Abstract
DNA double-strand break (DSB) repair via the homologous recombination pathway is a multi-stage process, which results in repair of the DSB without loss of genetic information or fidelity. One essential step in this process is the generation of extended single-stranded DNA (ssDNA) regions at the break site. This ssDNA serves to induce cell cycle checkpoints and is required for Rad51 mediated strand invasion of the sister chromatid. Here, we show that human Exonuclease 1 (Exo1) is required for the normal repair of DSBs by HR. Cells depleted of Exo1 show chromosomal instability and hypersensitivity to ionising radiation (IR) exposure. We find that Exo1 accumulates rapidly at DSBs and is required for the recruitment of RPA and Rad51 to sites of DSBs, suggesting a role for Exo1 in ssDNA generation. Interestingly, the phosphorylation of Exo1 by ATM appears to regulate the activity of Exo1 following resection, allowing optimal Rad51 loading and the completion of HR repair. These data establish a role for Exo1 in resection of DSBs in human cells, highlighting the critical requirement of Exo1 for DSB repair via HR and thus the maintenance of genomic stability.
INTRODUCTION
DNA double-strand breaks (DSBs) can be induced by a variety of factors such as chemotherapeutic agents, ionising radiation (IR) and by the products of cellular metabolism, including replication fork collapse. In order to maintain genomic stability, cells possess a complex network of signalling pathways involved in the detection, signalling and repair of DNA damage. Defects in these DNA repair pathways can lead to human genomic instability syndromes, with increased cancer susceptibility, neurological syndromes and immunodeficiency. The resection of DSBs to produce 3′ single-stranded DNA (ssDNA) tracts is a critical step in the repair of DSBs by homologous recombination (1). The ssDNA at the break site is essential for activation of the ATR signalling cascade which re-enforces ATM-induced cell cycle checkpoints (2). The MRN (MRE11, Rad50 and NBS1) complex, in association with CtIP, has been previously reported to be important for DSB resection (3). However, recent reports clearly indicate that yeast MRX (Mre11, Rad50 and XRS1) is involved only in limited resection at the break sites while extensive resection requires additional, redundant nucleases such as Exonuclease 1 (Exo1) and/or DNA2 (4).
Exo1 was first identified in Schizosaccharomyces pombe as a nuclease that is induced during meiosis (5). Exo1 belongs to the RAD2 family of nucleases and possesses 5′–3′ nuclease activity and 5′-flap endonuclease activity (6,7). Alternate splicing leads to two isoforms of Exo1 (a and b). The isoforms differ at the C-terminus, with Exo1b having an additional 48 amino acids. Exo1 is known to interact with several other proteins involved in replication and DNA repair including PCNA and mismatch repair (MMR) proteins (8). Exo1 is implicated in several DNA repair pathways including MMR, post-replication repair, meiotic and mitotic recombination (9–11). The involvement of Exo1 in DNA repair pathways including MMR suggests it may also be a target for mutation in tumourigenesis. Consistent with this, a cancer-prone phenotype can be observed in Exo1-deficient mice including increased susceptibility to lymphoma development (12). In addition, patients with atypical human non-polyposis colon cancer and other forms of colorectal cancer have been found to have germ-line variants of Exo1, which affect nuclease function and MMR protein interactions (13,14). Exo1 has been shown to participate in the formation of ssDNA and activation of ATR in response to telomere dysfunction in mice, suggesting that it may respond to uncapped teleomeres in mammalian cells (15). Recently we have also shown that Exo1 is required for DNA-damage-induced apoptosis (16) and ATR-mediated cell cycle checkpoint activation (17) following cellular exposure to DNA damaging agents thereby highlighting it’s role in maintaining genomic stability.
Here we show that depletion of Exo1 leads to cellular sensitivity to IR and defects in both HR-dependent DSB repair and in the accumulation of RPA34 and RAD51 at sites of damage, indicating that Exo1 plays a role in optimal generation of ssDNA and resection of DSBs. Consistent with this interpretation, the nuclease activity of Exo1 is necessary for its function in HR. We also demonstrate that Exo1 is phosphorylated after DNA damage and that this event is required for the subsequent recruitment of other DNA repair proteins and HR.
MATERIALS AND METHODS
Reagents, antibodies and cell lines
All cell lines were grown in DMEM supplemented with 10% FCS. Antibodies used were as follows: mouse anti-H2AX S139 (Millipore), goat anti-Exo1, mouse anti-cyclin B, goat anti-ATR and rabbit anti-Rad51 (Santa Cruz), mouse anti-RPA34 (AbCam), rabbit anti-γ-tubulin, mouse anti-β-actin, mouse anti-flag M2 antibody and rabbit anti-MRE11 (Sigma), rabbit anti-GFP (Molecular Probes), rabbit anti-Phospho (Ser/Thr) ATM/ATR substrate (SQ/TQ), mouse anti-ATM S1981 and rabbit anti-Chk2 Thr68 (Cell Signalling), mouse anti-ATM (Genetex). Antibodies against hSSB1 were raised in sheep as described previously (18). A polyclonal antibody recognizing Exo1 phosphorylated on S714 was raised using the synthetic peptide IKLD phospho-SQSDQTC conjugated to keyhole limpet hemocyanin (KLH) at the Institute of Medical and Veterinary Sciences, Adelaide, Australia. To inhibit ATM, cells were treated with 10µM KU55933 (Calbiochem) for 1 h prior to irradiation.
