Abstract
Epstein–Barr Virus (EBV) DNase (BGLF5) is an alkaline nuclease and has been suggested to be important in the viral life cycle. However, its effect on host cells remains unknown. Serological and histopathological studies implied that EBV DNase seems to be correlated with carcinogenesis. Therefore, we investigate the effect of EBV DNase on epithelial cells. Here, we report that expression of EBV DNase induces increased formation of micronucleus, an indicator of genomic instability, in human epithelial cells. We also demonstrate, using γH2AX formation and comet assay, that EBV DNase induces DNA damage. Furthermore, using host cell reactivation assay, we find that EBV DNase expression repressed damaged DNA repair in various epithelial cells. Western blot and quantitative PCR analyses reveal that expression of repair-related genes is reduced significantly in cells expressing EBV DNase. Host shut-off mutants eliminate shut-off expression of repair genes and repress damaged DNA repair, suggesting that shut-off function of BGLF5 contributes to repression of DNA repair. In addition, EBV DNase caused chromosomal aberrations and increased the microsatellite instability (MSI) and frequency of genetic mutation in human epithelial cells. Together, we propose that EBV DNase induces genomic instability in epithelial cells, which may be through induction of DNA damage and also repression of DNA repair, subsequently increases MSI and genetic mutations, and may contribute consequently to the carcinogenesis of human epithelial cells.
INTRODUCTION
Nucleases which break down DNA molecules are distributed ubiquitously in eukaryotic cells and microorganisms and some viruses also express nucleases during their life cycle. In the prokaryotic viruses, λ exonuclease (Redα) encoded by λ phage was shown to be important for processing the viral genome (1). In eukaryotic viruses, the best-studied nucleases are the alkaline nucleases (ANs) of the Herpesviridae and Baculoviridae. ANs are defined as enzymes that degrade DNA under alkaline condition. The AN encoded by a baculovirus was found to be involved in the resolution of replication intermediates and genome maturation (2). In the herpesviruses, the AN of herpes simplex virus 1 (HSV-1) had been shown to be required for efficient processing of viral DNA replication intermediates (3) and for the efficient production of viral progeny (4). Aside from their role in the viral life cycle, however, the effects of these ANs on the host cells are less well understood.
Epstein–Barr virus (EBV), a member of the herpesviridae, has been associated with many human malignancies, including Burkitt’s lymphoma (BL) and nasopharyngeal carcinoma (NPC) (5). EBV DNase (BGLF5) is an AN encoded by the BGLF5 open reading frame of EBV. The EBV life cycle has two stages, latency and the lytic cycle. EBV DNase is expressed in the early stage of the lytic cycle and is classified as an early lytic protein. EBV DNase had been shown to be important for the generation and processing of linear viral genomes (6). Biochemically, it exhibits both exonuclease and endonuclease activities, a requirement for divalent cations, and a preference for alkaline conditions (7–10). As substrates, dsDNA is digested processively but ssDNA, distributively (11). The endonuclease activity of EBV DNase seems to have a DNA structural preference but no sequence specificity. The exonuclease degrades DNA from 5′- to 3′-direction, generating 5′-monophosphate nucleosides (11). In contrast to the well-studied functions in vitro, the effects of EBV DNase on cells have been elucidated less clearly. Serological studies indicated that NPC patients have higher titers of antibody against EBV DNase than normal controls (12) and antibody levels may be raised prior to the appearance of the clinical symptoms of NPC (13). In histopathological studies, significant amounts of EBV DNase protein and nuclease activity were demonstrated in both fresh biopsies and transplanted tumor lines (14). Based on these observations, EBV DNase seems to play an important role in NPC carcinogenesis. However, the question of how EBV DNase contributes to carcinogenesis is not very clear.
