Abstract
Protein–protein interactions (PPIs) are ubiquitous in Biology, and thus offer an enormous potential for the discovery of novel therapeutics. Although protein interfaces are large and lack defining physiochemical traits, is well established that only a small portion of interface residues, the so-called hot spot residues, contribute the most to the binding energy of the protein complex. Moreover, recent successes in development of novel drugs aimed at disrupting PPIs rely on targeting such residues. Experimental methods for describing critical residues are lengthy and costly; therefore, there is a need for computational tools that can complement experimental efforts. Here, we describe a new computational approach to predict hot spot residues in protein interfaces. The method, called Presaging Critical Residues in Protein interfaces (PCRPi), depends on the integration of diverse metrics into a unique probabilistic measure by using Bayesian Networks. We have benchmarked our method using a large set of experimentally verified hot spot residues and on a blind prediction on the protein complex formed by HRAS protein and a single domain antibody. Under both scenarios, PCRPi delivered consistent and accurate predictions. Finally, PCRPi is able to handle cases where some of the input data is either missing or not reliable (e.g. evolutionary information).
INTRODUCTION
In order to fulfill their function, proteins must interact with one another and with other biomolecules, thus offering an enormous potential for the discovery of novel therapeutic agents able to act either as antagonist or agonist of protein–protein interactions (PPIs). Crystallographic studies have shown that proteins interact through large, typically 150–300 nm2 (1,2) [60 nm2 is the minimum area required to make a water-tight seal around a critical set of energetically favorable interactions (3)], and relatively featurelessness surfaces. Given these large interfacial areas, one school of thought considers that small-molecule inhibitors require binding-pockets or ‘clefts’ at the protein–protein interface, in order to attain the required affinities (4). However, as discussed by Wells and McLendon (5), this and a number of other objections to target the disruption of protein–protein interfaces have recently been challenged by new data [reviewed by Yin and Hamilton (6) and Wells and McLendon (5)].
Many of these successes have been aided by the realization, following the pioneering work of Clackson and Wells (7), that the binding energy for many protein–protein associations can be ascribed to a small and complementary set of interfacial residues—a hot spot—of binding energy surrounded by weaker interactions providing specificity. Protein interaction interfaces include many intermolecular contacts, involving 10–30 side chains on average from each protein. However, a typical hot spot accounts for less that half of the contact surface (5). The experimental detection of residues located in hot spots can be achieved by Alanine scanning mutagenesis (8), Alanine shaving (9) and residue grafting (9). These techniques are very time consuming, labor-intensive and involve a high economic cost. Therefore, there is a great interest in computational tools that can predict critical residues with high accuracy and thus, be used to aid and complement experimental efforts.
Several computational tools have been described in the past. These can be categorized in three groups depending on the information used for the prediction. Thus, methods that estimated the energetic contribution of each individual residue to the global binding energy, so called computational Alanine scanning (10–12), or by free-energy decomposition (13), have been proposed. Other methods exploit the structural features that are characteristic of hot spots such as solvent accessibility (14–16), atomic contacts (17), structural conservation (18), restricted mobility (19), and location in the interaction patch (20). Finally, a third type of method accounts for evolutionary information such as sequence conservation (15,21–23), sequence environment and evolutionary profile (24), and pattern mining (25). While theses attributes are informative, it has been found that individually they cannot unambiguously define hot spots (26).
In this article, we present a novel probabilistic method, Presaging Critical Residues in Protein interfaces (PCRPi), that combines these three main sources of information, namely energetic, structural and evolutionary determinants by using Bayesian Networks (BNs) (27,28). Many applications in Bioinformatics and Computational Biology use BNs (29–35) offering several clear advantages over alternative modeling approaches as it has been described elsewhere (36). We have developed and extensively benchmarked several BNs in a dataset of experimentally confirmed hot spots residues. Results show that PCRPi delivers robust and reliable predictions with a high accuracy even in cases where evolutionary information is missing or highly noisy. In addition, we have tested our prediction method with experimental Gly–Ala scanning analysis of a protein complex formed by HRAS and a single domain antibody (37). PCRPi predictions were highly accurate with the exception of one residue that was predicted as a critical but it was not confirmed by the mutagenesis analysis. Finally, when comparing with previously published methods (10,11,16,38), PCRPi showed a higher performance when the same dataset was analyzed.
MATERIAL AND METHODS
Dataset
The protein complexes used in this study were taken from four different sources: Alanine Scanning Energetics (ASEdb) (39) and Binding Interface (BID) (40) databases, and Kortemme and Baker’s (11) and Guerois et al.’s (10) works. We worked with 25 complexes (Table 1, Supplementary Data) summing up a total of 636 interface residues, 300 of which have been experimentally validated (78 experimentally confirmed hot spot residues, see next). We defined a residue as being critical or hot spot residue, if when mutated the difference in binding energy between the mutant and wild-type, unmutated complex is equal or larger than 2 Kcal mol−1. Finally, we derived two different datasets: Ab+, containing all 25 protein complexes and Ab− that does not include non-evolutionary related protein complexes (e.g. antigen–antibody). Both dataset, Ab+ and Ab−, were used for training and testing under cross-validation conditions. Additionally, the BID derived set used in Darnell et al. (38) and Tuncbag et al. works (16) was used for comparing PCRPi with previously described methods. The protein complexes 1dfj (K7; chain E), 1dzi (N154, Y157, Q215, D219, L220, T221, E256, H258; chain A), and 2nmb (Y2, I3; chain B) were excluded because the corresponding experimental data associated to the residues shown between parentheses could not be found in the BID database (40).