Cloning, site-directed mutagenesis and expression of Exo1 constructs
A full-length Exo1b clone was purchased from Origene. The full-length Exo1b and fragment of Exo1 encoding amino acids 1–514 were cloned into the ECOR-V and BamHI sites of the Flag-2 plasmid (Fermentas). Full-length Exo1b was cloned into the SalI and BamHI sites of pEGFP-C1 (Clontech). Mutation of S714A, S714E and D78A was carried out by site-directed mutagenesis using a Quikchange lightning site-directed mutagenesis kit as per manufacturer’s instructions (Stratagene). Expression constructs were transfected into cells using Lipofectamine 2000TM (Invitrogen) as per manufacturer’s instructions and samples were assayed 24 h after transfection.
siRNA
StealthTM Exo1 (UAGUGUUUCAGGAUCAACAUCAUCU) and control siRNA was purchased from Invitrogen. siRNA was transfected into HeLa cells using Lipofectamine 2000TM (Invitrogen) as per manufacturer’s instructions and samples were analysed 48 h after transfection. ATM siRNA was as described previously (18).
Laser micro-irradiation
Live cell imaging combined with laser micro-irradiation was carried out as described previously (19). Briefly, for time-lapse imaging, U20S cells were transfected with GFP-Exo1 constructs, seeded on glass bottom dishes (MatTek Cultureware) and maintained in CO2-independent medium (Invitrogen) at 37°C during imaging. Cells were micro-irradiated with a pulsed nitrogen laser (Spectra-Physics; 365 nm, 10 Hz) with the output set at 75% of the maximum. Time-lapse images were captured and the fluorescence intensities of micro-irradiated areas relative to non-irradiated areas within the cell nucleus were calculated using Axiovison Software (Carl Zeiss). For immunofluorescence studies, cells were seeded onto Premium coverglass (Fisher Scientific) and laser micro-irradiated 24 h later as described previously (17).
Homologous recombination assay
The homologous recombination assay was carried out as described previously (18).
Immunoblotting
Following indicated treatments cells were scraped from tissue culture plates and washed once in PBS. Cells were lysed in a buffer containing 20 mM Hepes pH 8, 150 mM KCl, 5% glycerol, 10 mM MgCl2, 0.5 mM EDTA, 0.02% NP-40, before use buffer was supplemented with 1 mM NaF, 1 mM Na3VO4 and protease inhibitors cocktail (Roche) and sonicated. Lysates were cleared by centrifugation and protein concentrations were estimated using the standard Bradford assay (Bradford reagent supplied by Bio-Rad). Typically 50 µg of protein lysate was separated on a 4–12% SDS-PAGE NOVEX gel (Invitrogen) and immunoblotted with the indicated antibodies.
Immunoprecipitation
Cells were treated and lysed as for immunoblotting. Following cell lysis, lysates were diluted in 600 µl of lysis buffer and precleared with 100 µl sepharose. The lysate was transferred to a new tube, to which 20 µl (total volume) of anti-FlagM2 beads (Sigma) were added. Lysates were incubated with beads for at least 2 h. Beads were then washed 3–4 times with lysis buffer. Loading buffer (Invitrogen) containing 10% β-mercaptoethanol was then added to bead pellets and samples were boiled for 5 min. Samples were analysed via immunoblotting (as above). Mapping of the ATM interaction was performed as described previously (20).
Immunofluorescence
HeLa cells were seeded onto coverslips the day before siRNA transfection. Following siRNA transfection cells were allowed to grow for 48 h before treatment or mock-treatment with the indicated DNA damaging agent. After treatment, cells were treated with an extraction buffer (21) for 5 min before fixation in 4% PFA. Cells were permeablised with 0.5% Triton-X for 5 min and blocked in 3% BSA for 30 min. Cells were incubated with indicated primary antibodies and Alexa-conjugated secondary antibodies (Molecular Probes) for 1 h each at room temperature. Cells were stained with DAPI before mounting onto slides. Cells containing over 10 foci were scored as positive for foci.
Colony-forming assay
Colony-forming assays were performed as described previously (22).
Chromosomal aberrations
Metaphase spreads to measure chromosomal aberrations were carried out as described previously (18,23). Briefly, cells were transfected with the indicated siRNA. After 72 h of transfection, cells were treated with 2 Gy and colcemid was added 6 h post-irradiation (for S/G2 cells) or 12 h post-irradiation (for G1 cells). Metaphases were analyzed as described previously. For each point, 50 metaphases were analyzed.
RESULTS
Exo1 is rapidly recruited to sites of DNA damage
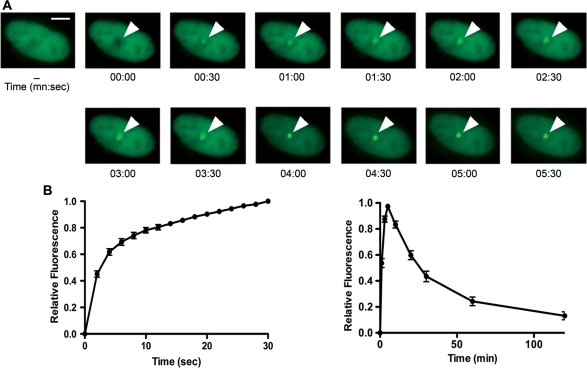
Exo1 has been implicated in resection of DNA at DSBs in both lower and higher eukaryotes (9,15,24). In order to further characterize the role of Exo1 in DNA DSB repair, we initially examined the recruitment of Exo1b (the predominant isoform present in mammalian cells) to sites of DNA damage. Laser micro-irradiation was used to induce a small area of DNA damage within the nuclei of U2OS cells expressing GFP-Exo1b as previously described (19). This treatment results in the generation of a mixture of different types of DNA damage, including DSBs. The spatiotemporal accumulation of GFP-Exo1b was determined by time-lapse imaging (Figure 1A and Supplementary Figure S1) and for quantification, the fluorescence intensity of the irradiated sites was measured, corrected for background and normalized to the preirradiation value as described previously (19). We found that GFP-Exo1b accumulated rapidly within 2 s after micro-irradiation (Figure 1B), showed a constant increase in accumulation at damage sites until 5 min and started to decline thereafter (Figure 1C). These findings suggest that Exo1 is rapidly and transiently recruited to sites of DNA damage.