Genomic instability appears to be a hallmark of cancers (15). It has been found in most types of cancers, including NPC, and correlated with the malignant levels of cancers (16–18). Therefore, genomic instability has been considered to be either a cause or the result of carcinogenesis (19,20). Generally, genomic instability is characterized by an increased frequency of genetic changes encompassing nucleotide-excision repair-associated instability, microsatellite instability (MSI), and chromosomal aberration-associated instability (19). Intrachromosomal genomic instability may result from increased rates of DNA damage overwhelming the ability of cellular repair systems to maintain genome integrity. In addition, impairment of repair systems also plays another major role in intrachromosomal genomic instability. There are five major repair systems to protect human cells from injury (21,22): (i) nucleotide excision repair (NER), (ii) base excision repair (BER), (iii) mismatch repair (MMR), (iv) homologous recombination (HR) and (v) non-homologous end-joining (NHEJ). When these repair pathways are interrupted, defective repairs are unable to cope with rates of DNA damage, leading to genomic instability. At the chromosome level, chromosomal instability results mainly from inappropriate segregation, DNA recombination and impaired mitosis. Some studies suggested that DNA double-stranded breaks (DSBs) also may cause chromosomal aberrations (23,24).
Increased rates of DNA damage and inhibition of DNA repair may arise from either internal (e.g. free radicals, replication stress) or external (e.g. microbial, UV, chemicals) sources (25). Recently, several human cancer-associated viruses, including human T-cell leukemia virus 1 (HTLV-1), hepatitis C viruses (HCV), and human papilloma viruses (HPV) have been proven to be involved in the induction of genomic instability or genetic aberrations (26–28). Among the herpesviruses, Kaposi’s sarcoma associated herpesvirus (KSHV) was found to induce genomic instability in endothelial cells (29). Latent infection of human B cells by EBV also has been shown to enhance genetic instability (30,31). Recently, we observed that recurrent chemical reactivations of EBV promote genomic instability and enhanced tumor progression of NPC cells (32). This recent observation implied that EBV could enhance genomic instability during the lytic cycle. In addition, we also found that EBV DNase had the most potent ability to induce genomic instability among several EBV lytic genes examined (32). It would be interesting to determine the effects of EBV DNase on cells, especially at the genomic level. Previously, Russo et al. demonstrated that DNA breakage could be enhanced by endonuclease and may contribute to genomic instability (33). Overexpression of RAG-1/2 endonuclease was found to cause genomic instability, which may be through the induction of DNA DSBs (34), and apurinic/apyrimidinic endonuclease (APE) was found to interfere with DNA repair, leading to genomic instability (35). Furthermore, hepatocellular carcinoma tissues have been shown to have overexpression of repair-related nucleases (36). All these results indicate that nuclease expression is correlated with human malignancy. On the other hand, ectopic expression of restriction endonucleases was found to induce DNA DSBs, chromosome aberration, oncogenic transformation and genetic mutation (37–40). Moreover, it was also demonstrated that several lytic proteins, including the AN of herpes simplex virus-2 (HSV-2), were involved in the induction of chromosomal aberrations (41). These studies suggested that nucleases, from either an intrinsic or extrinsic source, have the potential to cause genomic instability, although the detailed mechanisms remain unclear.
Combining these observations, we tried to address the question of whether a viral nuclease could induce genomic instability in the cells. In this communication, we first demonstrate that EBV DNase could induce genomic instability in human epithelial cells. We also provide evidence that EBV DNase may induce genomic instability through two mechanisms: directly, by damage to DNA, and indirectly, by inhibition of DNA repair. Moreover, we show that chromosomal aberrations developed when DNase was expressed in the epithelial cells. Finally, the genetic mutation assays show that EBV DNase could elicit mutation and MSI. These results reveal a novel mechanism through which AN induces genomic instability and also a new insight into the role of EBV in the carcinogenesis of NPC.
MATERIALS AND METHODS
Cell culture and antibodies
All of the following cell lines and their derivatives were maintained in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10% fetal calf serum. TW01, is a human NPC cell line (42). H1299, is a human lung carcinoma cell line (43). HEp2 cell, is a human laryngeal epithelial cell line (44).