Defining interface residues
A given residue is part of a protein interaction surface if it has atomic contacts with a residue(s) that belong to any other protein in the complex. The atomic interactions between residues were described using the CSU program (41) and includes any type of non-bonded interactions (i.e. polar, hydrogen bonds and hydrophobic interactions). Two types of interface residues were considered: first, residues that have been experimental validated either as critical or non-critical, i.e. ddGbinding ≥ 2.0 Kcal mol−1 or ddGbinding < 2.0 Kcal mol−1 when mutated, that are part of the training and testing datasets, plainly referred as interface residues; and second, mirror residues, residues that are part of the interface but belong to other protein in the complex.
Bayesian network attributes
Interaction engagement index
The interaction engagement (IE) index gauges the proportion of side chain atoms (including the main chain amino nitrogen and carboxyl oxygen) of a given interface residue i that have non-bonded interactions with mirror residues. Non-bonded atomic interactions were described using CSU. IE values range between 0 and 1 and were calculated using the following formula (1):
| 1 |
An IE index of 1.0 indicates that all atoms in the residue are actively engaged in atomic interactions with other proteins in the complex.
Topographical index (TOP)
The Topographical (TOP) index estimates the structural microenvironment of a given interface residue i and was calculated as the ratio between structurally neighbor residues and the average number of residues that a given residue type (e.g. Ala) interacts with when located at a protein interface (2):
| 2 |
By structurally neighbor residues is understood any mirror residues whose carbon alpha is enclosed in a sphere of 10 Å of radius centered on the carbon alpha of the given residue. The average number of contacts by residue type is shown in Table 2 (Supplementary Data), and it was calculated as follow: a non-redundant dataset of protein complexes was downloaded from the PiQSi database (42). Atomic contacts between protein subunits were assigned as shown in previous section. Interacting residues were grouped by residue type and statistical parameters were derived.
Table 2.
Comparison of different methods for the prediction of critical residues in protein interfaces using BID dataset
| Method | Precision (P) | Recall (R) | F1 score |
|---|---|---|---|
| PCRPi (Ab+)a | 0.79 | 0.64 | 0.71 |
| FoldXb | 0.75 | 0.36 | 0.49 |
| Robetta-Alac | 0.63 | 0.57 | 0.60 |
| KFCc | 0.51 | 0.36 | 0.42 |
| KFC-Ac | 0.53 | 0.48 | 0.51 |
| LDAc | 0.72 | 0.57 | 0.64 |
| Tuncbag et al. (16)c | 0.73 | 0.59 | 0.65 |
aPredictions were performed on the default expert BN (Figure 2A) using the Ab+ dataset as training set previous exclusion of 1fcc protein complex.
bValues were obtained running FoldX(10) with default parameters and a ddGbinding cut-off of 2.0 Kcal mol−1 (i.e. residues were considered critical if predicted ddGbinding ≥ 2.0 Kcal mol−1).
cPrecision, recall and F1 score values taken from Tuncbag et al. (16).
Sequence conservation index (CON and ANCCON)
The CON index refers to the conservation of mirror residues that are in contact with a given interface residue, whereas ANCON index is the conservation of the interface residue. To analyze the conservation, sequence profiles were derived as described in our previous work (43). In short, homologous sequences were culled from NR database of NCBI (44) using five iterations of PSI-BLAST (45) with an E-value of 0.0001. The homologous sequences were then filtered using ParseBlast (46) with default parameters to maximize the sequence sampling avoiding bias toward overrepresented protein families. The resulting sequence profile was given to al2co program (47) as an input and the al2co sequence conservation score was assigned to each residue.
For a given interface residue i, the CON index measure the ratio between mirror residues with an al2co_score ≥1.0 and the total number of mirror residues (#mirror_res) in contact with residue i (3).
| 3 |
The ANCCON index refers to the raw al2co sequence conservation score applied to each individual interface residue.
3D regional conservation index (3DCON and ANC3DCON)
The 3D regional conservation score was calculated using the same sequence profiles but using the normalized (Z-score) regional conservation score (CR) as defined by Landgraf et al. (48) as a measure of conservation. For a given interface residue i, the 3D regional conservation index is the ratio between mirror residues with a CR score ≥1.0 and the total number of mirror residues (#mirror_res) in contact with the given interface residue i (4).
| 4 |
The ANC3DCON index is the CR score of the given interface residue as derived from the multiple sequence alignment.
Computational alanine scanning (BE)
The last attribute used was the difference in estimated binding energy upon mutation to Alanine, i.e. computational Alanine scanning. Each interface residue was mutated to Alanine and the effect of such mutation in the stability of the protein complex was estimated using FoldX (10). Crystallographic waters, if any, were kept during the free energy calculations. For a given interface residue i, BE reflects the difference in binding free energy between the unmutated (wild-type) and mutated complex (5).
| 5 |
The use of Rosetta to estimate binding energy was also explored during the developmental phases of the project. There were however, in terms of performance, not big differences between Rosetta and FoldX when used as inputs to our BNs.
Bayesian Networks
The BNs were implemented using the Bayesian network toolbox for Matlab (BNT) (49). Additionally, the R package ‘Deal’ was used to learn the structure of expert BNs (50).
Training phase
Naïve and expert BNs were trained using the two datasets considered in this study: Ab+ and Ab−. In a naïve BN, all input variables (or attributes) are assumed to be independent and directly connected to the predictor, or class node, whereas in an expert BN, attributes are not assumed to be independent and conditional dependence between attributes is allowed. The maximum likelihood estimation (MLE) (51,52) and the expectation maximization (EM) (53) were used to learn the parameters of the BNs. Bayesian network structures (i.e. expert BNs) were learnt using a score-based approach implemented in R package (Deal) (50). The different BNs that were explored during this study can be found inTables 3 and 4 of the Supplementary Data.