Figure 1.
Recruitment of Exo1 to DNA DSBs. (A) U2OS cells expressing GFP-Exo1b fusion protein were micro-irradiated and images taken at the indicated times after damage. The scale bar represents 5 µm. (B) Graphical representation of Exo1 recruitment to sites of DNA damage from 0 to 30 s after damage induction. (C) Graphical representation of Exo1 recruitment and dissociation from sites of DNA damage from 0 to 120 min after damage induction. The data shown represents the mean and standard deviation (SD) of three independent experiments.
Exo1-knockdown causes enhanced cell killing, increased chromosome aberrations and reduced γH2AX clearance after IR exposure
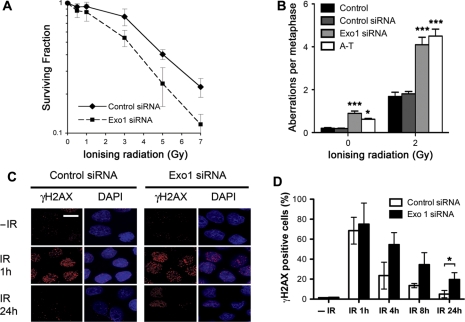
Since Exo1 is rapidly recruited to DSBs, we then depleted Exo1 in HeLa cells using siRNA (Supplementary Figure S2) and examined their sensitivity to IR via a colony-formation assay. Exo1-deficient cells were found to be significantly more sensitive to killing by IR compared to control siRNA-transfected cells, supporting the notion that Exo1 is required for DNA DSB repair (Figure 2A).
Figure 2.
DNA repair defects in Exo1-depleted cells. (A) Colony survival in HeLa cells transfected with control and Exo1 siRNA and treated with the indicated dose of IR. (B) Frequencies of spontaneous and IR (2 Gy) induced chromosome and chromatid breaks and gaps in control and Exo1-deficient cells are indicated. Chromosomal fragments are consequences of breaks either at the chromosome level or at the chromatid level. A-T cells are used as positive control. Fifty metaphases for each sample were analyzed. (C) and (D) HeLa cells were transfected with control or Exo1 siRNA and treated with 2 Gy IR before fixation and staining with γH2AX antibodies at the indicated times. Representative images (C) and graphical representation of data (D) are shown. The scale bar represents 10 µm. The results shown are the average of three independent experiments and error bars indicate the SD. One star and three stars designate statistical significance in a two-way ANOVA. *P < 0.05 and ***P < 0.001.
Next, we performed analysis of chromosome aberrations at metaphase in HEK293 cells with and without depletion of Exo1. This provides a sensitive method to evaluate the efficiency of DSB repair, in comparison to cellular survival assays, and provides a direct measurement of individual unrepaired DSBs. We found that depletion of Exo1 had no significant impact on the occurrence of chromosomal aberrations following IR in G1 phase cells (data not shown). In contrast, Exo1-deficient cells displayed a higher frequency of chromatid type aberrations in cells irradiated in the S and G2 phases (see ‘Materials and methods’ section for further details), compared with control siRNA-transfected cells. Since HR is carried out in S and G2 this suggests a role of Exo1 in HR (Figure 2B).
Next, we examined whether Exo1 deficiency influenced the kinetics of IR-induced γH2AX foci. The phosphorylation of H2AX (designated γH2AX once phosphorylated) is one of the earliest events to occur following DNA DSB induction and is associated with the formation of nuclear foci containing factors that are essential for DNA repair, replication and cell cycle regulation (25). H2AX phosphorylation occurs at sites of DSBs, irrespective of their origin and is critical for protecting the genome from spontaneous DSBs, as well as those induced by IR or V(D) J recombination (25–27). HeLa cells depleted of Exo1 using specific siRNA were exposed to 2 Gy of IR and monitored for γH2AX foci formation and disappearance at various time-points following IR. Maximal γH2AX foci were observed in both control and Exo1-depleted cells at 1 h after irradiation. In control siRNA-transfected cells, γH2AX foci were present in only ∼20% of cells by 4 h after damage and returned to predamage levels by 24 h (Figure 2C and D). In contrast, Exo1-depleted cells retained γH2AX foci in ∼50% of cells by 4 h and in ∼20% of cells by 24 h after DNA damage induction. The persistence of γH2AX foci in Exo1-depleted cells is clearly indicative of a DNA repair defect in these cells. These findings demonstrate that Exo1 plays a functionally important role in allowing cells to repair genotoxic damage and maintain chromosome stability during the S and G2 phase of the cell cycle.
