mice deficient for SENP1, the stabilization of HIF-1α during hypoxia was reduced, resulting in embryonic lethality due to severe fetal anemia. We have addressed whether SENP1 and RSUME also regulate HIF-2α activity. Knockdown of SENP1 increased the amount of HIF-2α-SUMO-2 conjugates, whereas knockdown of RSUME did not influence HIF-2α sumoylation.
Conjugated SUMOs can also act as a binding site for repressor proteins such as histone deacetylases (HDACs) (38,39). It appears that HDACs control HIF-2α since the HDAC-inhibitor TSA increased the activity of HIF-2α (Supplementary Figure S3). However, this was not dependent on HIF-2α sumoylation since the E396A mutant was similarly affected by TSA.
In contrast to the K394R and E396A mutations, the K497R and E499A mutations did not appear to reduce sumoylation of HIF-2α. Nevertheless, both sumoylation consensus sites are well conserved from zebrafish to human. Three consensus sumoylation sites are present in HIF-1α and two of these sites are situated in sequences which show a high degree of homology to sequences surrounding the consensus sites found in HIF-2α (Figure 1). Interestingly, the K391 sumoylation site in HIF-1α and the K394 sumoylation site in HIF-2α show a high degree of sequence similarity and both are used for SUMO conjugation. HIF-1α K532 and HIF-2α K497 also lie in highly similar sequence stretches and are both not used for SUMO conjugation. The presence of a second sumoylation site that is well conserved but appears not to be used for SUMO conjugation is puzzling. The site could potentially be inaccessible for the SUMO conjugation machinery. Alternatively, specific E3 factors might be required that mediate the conjugation of SUMOs to K497 but are missing in our assay. Furthermore, it is possible that sumoylation of K497 is restricted to specific stages of embryonic development or to specific cell types. The third HIF-1α consensus site, K477, has been shown to be used for SUMO conjugation, but no equivalent site is present in HIF-2α (6,25).
Rapidly growing cancer cells induce a hypoxic environment. Survival of these cells in such an environment is enabled, at least in part, by the HIF system. The HIF pathway is activated in many solid tumors and is therefore a major drug target for anti-cancer therapy (40). Overexpression of HIF-2α has been shown in several different tumor types and signifies an advanced tumor stage and/or poor patient survival outcome. By inducing the transcription of target genes, it contributes to vascularization of these tumors and thus to tumor growth and malignant behavior (14). Our results indicate that sumoylation inhibits HIF-2α. Inhibiting HIF-2α activity is expected to limit the growth rate of solid tumors. Potentially, stimulating the sumoylation or inhibiting the desumoylation of HIF-2α in tumors could be a novel strategy to limit tumor progression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Netherlands Organisation for Scientific Research (NWO).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Groot, Duyndam and Palvimo for generously providing reagents. They also thank M. Rabelink for assistance with lentivirus production and Dr Th. van Laar for insightful discussions.
REFERENCES
- 1.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 2.Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- 3.Chen A, Mannen H, Li SS. Characterization of mouse ubiquitin-like SMT3A and SMT3B cDNAs and gene/pseudogenes. Biochem. Mol. Biol. Int. 1998;46:1161–1174. doi: 10.1080/15216549800204722. [DOI] [PubMed] [Google Scholar]
- 4.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 5.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Berta MA, Mazure N, Hattab M, Pouyssegur J, Brahimi-Horn MC. SUMOylation of hypoxia-inducible factor-1alpha reduces its transcriptional activity. Biochem. Biophys. Res. Commun. 2007;360:646–652. doi: 10.1016/j.bbrc.2007.06.103. [DOI] [PubMed] [Google Scholar]
- 7.Wiesener MS, Turley H, Allen WE, Willam C, Eckardt KU, Talks KL, Wood SM, Gatter KC, Harris AL, Pugh CW, et al. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1alpha. Blood. 1998;92:2260–2268. [PubMed] [Google Scholar]
- 8.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin W.G., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 9.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 10.Bardos JI, Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim. Biophys. Acta. 2005;1755:107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro-oncology. 2005;7:134–153. doi: 10.1215/S1152851704001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp. Mol. Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 13.Hara S, Kobayashi C, Imura N. Nuclear localization of hypoxia-inducible factor-2alpha in bovine arterial endothelial cells. Mol. Cell Biol. Res. Commun. 1999;2:119–123. doi: 10.1006/mcbr.1999.0160. [DOI] [PubMed] [Google Scholar]
- 14.Lofstedt T, Fredlund E, Holmquist-Mengelbier L, Pietras A, Ovenberger M, Poellinger L, Pahlman S. Hypoxia inducible factor-2alpha in cancer. Cell Cycle. 2007;6:919–926. doi: 10.4161/cc.6.8.4133. [DOI] [PubMed] [Google Scholar]
- 15.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng J, Zhang L, Drysdale L, Fong GH. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc. Natl Acad. Sci. USA. 2000;97:8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compernolle V, Brusselmans K, Acker T, Hoet P, Tjwa M, Beck H, Plaisance S, Dor Y, Keshet E, Lupu F, et al. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat. Med. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- 19.Gort EH, van Haaften G, Verlaan I, Groot AJ, Plasterk RH, Shvarts A, Suijkerbuijk KP, van Laar T, van der Wal E, Raman V, et al. The TWIST1 oncogene is a direct target of hypoxia-inducible factor-2alpha. Oncogene. 2008;27:1501–1510. doi: 10.1038/sj.onc.1210795. [DOI] [PubMed] [Google Scholar]
- 20.van de Sluis B, Muller P, Duran K, Chen A, Groot AJ, Klomp LW, Liu PP, Wijmenga C. Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Mol. Cell Biol. 2007;27:4142–4156. doi: 10.1128/MCB.01932-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vellinga J, Uil TG, de Vrij J, Rabelink MJ, Lindholm L, Hoeben RC. A system for efficient generation of adenovirus protein IX-producing helper cell lines. J. Gene Med. 2006;8:147–154. doi: 10.1002/jgm.844. [DOI] [PubMed] [Google Scholar]
- 22.Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 23.Jaffrey E, Hay R. Detection of modification by ubiquitin-like proteins. Methods. 2006;38:35–38. doi: 10.1016/j.ymeth.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 24.Bae SH, Jeong JW, Park JA, Kim SH, Bae MK, Choi SJ, Kim KW. Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem. Biophys. Res. Commun. 2004;324:394–400. doi: 10.1016/j.bbrc.2004.09.068. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao R, Zhang FP, Tian F, Anders FP, Wang X, Sjoland H, Billig H. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1alpha in adult mouse brain and heart in vivo. FEBS Lett. 2004;569:293–300. doi: 10.1016/j.febslet.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 27.Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 29.Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, Andersen JS, Vertegaal AC. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell Proteomics. 2008;7:2107–2122. doi: 10.1074/mcp.M800025-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, Holsboer F, Arzt E. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 33.Brahimi-Horn MC, Pouyssegur J. HIF at a glance. J. Cell Sci. 2009;122:1055–1057. doi: 10.1242/jcs.035022. [DOI] [PubMed] [Google Scholar]
- 34.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo B, Sharrocks AD. Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol. Cell Biol. 2009;29:3204–3218. doi: 10.1128/MCB.01128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 37.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 38.Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 39.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 40.Hewitson KS, Schofield CJ. The HIF pathway as a therapeutic target. Drug Discov. Today. 2004;9:704–711. doi: 10.1016/S1359-6446(04)03202-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.