Prediction phase
Some of the most promising trained BNs were then used as predictors. A 10-fold cross-validation experiment was used to assess the accuracy of the predictions. Each of the interface residues in our datasets was randomly assigned to one to the 10 subsets, where one of the subsets was selected as validation data while the remaining 9 subsets were used as a training set. Additionally, a leave-one-out cross-validation experiment was also performed. Each individual protein complex was selected as validation data, whereas the rest of protein complexes were used as training set.
Assessing the performance of the BNs
The datasets contain positives (P; i.e. experimentally confirmed critical residue) and negatives (N; i.e. experimentally confirmed non-critical residue) cases. The BNs assign a probability score between 0.0 and 1.0 to each residue under test. Consequently, classification performance depends on the probability threshold (any value between 0.0 and 1.0) above which a residue is predicted as critical and below is predicted as non-critical. The performances of the BNs were evaluated in terms of sensitivity, specificity, precision, F1 score and accuracy. A more extensive explanation of these statistical metrics is given in the Supplementary Data.
Using sensitivity and specificity values, the receiver operating characteristic (ROC) curves were plotted and subsequently the area under ROC curves (AUC) was calculate to evaluate model performance. A ROC curve plots sensitivity versus (1-specificity) across a range from 0.0 to 1.0 of probability thresholds. The AUC represents the area beneath the ROC curve, where a value 1.0 being indicative of a perfect classifier and of 0.5 as a classifier that is no better than random.
Gly–Ala mutagenesis analysis of anti-RAS VH–HRAS complex
A detailed explanation of reagents and site-directed mutagenesis procedure can be found at the Supplementary Data. The effect of mutations in the ability of anti-RAS VH single domain antibody to bind HRAS was estimated using a mammalian two-hybrid luciferase assay as follow. Chinese hamster ovary (CHO) cells were grown in DMEM medium with 10% fetal calf serum containing penicillin and streptomycin. A Firefly luciferase reporter CHO cell line was established by co-transfecting CHO cells with linearized pG5-Fluc (a plasmid with a minimal promoter linked to five copies of the GAL4 DNA binding sequence) (Promega) and pPGK-puro (54) plasmids using Lipofectamine 2000 (Invitrogen). Stably transfected cells were selected for 7 days using 10 µg/ml puromycin (Sigma). The CHO-Luc stable clone 15 (CHO-Luc15) was chosen for further assays. For luciferase assays, the CHO-Luc15 were seeded in 12-well culture plates the day before transfection and grown until more than 90% confluent. One microgram triplex vector was transfected to obtain co-expression of a mutant form of VH#6-VP16 fusion (prey) with GAL4DBD-HRASG12V (or control) bait and the Renilla luciferase for normalising transfection efficiencies using 2 µl Lipofectamine 2000 according to the manufacturer’s instructions. After 48 h, the cells were harvested, lysed and assayed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. The data represent a minimum of three experiments for each point and each of which was performed in duplicate. Values are normalised for stimulated Firefly luciferase levels compared with levels for transfected Renilla luciferase.
RESULTS
Rationale behind individual measurement and Ab+Ab− dataset
PCRPi exploits seven measures of different nature to predict whether a given residue is going to be critical in a specific PPI. Two of the measures, IE and TOP, utilize structural information. IE is a simple measure that gauges the fraction of atoms of a given residue that are actively engaged in atomic contacts with the other proteins in the complex, whereas the TOP index quantifies whether an interface residue is interacting intimately with partner proteins or is located in a more flat or unprotected region. The second group of variables account for evolutionary information. The CON and 3DCON reveal conservation in mirror residues and therefore reward interface residues that interact with conserved mirror residues (CON) or conserved patches (3DCON), whereas ANCCON and ANC3DCON gauge the conservation of the individual interface residues. Finally, BE determines the predicted energetic contribution of each individual interface residue to the strength of the interaction. Whether mutations in specific residues would result in a less stable complex is estimated by this last metric.
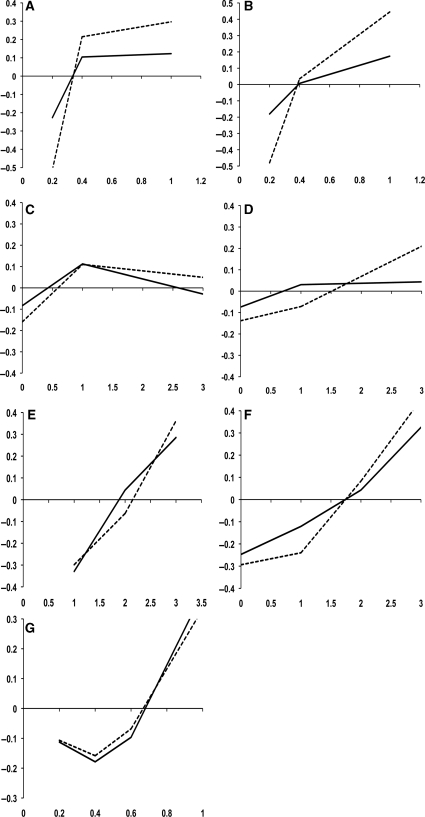
Overall, all but evolutionary-based measures are good discriminating between critical and non-critical residues (Figure 1). It is clear that as TOP, IE and BE values increase (Figure 1E–G; solid line), the difference between experimentally critical and non-critical residues becomes more positive, i.e. critical residues show a distribution of TOP, IE and BE values skewed toward high values as compared with non-critical residues. This trend is not observed in evolutionary-based variables (Figure 1A–D; solid line). A careful analysis of the protein complexes in the dataset revealed the presence of what we are going to term non-evolutionary related protein complexes. By non-evolutionary related protein complexes it is understood protein complexes that do not have a common evolutionary history. The best example of non-evolutionary related protein complexes is the antigen–antibody. Antibodies undergo an accelerate evolution adjusting the amino acid composition at the complementarity determining regions (CDRs) to improve the binding to a given antigen. On the other hand, protein complexes that have a common evolutionary history, mutually adjust changes in the protein sequence in order to preserve the interaction (55).