Exo1 is required for homologous recombination repair of DSBs
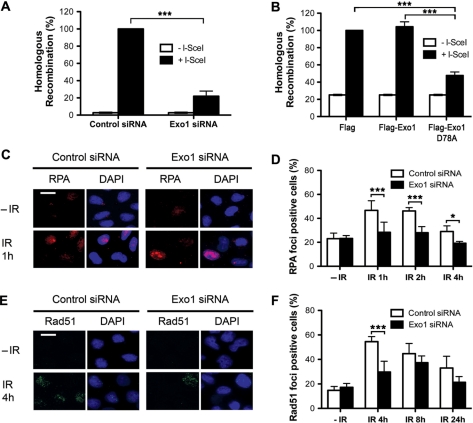
We next quantified HR in cells depleted of Exo1 using an assay for the reconstitution of a green fluorescent protein reporter gene (pDR-GFP) within a chromosomally integrated plasmid substrate (28,29). To detect HR repair of an induced chromosomal DSB, the I-SceI expression vector was transiently transfected into MCF7 cells containing a stably integrated pDR-GFP plasmid (MCF7 DR-GFP cells). Flow cytometry was used to quantify GFP positive cells. When I-SceI was transfected into Exo1-depleted cells, significantly less GFP positive cells were detected compared to control cells (Figure 3A). This difference in HR induction was not due to differences in I-SceI transfection efficiency, as expression of a control plasmid, EGFP, containing the full-length cDNA of GFP, was comparable between Exo1-depleted and control cells (data not shown). These data suggest that Exo1 is required for efficient homologous recombination repair.
Figure 3.
Recruitment and repair defects in Exo1-deficient cells. (A) HR repair is reduced in cells transfected with Exo1 siRNA. Twenty-four hours after siRNA transfection MCF7-DRGFP cells were transfected with I-SceI plasmid (pCBSCE). Fourty-eight hours after pCBSCE transfection FACS analysis was carried out to detect GFP positive cells. (B) HR repair is reduced in cells expressing nuclease-deficient Exo1. Twenty-four hours after transfection with empty vector, wild-type Exo1 or D78A, MCF7-DRGFP cells were transfected with I-SceI plasmid (as above). Forty-eight hours after pCBSCE transfection FACS analysis was carried out to detect GFP positive cells. The results shown are the average of three independent experiments and error bars indicate the SD (A) and (B). (C)–(F) HeLa cells were transfected with control or Exo1 siRNA and treated with 5 Gy IR before fixation and staining with RPA34 (C) and (D) or Rad51 (E) and (F) antibodies. The scale bar represents 10 µm. The mean and SD from two-four independent experiments is depicted (D) and (F). One star and three stars designate statistical significance in a two-way ANOVA. *P < 0.05 and ***P < 0.001.
In order to further examine the role of Exo1 in HR, we subsequently investigated whether the nuclease activity of Exo1 was required for HR by overexpressing a nuclease-deficient Exo1b in which Aspartic acid at amino acid 78 was mutated to Alanine (D78A). This mutation has previously been shown to severely reduce the exonuclease activity of Exo1 (6). MCF7 DRGFP cells, containing the HR reporter construct were transfected with Flag vector, wild-type Flag-Exo1b or nuclease-deficient D78A Exo1b, 24 h before transfection with the I-SceI plasmid. GFP-expression was analysed via FACS (as above). Cells expressing Exo1 D78A showed significantly less HR induction than cells expressing either Flag vector alone or Flag-Exo1b (Figure 3B and Supplementary Figure S3), providing direct evidence that Exo1 nuclease activity is required for normal HR.
Recruitment of HR proteins is defective in Exo1-depleted cells
To provide some insight into the role of Exo1 in HR, we analyzed the recruitment of HR proteins to repair foci in Exo1-depleted cells. The resection of DSBs is followed by the recruitment of replication protein A to the ssDNA tracts (30). Since Exo1 has been implicated in resection of DSBs (24,31,32), we next investigated whether RPA is still recruited to sites of DSBs in the absence of Exo1. We found that fewer RPA34 foci were formed in Exo1-depleted cells following IR treatment when compared to control siRNA-transfected cells (Figure 3C and D), while Exo1 depletion had no global effect on the total amount of RPA34 (Supplementary Figure S4) or on cell cycle progression (data not shown). We next examined the formation of IR-induced Rad51 foci. We observed that depletion of Exo1 by siRNA treatment significantly decreased the number of Rad51 foci (Figure 3E and F). In support of this we also found that Exo1 was recruited to laser tracks earlier than Rad51 (Supplementary Figure S5). Collectively, our data indicates that Exo1 may promote optimal resection of DSBs to allow efficient loading of RPA and oligomerization of Rad51.
Recruitment of MRN and activation of ATM is normal in Exo1-depleted cells
The recruitment of MRX (MRN in humans) to DSBs is one of the earliest events in the DNA damage response and facilitates both ATM activation as well as limited DSB resection (33). To investigate whether Exo1 is required for the recruitment of the MRN complex in human cells, we next looked at DSB-induced foci formed by the nuclease component of the MRN complex, Mre11 following IR treatment in Exo1-depleted cells. The extent of Mre11-foci formation was found to be similar in control and Exo1-depleted cells (Supplementary Figure S6). Another protein that acts early in the DNA damage response, hSSB1 (18) was also recruited to foci in the absence of Exo1 (Supplementary Figure S6). This suggests that Exo1 is not required for the recruitment of Mre11 or hSSB1 to DSBs and possibly acts downstream or in a separate pathway. Furthermore, consistent with other studies we found that Exo1-deficient cells show normal ATM activation and activity as well as normal ATM-dependent checkpoint pathway activation after IR suggesting that Exo1 is not required for efficient DSB signaling (Supplementary Figure S4) (24). Similarly, cells-deficient in the Mre11 nuclease, although markedly defective in DSB repair, efficiently activate the ATM signaling pathway after exposure to IR (34), suggesting that DSB processing is dispensable for activation of the ATM-dependent signaling pathway.