Antibody to EBV DNase, anti-DNase 311H, was generated in our laboratory previously (45) and anti-β-actin and GAPDH antibodies were purchased from Sigma-Aldrich Co. The antibody of γH2AX was purchased from Upstate Co. The specific antibodies of MMR-related proteins, including MSH2, MSH6, MLH1 and PMS2, were purchased from BD, Biosciences.
DNA constructs
The EBV DNase expression plasmids were constructed by cloning the respective coding sequences into the pEGFP-CPO-IRES-puro vector, which was derived from pEGFP-C1, with insertion of an IRES-puro cassette, by CPO site ligation. Plasmid of Mutant 1 encoded the sequence of EBV DNase with F452A point mutation (46) and subcloned into pEGFP-CPO-IRES-puro vector. Similarly, Mutants 2 and 3 encoded the sequences of EBV DNase with E225A/K227A double mutations and E225A/K227A/F452A triple mutations which were generated by site-directed mutagenesis and subcloned into pEGFP-CPO-IRES-puro vector. The coding sequences of DNase and the F452A mutant (46) were inserted into the HindIII site of the p3′SS vector (Stratagene), to generate pIn-DNase and pIn-F452A. For MSI assay, the DNA fragment TAG-Bsd, which contains a stop codon TAG replacing the start codon ATG in front of the blasticidin resistance gene coding sequence, were generated by site-directed mutagenesis. The DNA fragment CA16-Bsd, containing 16 dinucleotide repeats (CA) downstream from the initiation codon of the blasticidin resistance gene, was generated by PCR using the upstream primer (5′-cacggtccgcacacacacacacacacacacacacacacacattgccaagcctttgtctcaagaag-3′) with an artificial CpoI site, downstream primer (5′-cacggaccgttagccctcccacacataacc-3′) with an artificial CpoI site and pLenti6/TR (Invitrogen) as the template. The DNA fragment TAG-Bsd and CA16-Bsd were inserted into a modified pcDNA3.1 (with an HA tag and a CpoI site) to generate pBsd2-puro and pBsd4-puro. The construct was verified by DNA sequencing. The pBsd4-puro plasmid contains 16 dinucleotide repeats (CA) downstream of the initiation codon of the blasticidin resistance gene, so that the blasticidin resistance gene coding sequence is out of frame. These plasmids contain a puromycin resistance gene which was used to establish stable transfectants.
Western blotting analysis
The cellular extracts were separated by SDS–PAGE and transferred to Hybond-C super membrane (Amersham Biosciences Ltd.). The blot was incubated with blocking buffer (10 mM Tris–HCl, pH 8.0, 0.9% NaCl and 4% skim milk) for 1 h and reacted with indicated antibodies for 1 h at room temperature. After washing three times with washing buffer, the blot was incubated with horseradish peroxidase-labelled goat anti-mouse IgG (Amersham Biosciences Ltd) diluted 1 : 2500 with blocking buffer for 1 h at room temperature. After incubation, the blot was washed three times in washing buffer and once with water, and then developed with a freshly prepared substrate (Amersham Biosciences Ltd.). The luminescence was detected by a short exposure to X-ray film.
Immunofluorescence staining
HEp2 cells were grown on the cover slides and transfected with indicated plasmids for 24 h. For γH2AX staining, cells were washed one time with phosphate buffered saline (PBS) and fixed with 2% formaldehyde for 10 min. Then, fixed cells were permeabilized with 0.4% Triton X-100 in PBS for 5 min. After washed with 4% FCS in PBS (4% FCS–PBS) for three times, cells were blocked in 4% FCS–PBS for 30 min. The cells were incubated with 2 µg/ml of anti-phospho-Histone H2A.X (Ser139) (Upstate, Charlottesville, VA) in 4% FCS-PBS at 37°C for 1 h, and then washed three times with 4% FCS–PBS. The secondary antibody was Rhodamine-conjugated goat antimouse IgG, diluted 1 : 100 in 4% FCS–PBS. After incubation with secondary antibody at 37°C for 1 h, the cells were washed three times with 4% FCS–PBS and visualized with fluorescence microscope. Nucleus was visualized by DAPI staining. Cells were treated with bleomycin (1 µM) for 24 h as the control of DNA-strand breaks.