Figure 1.
(A–G) Discriminative power of each individual measure used as inputs to the BNs. Y-axis shows the difference between the frequency of critical and non-critical residue at a given score point (X-axis). Negatives values indicate that the frequency of non-critical residues is higher than the frequency of critical residues for the given score of the particular measure: CON (A), 3DCON (B), ANCCON (C), ANC3DCON (D), TOP, (E), BE (F) and IE (G) on Ab+ dataset (solid line) and Ab− dataset (dashed line). For frequency calculations, IE, CON and 3DCON scores were binned by increments of 0.2, whereas TOP, BE, ANCCON and ANC3DCON scores, a 1.0 bins were used.
Based on the previous observation, the initial dataset was divided in two: Ab+ (effectively the initial set) and Ab− dataset. The difference between Ab+ and Ab− datasets is the presence or absence of non-evolutionary related complexes, respectively (Table 1, Supplementary Data). Using the Ab+ dataset, CON, ANCCON, 3DCON and ANC3DCON measures are unable to distinguish between critical and non-critical residues. Even at very high scores (i.e. high sequence conservation) the difference between frequency of critical and non-critical residues was close to 0; thus, critical and non-critical residues were indistinguishable (Figure 1A–D; solid line). However, when scores were re-calculated and plotted using the Ab− dataset a clear improvement was observed: evolutionary-based metrics were able to distinguish between critical and non-critical residues (Figure 1A–D; dashed line). Furthermore, the attributes that are sequence conservation-independent behave similarly regardless of the dataset used (Figure 1E–G; solid and dashed line). Ab+ dataset was still used both during the training and testing phases to emulate cases where BNs have to deal with noisy or missing input data (meaningless evolutionary-based data in this particular case).
Training phase
Both the Ab+ and Ab− datasets were used to train 60 naïve and 25 expert BNs originating from the combination of one, two, three, four, five, six and seven attributes. The performance of the BNs was assessed in terms of area under the ROC curves (AUC) values. The summary of BNs that were trained and their respective performances are shown in Tables 3 and 4 (Supplementary Data).
Using naïve BNs, the largest AUC correspond to the BN that combines TOP, BE, 3DCON and ANC3DCON measures (Table 3, Supplementary Data). More complex BNs (i.e. use more input variables) showed marginal differences when compare with top performer BN. There was however a clear improvement when comparing with BNs with only one attribute. Thus, individual measures alone showed a reasonable prediction power but when combined the performance was much improved. In the case of expert BNs, the largest AUC is observed when all the measures are combined (Table 4, Supplementary Data). AUC values were higher than the top naïve BN. In addition, expert BNs seemed more robust, since the difference in AUC using Ab+ and Ab− databases were smaller (i.e. more similar AUC values).
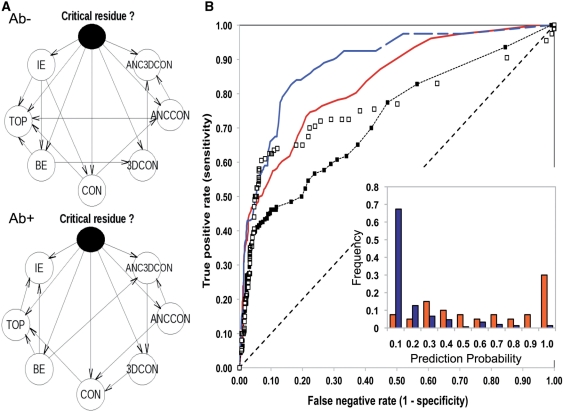
Given the differences observed between naïve and expert BNs and the better performance of expert BNs under both Ab+ and Ab− datasets; two expert BNs (tailored for the Ab+, Ab− datasets) that combine all seven measures were selected as default predictors (Figure 2A for schematic representation of the BNs’ architectures), and used in all subsequent analyses.
Figure 2.
Predictive performances of default BNs combining IE, TOP, CON, ANCCON, 3DCON, ANC3DCON and BE measures. (A) Schematic representation of the default BNs’ architectures. Nodes represent each of the variables and the arrows represent conditional dependence relationships. The predictor or class node is shown as a solid node. (B) ROC curves default BNs during 10-fold cross-validation prediction on the Ab+ (red solid line) and the Ab− (blue dashed line) datasets. Resulting ROC curves using only BE are also shown: solid and empty squares for Ab+ and Ab− datasets respectively. Inset shows the distribution of prediction probabilities obtained using the default BN in the Ab− dataset as a function of nature of interface residues, i.e. critical (red) and non-critical (blue).
Prediction phase
The expert BNs that were explored and selected during the training phase were taken forward to the prediction phase (Figure 2A). In addition, the naïve version of default BNs (i.e. seven attributes directly connected to the class node and without connections between them) and single attributes BNs were used as control. The predictive performance of the BNs was evaluated in a 10-fold and in a leave-one-out cross-validation experiments reporting AUC and accuracy. Eight and two naïve and expert BNs, respectively were examined during the cross-validation experiments.