Exo1 is phosphorylated in response to DSBs
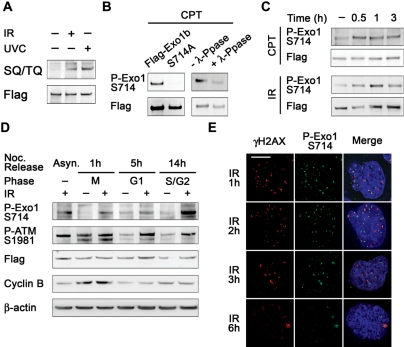
Since the activity of Exo1 has been suggested to be regulated by phosphorylation in budding yeast (35), we next investigated whether the phosphorylation of Exo1 contributes to the regulation of its nuclease activity during HR. ATM and ATR are the major kinases responsible for signaling and repair events following DNA damage and phosphorylate their substrates on a SQ/TQ motif (36). We performed an immunoprecipitation using exogenous Flag-Exo1b and immunoblotted with an antibody that specifically recognizes phosphorylated SQ/TQ residues in ATM/ATR substrates. Exo1b was found to be phosphorylated on SQ residues (there are no TQ motifs in the amino acid sequence of Exo1b) following cellular exposure to UVC or IR (Figure 4A). Notably, a large scale phospho-proteomics study that identified ATM/ATR targets after IR and UV revealed that human Exo1 was phosphorylated on an SQ motif, Serine 714 (S714) following DNA damage, so we decided to further characterize this phosphorylation event in human Exo1b (36).
Figure 4.
Exo1 is phosphorylated on S714 after DSB induction. (A) HeLa cells were transfected with Flag-Exo1 and treated with 6 Gy IR or 40 J/m2 UVC and extracts taken after 1 h. Extracts were immunoprecipitated with Flag M2 beads and immunoblotted with an antibody that specifically recognises phosphorylated SQ/TQ residues and Flag antibodies. (B) HeLa cells were transfected with Flag-Exo1 or Flag-S714A and treated with 1 µM CPT; extracts were taken after 1 h. Extracts were immunoprecipitated [as in (A)] and immunoblotted with the indicated antibodies. Second panel, cell extracts were incubated in the presence of λ-phosphatase where indicated. Immunoprecipitates were immunoblotted with the indicated antibodies. (C) HeLa cells were transfected with Flag-Exo1 and treated with 1 µM CPT (top panels) or 6 Gy IR (bottom panels). Extracts were taken after the indicated time and immunoprecipitated as in (B). Immunoprecipitates were immunoblotted with the indicated antibodies. (D) HeLa cells were transfected with Flag-Exo1 and synchronised using nocodazole. Cells were treated with 6 Gy IR at the indicated stage of the cell cycle and extracts were taken after 1 h. Extracts were immunoblotted with the indicated antibodies. (E) HeLa cells were treated with 1 Gy IR and fixed after 1 h. Immunofluorescence was carried out using the indicated antibodies. The scale bar represents 5 µm.
We raised a rabbit polyclonal antibody against the S714 phosphorylation site. This antibody specifically recognized wild-type Exo1 after exposure to camptothecin (CPT), a topoisomerase I poison that generates DNA replication-associated DSBs. The antibody did not recognize Exo1 when S714 was mutated to alanine in Flag-Exo1 S714A (Figure 4B). Treatment of cell lysates with λ-phosphatase after CPT treatment also prevented the detection of Exo1 with anti-phospho S714 antibody (Figure 4B). Exo1b was phosphorylated within 30 min following DSB induction by CPT and IR (Figure 4C). Next, we wanted to investigate whether the phosphorylation of Exo1 on S714 changed during the cell cycle, potentially indicating a role in a specific phase of the cell cycle. HeLa cells, transiently expressing Flag-Exo1b were synchronised using a nocodozole block and release protocol (37) and exposed to IR. Although phosphorylation of S714 was induced after IR during all the phases of the cell cycle, it was significantly prominent in S/G2 phases (Figure 4D and Supplementary Figure S7) indicating that phosphorylation may be important for regulation of HR, which occurs predominantly in the S/G2 phases.
Indeed, using immunofluorescence microscopy analysis with the same antibody, we found that the antibody could specifically recognise endogenous Exo1 phosphorylated on S714 following DNA damage (Supplementary Figure S8). Exo1 phosphorylated on S714 was found to co-localise with γH2AX, a known marker of sites of DSBs following IR. The kinetics of S714 foci were also similar to that of γH2AX, with maximal foci observed 1 h after IR exposure and the majority of foci dissipated after 6 h (Figure 4E). Similar kinetics of foci resolution were also observed for S714 and γH2AX following release from CPT (Supplementary Figure S9). Moreover, the phosphorylation of endogenous Exo1 occurred within 5 min in laser micro-irradiation experiments (Supplementary Figures S10 and 11). Upon transfection of GFP-Exo1 into U2OS cells followed by laser micro-irradiation, we observed that Exo1 was recruited within 1 min whereas S714 phosphorylation did not occur until 5 min after damage (Supplementary Figure S11). This indicates that Exo1 phosphorylation might occur at the sites of DSBs following its recruitment.