Detection of the formation of micronucleus
Detection of micronucleus (MN) formation was performed as described previously (47). Cells were seeded onto coverslips and allowed to adhere for 24 h prior to transfection or IPTG treatment. After 24 h transfection or 48 h IPTG treatment, the culture medium was removed and the cells were washed twice in PBS. Cells were fixed with ice methanol for 15 min. After washing twice in phosphate-buffered saline (PBS, pH 7.4), the cells were stained with Hoechst (0.2 μg/ml, Sigma-Aldrich, St Louis, MO) for 15 min. Micronuclei were judged using a fluorescence microscope.
Nuclease activity assay
Appropriate number of cells (5 × 106 to 5 × 107 cells/ml) at each condition was collected. Then the cells were resuspended in extraction buffer (50 mM Tris–HCl, pH 7.5, 0.3 M KCl, 5 mM 2-mercaptoethanol, 1 mM PMSF, 20% glycerol).After centrifugation at 9170 g, 4°C for 10 min, supernatants were diluted with dilution buffer (50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 10 mM 2-mercaptoethanol, 0.05% BSA) (100–200 μg/ml). The 10 μl diluted solutions were incubated with 90 μl of 1μg/ml salmon sperm DNA for 1 h at 37°C. Then samples were added with 100 μl picogreen dye (Molecular Probe, 200-fold diluted in 10 mM Tris–HCl, pH 8.0, 10 mM EDTA), mixed well, and fluorescence (Ex: 480 nm, Em: 520 nm) detected with a microplate reader. Enzyme activity is defined as percentage of hydrolyzed DNA which was digested by EBV DNase.
γH2AX detection by flow cytometry
To evaluate the DNA DSB, cells were treated and harvested, fixed in 70% ethanol. The fixed cells were permeabilized with 1% Triton X-100 and 4% FBS, and then followed by resuspension with anti-γH2AX antibody (dilution 1 : 500, Upstate) and incubation for 2 h. Cells were washed with PBS and incubated with a 1 : 1000 dilution of goat anti-mouse IgG Rhodamine-conjugated antibody for 1 h. Cells were washed and analyzed using a Becton Dickinson FACScan flow cytometer (BD Biosciences, San Jose, CA). Each experiment was in duplicate with 10 000 cells.
Comet assay
A single-cell gel electrophoresis (comet assay) kit was employed for evaluating DNA damage (Trevigen, Inc.). The comet assay is an effective method for detection of DNA damage in cells. This assay is based upon the ability of denatured, cleaved DNA fragments migrate faster than undamaged DNA. After 24 h transfection of effector plasmids, cells were resuspended in 1× PBS and combined with molten low melting agarose (at 37°C) at a ratio of 1 : 10 (v/v) and immediately pipetted 75 μl onto CometSlideTM. After cell lysis, samples were treated with NaOH solution (300 mM NaOH, 1 mM EDTA) to unwind and denature the DNA. The samples were applied to electrophoresis, stained with SYBR Green DNA dye, and observed by a fluorescence microscope to score with comet appearance (48). One hundred comets were scored randomly, and each comet assigned a value of 0 to 4 according to its class. The undamaged cells are classified as grade 0 (most intense centre and no tail), and the most damaged cells as grade 4 (disintegrated centre and longest tail). The standard scheme of scoring is described in Supplementary Figure S1. The total score was represented by comet scores to correlate with levels of DNA damage. The results were averaged with at least three independent experiments to calculate the mean and the standard deviation.