As observed during the training phase, the combination of attributes resulted in a better performance, and the best predictions, in terms of accuracy and AUC values, were achieved with default BNs (Table 1). A slightly decrease in performance is observed in those cases were sequence conservation measures are used on the Ab+ set, again highlighting the robustness of the BNs. This property is specific of BNs and it is difficult to emulate with other learning methods. Figure 2B shows the ROC curves based on the probabilities calculated with the default BNs. The AUC and accuracy of prediction of default BNs in the case of the Ab+ and Ab− datasets were 0.83, 0.89, 0.81 and 0.84, respectively (Table 1). Comparing with the AUC values with the best individual measure: BE, default BNs achieved higher sensitivity for similar a range of false negative rate values, both in Ab+ and Ab− datasets (Figure 2B). Finally, when the distribution of prediction probabilities is plotted as function of the importance of the interface residue, i.e. critical or non-critical, a clear correlation can be observed, i.e. residues that are predicted with high probability are more likely to be important in the interaction, i.e. critical (Figure 2B, inset).
Table 1.
Accuracy and AUC values for different BNs (naïve and expert) tested during 10-fold cross validation process on Ab+ and Ab− datasets
| Naïve BNs |
Expert BNs |
|||||||
|---|---|---|---|---|---|---|---|---|
| Measuresa | AUC |
Accuracy |
AUC |
Accuracy |
||||
| Ab+ | Ab− | Ab+ | Ab− | Ab+ | Ab− | Ab+ | Ab− | |
| IE | 0.75 | 0.72 | 0.76 | 0.74 | – | – | – | – |
| TOP | 0.74 | 0.72 | 0.71 | 0.77 | – | – | – | – |
| BE | 0.72 | 0.76 | 0.71 | 0.85 | – | – | – | – |
| CON | 0.52 | 0.69 | 0.72 | 0.76 | – | – | – | – |
| 3DCON | 0.51 | 0.66 | 0.68 | 0.71 | – | – | – | – |
| ANCCON | 0.44 | 0.49 | 0.51 | 0.56 | – | – | – | – |
| ANC3DCON | 0.58 | 0.61 | 0.71 | 0.77 | – | – | – | – |
| IE, TOP, BE, CON, 3DCON, ANCCON, ANC3DCON | 0.82 | 0.88 | 0.79 | 0.86 | 0.83 | 0.89 | 0.81 | 0.84 |
aBN attributes as described in the ‘Material and Methods’ section.
A leave-one-out cross-validation was also performed to evaluate the predictive performance of the method. Given the large differences in mutational information that is available for each of the protein complexes and in order to avoid the bias that would be introduced by these complexes contributing the most to the statistical analysis, the performance of the method was assessed in terms of the ability of recover critical residues as a function of screened residues. A critical residue was considered to be ‘recovered’ if the prediction probability was equal or higher than 0.8. Figure 1 (Supplementary Data) shows the percentage of recovered critical residues upon prediction by using the default expert BN (Figure 2A, Ab+) and the naïve version. On average, default BN was able to recover up to 75% of critical residues, i.e. 75% of the actual critical residues were predicted with a probability equal or higher than 0.8 (using a naïve BN the percentage of recovery was 68%).
Prediction examples
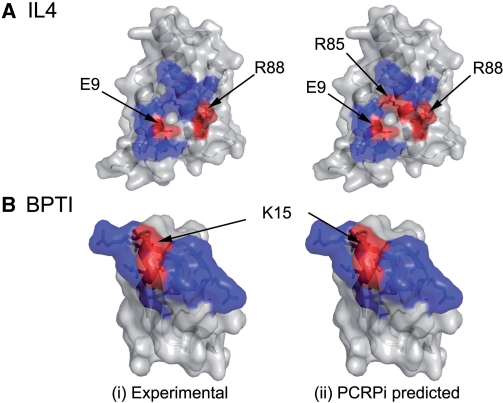
The protein complex formed by interleukin 4 and receptor alpha (PDB identification code 1iar) is one of the complexes that was analyzed (56). Two out of 20 residues that mediate the interaction between interleukin 4 and the receptor alpha have been experimentally verified as critical residues. PCRPi prediction was highly accurate when compared with the available experimental data (10) (Figure 3A). Arg85 (following the PDB numbering) was also predicted as a critical residue, although the decrease in binding energy according to the available mutational data is only 0.41 Kcal mol−1. As it is shown in Figure 3A, Arg85 is located in the center of the interaction patch, flanked by two important residues Arg88 and Glu9. Arg88 is a polar residue with a long side chain that protrudes the surface and makes extensive atomic contacts with the receptor alpha, hence a priori a clear candidate to be an important residue in the interaction.
Figure 3.
Comparison between experimentally determined and predicted critical residues. (A and B) The surface representation of the interaction surface of the human interleukin 4 (PDB code 1iar, chain A), and the bovine basic pancreatic trypsin inhibitor (BPTI) (PDB code 1cbw, chain D), respectively. Residues depicted in red were either experimentally determined (ddGbinding ≥ 2.0 Kcal mol−1 when mutated) (i) or predicted (prediction probability ≥ 0.8) as critical residues (ii). Conversely, residues depicted in blue are either non-critical according to experimental data (i) or were predicted as non-critical (ii).
A second example is the protein complex formed by chymotrysin and the basic pancreatic trypsin inhibitor (BPTI) (PDB identification code 1cbw) (57). BPTI interacts with chymotrypsin through an interface that includes 15 residues, one of which, Lys15, was proved to be critical for BPTI binding (57). As it is shown in Figure 3B, PCRPi prediction was 100% accurate, since all validated critical and non-critical residues were predicted as such.