Exo1 is a substrate of the ATM kinase
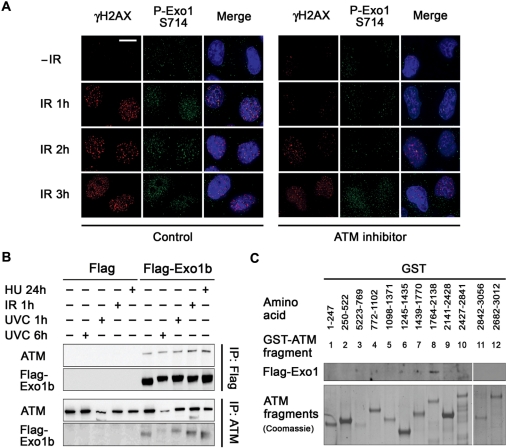
Since Exo1 is phosphorylated on an SQ motif we next decided to investigate whether Exo1 is phosphorylated by ATM. Cells treated with a specific ATM inhibitor, KU55933 prior to irradiation displayed greatly reduced numbers of IR-induced P-Exo1 S714 nuclear foci at 1 and 2 h after IR treatment (Figure 5A). It should be noted however that at 3 h after IR treatment P-Exo1 S714 nuclear foci were formed, suggesting that other kinases such as ATR or DNA-PK can phosphorylate this site at later times after damage. In addition, transfection of cells with ATM siRNA greatly reduced the number of IR-induced P-Exo1 S714 nuclear foci (Supplementary Figure S12). Similarly, laser-micro-irradiation showed that while Exo1 was rapidly phosphorylated at the sites of laser-induced DNA damage, this phosphorylation was significantly delayed in cells treated with KU55933 (Supplementary Figure S10). We also found that ATM activity was not required for recruitment of Exo1 to laser tracks as GFP-Exo1 was recruited to laser tracks normally in cells treated with KU55933 (data not shown). Taken together these data suggest that although Exo1 is recruited to DSBs independently of ATM, ATM is required for rapid phosphorylation of Exo1, though additional kinases such as ATR or DNA-PK may compensate for ATM at later times. In light of the above, we next investigated whether Exo1 interacted with ATM. Co-immunoprecipitation (Figure 5B) and GST-ATM fragment pull-down assays (20) (Figure 5C) indicated that the interaction between Exo1b and ATM was direct, mediated by ATM fragment 8 (aa 1764–2138) and was not influenced by various forms of DNA damage.
Figure 5.
Exo1 interacts with ATM and phosphorylation of Exo1 on S714 is dependent upon ATM. (A) HeLa cells were pretreated or mock-treated with the ATM inhibitor, KU55933 (10 µM) 1 h prior to treatment with 6 Gy IR and fixed at the indicated time points. Cells were immunostained with the indicated antibodies. The scale bar represents 10 µm. (B) Top panels, HeLa cells were transfected with Flag-Exo1 and treated with 40 J/M2 UVC, 6 Gy IR or 2 mM HU and extracts taken after the indicated time. Immunoprecipitations were carried out using Flag M2 Beads. Immunoprecipitates were immunoblotted with the indicated antibodies. Bottom panels, HeLa cells were treated as above and immunoprecipitated using ATM antibodies. Immunoprecipitates were immunoblotted with the indicated antibodies. (C) Exo1 interacts with GST-ATM fragment 8 (amino acids 1764–2138). Recombinant GST-ATM fragments representing the full length of ATM were used in pull-down assays. Total cell extracts from HeLa cells expressing Flag-Exo1 were mixed with glutathione agarose beads containing GST-ATM fusion proteins. Bound proteins were analysed by immunoblotting with Flag antibodies (top panel) and levels of GST-ATM fragments were detected by coomassie staining (bottom panel).
Phosphorylation of Exo1 is required for recruitment of DNA repair proteins
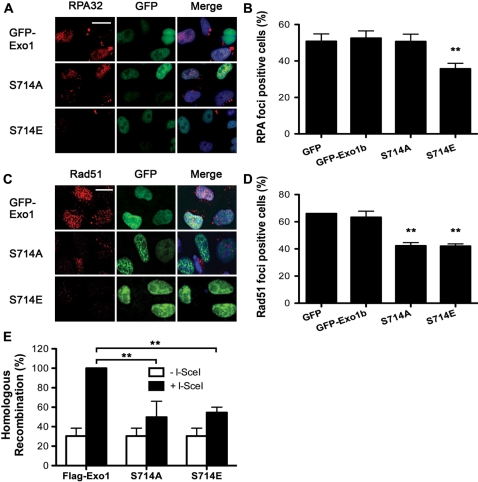
In order to further investigate the role of Exo1 phosphorylation we next looked at the recruitment of RPA34 to IR-induced foci in cells expressing wild-type GFP-Exo1b, non-phosphorylatable mutant Exo1b (GFP-S714A) and phospho-mimic mutant Exo1b (GFP-S714E) in order to estimate the extent of DSB resection in these cells. We found that expression of S714E significantly inhibited RPA34 foci formation compared to cells expressing the GFP vector alone, wild-type Exo1b or S714A (Figure 6A and B). The normal DSB resection observed in cells expressing the S714A mutant indicates that phosphorylation of Exo1 at this site is not required for its resection activity. In fact, reduced DSB resection by the phospho-mimic S714E mutant indicates that phosphorylation at this site may actually attenuate the exonuclease function of this protein. Consistent with this expression of either S714A or S714E impaired the recruitment of Rad51 to foci when compared with GFP vector alone or wild-type Exo1b (Figure 6C and D). The recruitment defects observed in cells expressing Exo1 mutants were not attributable to an alteration in the cell cycle distribution of the transfected cell population (Supplementary Figure S13). Furthermore, cells expressing either type of phospho-mutant showed diminished efficiency of HR in the DR-GFP HR substrate (Figure 6E). Taken together these data suggest that the phosphorylation of Exo1 on S714 may occur after the recruitment of RPA34 but before the recruitment of Rad51 and that possible inhibition of the exonuclease activity by phosphorylation might prevent the generation of substrates unsuitable for Rad51 loading and HR.
Figure 6.
Exo1 phosphorylation is required for recruitment of Rad51 and DSB repair by HR. (A) and (B) HeLa cells were transfected with the indicated constructs. Cells were treated with 5 Gy IR and fixed 2-h after treatment. Immunofluorescence was carried out using RPA34 antibodies. Cells expressing GFP were scored as positive or negative for RPA34 foci. Representative images are shown. Two stars designate significant statistical difference to the control in a t-test, **P = 0.0032. (C) and (D) HeLa cells were transfected with the indicated constructs. Cells were treated with 5 Gy IR and fixed 4-h after treatment. Immunofluorescence was carried out using Rad51 antibodies. Cells expressing GFP were scored as positive or negative for Rad51 foci. Representative images are shown. Two stars designate significant statistical difference to the control in a t-test, **P < 0.005. The scale bar represents 10 µm. (E) HR repair is reduced in cells expressing S714A and S714E. Twenty-four hours after transfection with wild-type Exo1, S714A or S714E, MCF7-DRGFP cells were transfected with I-SceI plasmid (pCBSCE). Forty-eight hours after pCBSCE transfection FACS analysis was carried out to detect GFP positive cells. Two stars designate statistical significance in a two-way ANOVA. **P < 0.001. (B), (D) and (E) The results shown are the average of three independent experiments and error bars indicate the SD.
DISCUSSION
DSB repair via HR is crucial to maintain genomic stability and prevent tumour development. One of the first stages of HR is the resection of DNA surrounding the DSB to expose ssDNA, which is necessary for Rad51-mediated strand invasion to occur. Here, we suggest that Exo1 plays a critical role in the HR pathway functioning to allow correct resection of DSBs. In support of this we show that Exo1 like other early participants of the HR process is rapidly recruited to sites of DSBs (within 10 s). We also demonstrate that Exo1-deficient cells are hypersensitive to IR, display defective clearance of IR-induced γH2AX foci and exhibit both spontaneous and IR-induced chromosomal aberrations. These data indicate that Exo1 functions in chromosomal damage repair via HR. Our findings are consistent with recent reports showing that yeast Exo1 is required for resection of DSBs in order for HR to occur and indeed further results discussed here support this notion. Firstly, Exo1-deficient cells were found to be defective in HR of I-SceI-induced DSBs. Secondly, we also observed defective recruitment of RPA34, a ssDNA-binding protein required for HR at DSBs in Exo1-deficient cells. The loss of RPA foci formation following DNA damage suggests that the formation of its ssDNA substrate is impaired in Exo1-deficient cells. This was further backed by a defect in Rad51 foci formation at DSBs. Thirdly, we also revealed that HR was dependent upon Exo1 nuclease activity, providing direct evidence that Exo1 promotes resection of DSBs.
Recent studies in yeast have also implicated Exo1 as having a role in resection of DSBs (24,31,32). One such study suggests that the MRX complex is responsible for the initial resection of DSBs, while Exo1 (along with Sgs1) are required for the extensive resection needed for Rad51 loading and strand exchange during HR (31). Our data supports this model as Mre11 foci are still formed in Exo1-deficient cells suggesting Exo1 functions downstream of Mre11 or in a separate pathway. In vitro biochemical analysis using human proteins has shown that Exo1 can generate substrates suitable for Rad51-mediated DNA pairing and that the yield of this reaction can be further increased by the addition of BLM (38). In the above-mentioned study human Exo1 was absolutely required for homologous DNA pairing and therefore Rad51-mediated strand exchange, supporting our data that in human cells Exo1 is required for HR even in the presence of BLM (38). In agreement with this, another study found a mild but reproducible defect in RPA34 foci formation in Exo1-deficient cells following induction of DSBs by CPT treatment, suggesting that resection of specific subsets of DSBs is completely dependent upon Exo1 (24). It should be noted, however, that in cells depleted of both Exo1 and BLM the percentage of cells containing RPA34 foci was reduced even further, although this may reflect the nature of the breaks induced at replication forks.
To further characterize the role of Exo1 in the HR process we have now identified that Exo1 is phosphorylated at S714 in an ATM specific manner following induction of DSBs. This site has previously been mapped during another study and was shown to be phosphorylated in an ATR dependant manner in response to the replication fork-stalling agent hydroxurea. It was suggested that this event (in addition to two other phosphorylation sites) causes degradation of Exo1 (39). Since we have never observed Exo1 degradation following IR treatment and the S714E and A-mutant Exo1 displayed comparable levels of expression, we conclude that the destabilization of phosphorylated Exo1 observed previously is unlikely to be the function of S714 phosphorylation after IR treatment. However, we cannot exclude the possibility that another mechanism exists to stabilize Exo1 after IR treatment and prevent its degradation.
We have shown here that following DSB-induction Exo1 is initially phosphorylated by an ATM-dependent mechanism, however, at later time points Exo1 phosphorylation is independent of ATM suggesting that it is phosphorylated redundantly, possibly by ATR or DNA-PK. We show that while RPA34 recruitment to foci was normal in cells expressing the non-phosphorylatable mutant of Exo1 (S714A), Rad51 recruitment to foci was defective. This suggests that the phosphorylation event at S714 is not required for the generation of the ssDNA tracks but is possibly required to allow Rad51 loading. Interestingly we observed that cells expressing a phospho-mimic Exo1 (S714E) are deficient in generation of ssDNA/DNA resection as measured by the recruitment of RPA34 and Rad51 to foci. This phospho-mimic mutant also has impaired HR activity. At present we can only speculate as to the specific function of the phosphorylation event, however a recent paper examining four phosphorylation sites in yeast Exo1 suggested that phosphorylation probably inhibits the activity of Exo1 (35).