Validating the PCPRi predictions in the VH–Anti-RAS–HRAS complex
PCRPi was also used to predict the critical residues mediating the interaction between the VH domain of an Fv antibody fragment that binds to mutant HRAS with high affinity and specificity (37). This was a blind prediction and not just validated data compiled from the scientific literature, and a likely application for the computational tool that is presented here. HRAS and anti-RAS VH interact through a large surface of ∼800 Å2 that comprises residues located across all three CDR loops of the VH domain.
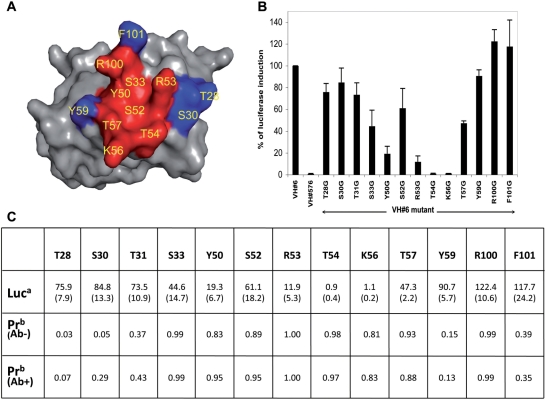
PCRPi predicted eight residues as forming a critical cluster around the central region of the interface (Figure 4A). As an approach to verify the PCPRi predictions, residues in the VH CDR loops were mutated individually into Gly/Ala and the effect of mutations were determined in a mammalian cell transfection assay using luciferase production as a reporter of interaction of the anti-RAS VH with HRAS. The effects of these mutations on the ability of the VH to interact with HRAS are shown in Figure 4B, where for example R53G/A, T54G/A and K56G/A mutations proved ablative of binding. Considering a residue as critical to the interaction if when mutated the percentage of luciferase induction fell below 60% of that seen with unmutated anti-RAS VH, the PCRPi method was able to predict all critical and non-critical residues shown by mutation with the exception of Arg100 that was predicted as critical when in fact its mutation showed no effect in luciferase induction (Figure 4B and C).
Figure 4.
Analysis of PCRPi predictions and experimental verification on the anti-RAS VH-HRAS complex (36). (A) Surface representation of the interaction surface of the anti-RAS VH. Depicted in red and in blue respectively, residues predicted as critical and non-critical by PCRPi (at prediction probability cut-off of 0.8). (B) Fold luciferase induction in CHO-luc cells transfected with DBD-HRASG12V and VH#6-VP16 in which the mutation of the indicated VH amino acids was done. Anti-LMO2 VH#576 (T.T. and T.H.R., manuscript in preparation) is presented as negative control as it does not interact with HRAS. Error bars represent SD. (C) Comparison of the average change in luciferase induction: Luc (standard deviation shown in brackets), and prediction probability: Pr using default BNs trained in Ab+ and Ab− datasets. Mutated residues that resulted in a drop in luciferase induction below 60% were considered as critical for the interaction.
DISCUSSION
The PCRPi method
In the present work, we have devised a novel method, PCRPi, to predict critical residues in protein interfaces. We have characterized residues located in protein interfaces using seven different measures or attributes. Each of the attributes account for a different aspect of the nature of interface residues. We have trained a number of BNs to distinguish between experimentally verified critical and non-critical residues from a benchmark dataset of 25 protein complexes. The results have shown that the prediction accuracy improves as the number of measures that are combined increases. Also, expert BNs showed better performance than naïve BNs.
PCRPi is also able to handle situations where some data are missing and/or not reliable as in these the overall performance was comparable to that of those where full reliable data is given. This fact is portrayed in the study of the Ab+/Ab− datasets. As explained before, some of the protein complexes included in Ab+ are non-evolutionary related, such as antigen–antibody complexes (Table 1, Supplementary Data). For seven of these complexes, mutational data comes from residues changes in the antigen, therefore, the CON and 3DCON data refers to sequence conservation in antibodies; in the remaining complexes the available mutational data comes from mutations in the antibody. In either way, evolutionary-based measures were showing not conservation for critical residues, either themselves (ANCCON and ANC3DCON) or in the mirror residues (CON and 3DCON). However, as data shows, PCRPi is a robust predictor that compensate for the lack of evolutionary information, as the performance achieved was comparable to that of Ab− dataset.
Besides the benchmark of PCRPi using a validated set, PCRPi has been also applied to the study of the protein complex formed by HRAS and a single domain antibody (37) to which we have direct access. As it is shown in Figure 4, all residues predicted as critical (with the exception of one) and non-critical were subsequently experimentally confirmed, again proving the usefulness of the proposed method as a complement and guidance to experimentally driven research.
Comparison of PCPRi with existing approaches
Comparing with previously published works is difficult mainly because the heterogeneity of the datasets employed to benchmark the methods and sometimes the difficulty to access the methods themselves. However, we compared our results with related works that employs a predictive model based in decision trees (38) and a recently published method (16), plus two energy-based methods: Robetta-Ala (11) and FoldX (10) that are freely available, using a comparable dataset: the BID derived database.
In terms of sensitivity and specificity PCRPi delivers a higher recall and precision that the top ranking method (Table 2) with an overall accuracy of 0.75 (comparing to an accuracy of 0.63, 0.64, and 0.70 on Robetta-Ala, FoldX and Tuncbag et al. (16), respectively). Regarding the F1 scores, PCRPi predictions have the best balance between precision and recall rates (Table 2). The F1 score is a robust metric that gauges the relationship between precision and the recall rates, hence high F1 score means an equilibrate balance between precision and recall rates.
CONCLUSION
In this work, we developed a novel method, PCRPi, to predict critical residues in protein interfaces. PCRPi relies in the integration of seven different types of sources of information by using BNs. Each individual measure provides a different type of information, i.e. energetic, structure-based and sequence-based features. We trained a number of BNs to distinguish between critical and non-critical residues taken from a benchmark dataset of 25 protein complexes. We found that by adding new information to the BN and also by adding new connections between attributes, i.e. expert BNs, the prediction accuracy was improved. PCRPi delivers very consistent predictions both under benchmarking and real cases scenarios (anti-RAS VH–HRAS complex). We also emulated and tested the robustness of the method by incorporating noisy data (i.e. evolutionary-based information). The overall performance of the BN when handling missing or noisy evolutionary information (Ab+ dataset) was comparable to that of evolutionary information is reliable (Ab− dataset). Comparing to current methods, PCRPi delivers better predictions both in terms of precision and recall and also in terms of F1 scores that measure the balance between precision and recall.
Finally, PCRPi will be a great help in the study of protein complexes and understanding the individual residue contribution to the global interaction, as well as a tool to select candidate residues for mutagenesis studies and as a complement to experimental studies. Our approach can also contribute to protein design experiments by predicting not only the important residues (i.e. ‘untouchable’) but also those that play a less important role and therefore can be selected to improve and enhance the binding between proteins. Lastly, our approach can be invoked in the process of drug discovery of novel therapeutic agents to target PPI because it allows the highlighting of the important residues that are mediating such interactions to facilitate further computational manipulation of chemical mimics.
The PCRPi algorithm and datasets used in this study are available upon request to the corresponding author. A web-server is currently being developed to allow a remote access to the method.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Councils United Kingdom (RCUK) Academic Fellowship scheme (to N.F.F.); Medical Research Council (MRC to T.T. and T.H.R.); an internal grant awarded by the Leeds Institute of Molecular Medicine and a Wellcome ViP award (to S.A.S.). Funding for open access charge: Research Councils United Kingdom.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
N.F.F. thanks Dr E. Gendra for critical reading of the manuscript and Ms Martina F. Gendra for insightful and stimulating discussions during the initial phases of project. N.F.F. acknowledges constructive inputs and suggestions of one the two anonymous reviewers.
REFERENCES
- 1.Jones S, Thornton JM. Principles of protein-protein interactions. Proc. Natl Acad. Sci. USA. 1996;93:13. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 3.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 4.Blundell TL, Sibana BL, Montalvao RW, Brewerton S, Chelliah V, Worth CL, Harmer NJ, Davies O, Burke D. Structural biology and bioinformatics in drug design: opportunities and challenges for target identification and lead discovery. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 2006;361:413–423. doi: 10.1098/rstb.2005.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells JA, McLendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 6.Yin H, Hamilton AD. Strategies for targetting protein-protein interactions with synthetic agents. Ang. Chem. Int. Edn. 2005;44:2–35. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- 7.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 8.Wells JA. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 9.Jin L, Wells JA. Dissecting the energetics of an antibody-antigen interface by alanine shaving and molecular grafting. Protein Sci. 1994;3:2351–2357. doi: 10.1002/pro.5560031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 11.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl Acad. Sci. USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreira IS, Fernandes PA, Ramos MJ. Computational alanine scanning mutagenesis—an improved methodological approach. J. Comput. Chem. 2007;28:644–654. doi: 10.1002/jcc.20566. [DOI] [PubMed] [Google Scholar]
- 13.Lafont V, Schaefer M, Stote RH, Altschuh D, Dejaegere A. Protein-protein recognition and interaction hot spots in an antigen-antibody complex: free energy decomposition identifies “efficient amino acids”. Proteins. 2007;67:418–434. doi: 10.1002/prot.21259. [DOI] [PubMed] [Google Scholar]
- 14.Landon MR, Lancia DR, Jr, Yu J, Thiel SC, Vajda S. Identification of hot spots within druggable binding regions by computational solvent mapping of proteins. J. Med. Chem. 2007;50:1231–1240. doi: 10.1021/jm061134b. [DOI] [PubMed] [Google Scholar]
- 15.Guney E, Tuncbag N, Keskin O, Gursoy A. HotSprint: database of computational hot spots in protein interfaces. Nucleic Acids Res. 2008;36:D662–D666. doi: 10.1093/nar/gkm813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009;25:1513–1520. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Zhao B, Cui Z, Gan J, Sakharkar MK, Kangueane P. Identification of hot spot residues at protein-protein interface. Bioinformation. 2006;1:121–126. doi: 10.6026/97320630001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Keskin O, Ma B, Nussinov R, Liang J. Protein-protein interactions: hot spots and structurally conserved residues often locate in complemented pockets that pre-organized in the unbound states: implications for docking. J. Mol. Biol. 2004;344:781–795. doi: 10.1016/j.jmb.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 19.Yogurtcu ON, Erdemli SB, Nussinov R, Turkay M, Keskin O. Restricted mobility of conserved residues in protein-protein interfaces in molecular simulations. Biophys. J. 2008;94:3475–3485. doi: 10.1529/biophysj.107.114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keskin O, Ma B, Nussinov R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 21.Hu Z, Ma B, Wolfson H, Nussinov R. Conservation of polar residues as hot spots at protein interfaces. Proteins. 2000;39:331–342. [PubMed] [Google Scholar]