It is tempting to speculate that S714 phosphorylation is not required for the initiation of Exo1-mediated DNA resection, but rather that the phosphorylation is required to attenuate Exo1 activity and perhaps prevent the generation of substrates unsuitable for HR. Inactivation of a nuclease by phosphorylation after DNA damage is not without precedent. A recent paper has also shown that phosphorylation of Mre11 on SQ/TQ residues decreases its affinity for DNA, causes disassembly from chromatin and thereby results in its inactivation, it is possible that phosphorylation of Exo1 may function in a similar way (40). Our interpretations do not exclude additional functions of phosphorylated Exo1 in recombinational DNA repair. It is also possible that phosphorylation of Exo1 on S714 changes its binding partners. We have observed here that phosphorylation of Exo1 by ATM was dispensable for resection as revealed by normal RPA foci formation in non-phosphorylatable mutant (S714A) expressing cells; however these cells are likely to be defective in recombinational repair due to defective loading of Rad51. Indeed we also found that cells expressing the non-phosphorylatable mutant displayed defective I-SceI-induced HR. Taken together, our data suggests that the DNA substrates generated by action of non-phosphorylatable Exo1 cannot be used efficiently by Rad51 to form nucleoprotein filaments. Further studies will be required to address whether this is due to the over-resection by unregulated Exo1.
A recent study has shown that HR of IR-induced DSBs that arise during G2 is reduced in ATM-deficient cells (41). Since we have shown that ATM is required for the timely phosphorylation of Exo1 and that this phosphorylation event is required for the regulation of HR, this may further confirm a role for ATM in controlling recombinational repair of DSBs. However, the role of ATM in Exo1 phosphorylation appears to be redundant suggesting that other proteins such as ATR or DNA-PK can compensate albeit at a later time-point.
DNA DSBs induce a vast array of signaling networks, which can greatly influence DNA repair, cell cycle checkpoint activation and cell viability and therefore it is understandable that multiple levels of regulation may exist to inactivate destructive proteins such as nucleases. Exo1 activity may be attenuated by ATM-dependent phosphorylation following its role in resection to prevent the destruction of substrates required for HR. Given the importance of accurate recombination in preserving the genome, our findings clearly illustrate the role of Exo1 in maintaining genomic stability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Health and Research Council of Australia (442903 to K.K.K.); the National Institutes of Health (grant numbers CA129537 and CA123232 to T.K.P.) and National Aeronautics and Space Administration (grant number NNA05CS597G to S.B.). Funding for open access charge: National Health and Research Council of Australia.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem. Sci. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein KA, Rothstein R. At loose ends: resecting a double-strand break. Cell. 2009;137:807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J. Biol. Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 6.Lee Bi BI, Nguyen LH, Barsky D, Fernandes M, Wilson D.M., III Molecular interactions of human Exo1 with DNA. Nucleic Acids Res. 2002;30:942–949. doi: 10.1093/nar/30.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J. Biol. Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 8.Lee SD, Alani E. Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. J. Mol. Biol. 2006;355:175–184. doi: 10.1016/j.jmb.2005.10.059. [DOI] [PubMed] [Google Scholar]
- 9.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H., Jr, Kneitz B, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun X, Zheng L, Shen B. Functional alterations of human exonuclease 1 mutants identified in atypical hereditary nonpolyposis colorectal cancer syndrome. Cancer Res. 2002;62:6026–6030. [PubMed] [Google Scholar]
- 14.Wu Y, Berends MJ, Post JG, Mensink RG, Verlind E, van der Sluis T, Kempinga C, Sijmons RH, van der Zee AG, Hollema H, et al. Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology. 2001;120:1580–1587. doi: 10.1053/gast.2001.25117. [DOI] [PubMed] [Google Scholar]
- 15.Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, et al. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolderson E, Richard DJ, Edelmann W, Khanna KK. Involvement of Exo1b in DNA damage-induced apoptosis. Nucleic Acids Res. 2009;37:3452–3463. doi: 10.1093/nar/gkp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomimatsu N, Mukherjee B, Burma S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 2009;10:629–635. doi: 10.1038/embor.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- 19.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller SP, et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat. Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 21.Young DB, Jonnalagadda J, Gatei M, Jans DA, Meyn S, Khanna KK. Identification of domains of ataxia-telangiectasia mutated required for nuclear localization and chromatin association. J. Biol. Chem. 2005;280:27587–27594. doi: 10.1074/jbc.M411689200. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Bolderson E, Kumar R, Muniandy PA, Xue Y, Richard D, Seidman M, Pandita TK, Khanna KK, Wang W. hSSB1 and hSSB2 form similar multi-protein complexes that participate in DNA damage response. J. Biol. Chem. 2009;284:23525–23531. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. Involvement of human MOF in ATM function. Mol. Cell Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 26.Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, et al. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson CL. Chromatin: mysteries solved? Biochem. Cell Biol. 2001;79:219–225. [PubMed] [Google Scholar]
- 28.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 31.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 34.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27:2400–2410. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuoka S, Ballif BA, Smogorzewska A, McDonald E.R., III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 37.van der Horst A, Khanna KK. The peptidyl-prolyl isomerase Pin1 regulates cytokinesis through Cep55. Cancer Res. 2009;69:6651–6659. doi: 10.1158/0008-5472.CAN-09-0825. [DOI] [PubMed] [Google Scholar]
- 38.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl Acad. Sci. USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Shemerly M, Hess D, Pyakurel AK, Moselhy S, Ferrari S. ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res. 2008;36:511–519. doi: 10.1093/nar/gkm1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Virgilio M, Ying CY, Gautier J. PIKK-dependent phosphorylation of Mre11 induces MRN complex inactivation by disassembly from chromatin. DNA Repair. 2009;8:1311–1320. doi: 10.1016/j.dnarep.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.