- 22.Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl Acad. Sci. USA. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma B, Nussinov R. Trp/Met/Phe hot spots in protein-protein interactions: potential targets in drug design. Curr. Top Med. Chem. 2007;7:999–1005. doi: 10.2174/156802607780906717. [DOI] [PubMed] [Google Scholar]
- 24.Ofran Y, Rost B. Protein-protein interaction hotspots carved into sequences. PLoS Comput. Biol. 2007;3:e119. doi: 10.1371/journal.pcbi.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu CM, Chen CY, Liu BJ, Huang CC, Laio MH, Lin CC, Wu TL. Identification of hot regions in protein-protein interactions by sequential pattern mining. BMC Bioinformatics. 2007;8(Suppl. 5):S8. doi: 10.1186/1471-2105-8-S5-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 27.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of plausible inference. San Francisco: Morgan Kaufmann Publisher Inc; 1988. [Google Scholar]
- 28.Jordan MI. Boston, USA: Kluwer Academic Publishers; 1988. Learning in graphical models. [Google Scholar]
- 29.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 30.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc. Natl Acad. Sci. USA. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pudimat R, Schukat-Talamazzini EG, Backofen R. A multiple-feature framework for modelling and predicting transcription factor binding sites. Bioinformatics. 2005;21:3082–3088. doi: 10.1093/bioinformatics/bti477. [DOI] [PubMed] [Google Scholar]
- 32.Cai D, Delcher A, Kao B, Kasif S. Modeling splice sites with Bayes networks. Bioinformatics. 2000;16:152–158. doi: 10.1093/bioinformatics/16.2.152. [DOI] [PubMed] [Google Scholar]
- 33.Husmeier D. Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics. 2003;19:2271–2282. doi: 10.1093/bioinformatics/btg313. [DOI] [PubMed] [Google Scholar]
- 34.Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- 35.Bradford JR, Needham CJ, Bulpitt AJ, Westhead DR. Insights into protein-protein interfaces using a Bayesian network prediction method. J. Mol. Biol. 2006;362:365–386. doi: 10.1016/j.jmb.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 36.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. Inference in Bayesian networks. Nat. Biotechnol. 2006;24:51–53. doi: 10.1038/nbt0106-51. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka T, Williams RL, Rabbitts TH. Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J. 2007;26:3250–3259. doi: 10.1038/sj.emboj.7601744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darnell SJ, Page D, Mitchell JC. An automated decision-tree approach to predicting protein interaction hot spots. Proteins. 2007;68:813–823. doi: 10.1002/prot.21474. [DOI] [PubMed] [Google Scholar]
- 39.Thorn KS, Bogan AA. ASEdb: a database of alanine mutations and their effects on the free energy of binding in protein interactions. Bioinformatics. 2001;17:284–285. doi: 10.1093/bioinformatics/17.3.284. [DOI] [PubMed] [Google Scholar]
- 40.Fischer TB, Arunachalam KV, Bailey D, Mangual V, Bakhru S, Russo R, Huang D, Paczkowski M, Lalchandani V, Ramachandra C, et al. The binding interface database (BID): a compilation of amino acid hot spots in protein interfaces. Bioinformatics. 2003;19:1453–1454. doi: 10.1093/bioinformatics/btg163. [DOI] [PubMed] [Google Scholar]
- 41.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 42.Levy E. PiQSi: protein quaternary structure investigation. Structure. 2007;15:1364–1367. doi: 10.1016/j.str.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez-Fuentes N, Rai BK, Madrid-Aliste CJ, Fajardo JE, Fiser A. Comparative protein structure modeling by combining multiple templates and optimizing sequence-to-structure alignments. Bioinformatics. 2007;23:2558–2565. doi: 10.1093/bioinformatics/btm377. [DOI] [PubMed] [Google Scholar]
- 44.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rai BK, Madrid-Aliste CJ, Fajardo JE, Fiser A. MMM: a sequence-to-structure alignment protocol. Bioinformatics. 2006;22:2691–2692. doi: 10.1093/bioinformatics/btl449. [DOI] [PubMed] [Google Scholar]
- 47.Pei J, Grishin N. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 48.Landgraf R, Xenarios I, Eisenberg D. Three-dimensional cluster analysis identifies interfaces and functional residue clusters in proteins. J. Mol. Biol. 2001;307:1487. doi: 10.1006/jmbi.2001.4540. [DOI] [PubMed] [Google Scholar]
- 49.Murphy B. The Bayes Net Toolbox for Matlab. Comput. Sci. Stat. 2001;33:331–350. [Google Scholar]
- 50.Bottcher S, Dethlefsen C. Deal: a package for learning Bayesian networks. J. Stat. Software. 2003;8:1–40. [Google Scholar]
- 51.Sham P. Statistics in Human Genetics. London: Edward Arnorld; 1998. [Google Scholar]
- 52.Edwards A. What did Fisher mean by “inverse probability” in 1912–1922? Stat. Sci. 1997;12:177–184. [Google Scholar]
- 53.Mclachlan G, Krishnan T. The EM Algorithm and Extensions. New york, USA: John Wiley & Sons; 1997. [Google Scholar]
- 54.Tucker KL, Beard C, Dausmann J, Jackson-Grusby L, Laird PW, Lei H, Li E, Jaenisch R. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 1996;10:1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- 55.Pazos F, Valencia A. Protein co-evolution, co-adaptation and interactions. EMBO J. 2008;27:2648–2655. doi: 10.1038/emboj.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hage T, Sebald W, Reinemer P. Crystal structure of the interleukin-4/receptor alpha chain complex reveals a mosaic binding interface. Cell. 1999;97:271–281. doi: 10.1016/s0092-8674(00)80736-9. [DOI] [PubMed] [Google Scholar]
- 57.Scheidig AJ, Hynes TR, Pelletier LA, Wells JA, Kossiakoff AA. Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer's; amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): engineering of inhibitors with altered specificities. Protein Sci. 1997;6:1806–1824. doi: 10.1002/pro.5560060902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.