Transfection
For TW01, H1299 and HEp2 cells, plasmid DNA was transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer's; instructions. Cells were seeded one day before transfection and plasmid DNA and Lipofectamine 2000 were mixed in Opti-MEM medium (Invitrogen) and incubated for 20 min, then added to the culture well containing cells.
Host cell reactivation assay
The host cell reactivation (HCR) assay was as described previously (47). In brief, firefly luciferase reporter plasmid (pCMV-Luc), damaged with different methods or not, such as 2000 J/m2 of UV light, 10 μg/μl of cisplatin and 100 μM of MNNG pre-treatment, undamaged Renilla luciferase pRL-CMV reporter (Promega) as the internal control and effector plasmids (DNase and mutants) were cotransfected into cells with Lipofectamine 2000 (Invitrogen). At 24 or 48 h post-transfection, cells were lysed in 50 ml lysis solution (0.1M HEPES, pH 7.8, 1% Triton X-100, 1mM CaCl2 and 1mM MgCl2) and 25 ml aliquots of the lysates were taken for next assay. Firstly, 25 ml of Luciferase Assay Reagent II (Promega) was added to the samples for measurement of firefly luciferase activity using a luminescence counter (Packard), and then 25 ml of Stop & Glo Reagent (Promega) was added to the lysates. The luciferase activity was measured using a luminescence counter (Packard). The factor of repair conversion was defined as the firefly luciferase activity of each sample normalized to the Renilla luciferase activity. The relative fold of HCR was calculated by dividing the repair conversion of effector transfectants by that of vector transfectants. The results were averaged with at least three independent experiments to calculate the mean and the standard deviation.
In vitro microsatellite instability and gene mutation assay
This assay was established by Naganuma et al. (49) previously. Briefly, reporter plasmid pBsd2-puro has a stop codon TAG replacing the start codon ATG in front of the blasticidin resistance gene coding sequence, and pBsd4-puro contains 16 dinucleotide repeats (CA) downstream from the initiation codon of the blasticidin resistance gene which is out of frame, so that the blasticidin resistance proteins in these two reporters were not expressed. In protocol, the reporter plasmid pBsd2-puro or pBsd4-puro as transfected into Hp3′SS or HB7 cells, then transfectants were selected with puromycin (1 µg/ml). Puromycin-resistant clones emerged 2–3 weeks later. For gene mutation and MSI assay, cells harboring pBsd2-puro or pBsd4-puro were seeded in 100-mm culture dishes (2 × 103 or 1 × 104 cells per dish), incubated with blasticidin (5 µg/ml). After 14 days of incubation, the cells were stained with 0.05% p-iodonitrotetrazolium violet dye and then colonies larger than 1 mm were counted. The frequency of gene mutation and MSI were calculated by dividing the number of colonies by those of total seeded cells.
Quantitative RT–PCR
Amounts of mRNA expressed from different genes were detected by quantitative real time PCR. Total RNA was isolated from vector and DNase-transfected TW01 cells,and mock- or IPTG-treated Hp3′SS and HB7 cells using Trizol (Invitrogen) following the manufacturer’s protocol, then subjected to DNase I treatment and stored at –80°C. RNA purity was confirmed by determining the OD260/OD280 nm absorption ratio. Reverse transcription (RT) was performed with SuperScript II RNase H reverse transcriptase (Invitrogen), 1–2 µg RNA and oligo dT primers. Then RT products were used for real-time quantitative PCR (QPCR) reaction. The four MMR primers were designed with Primer 3 software (Infobiogen, Evry, France). Others were suggested by published literatures [ERCC1 and XPA (50); XPB and CSB (51); XPF (52); CSA (36); BRCA2 and Rad51 (53)]. The total primers of target genes are shown in Table 1. The standard products of each gene from RT-PCR were purified by gel extraction. The purified products were subjected to further PCR and purified again. After quantifying the cDNA concentration by absorbance at 260 nm and the copy number was calculated by using the molecular mass of the PCR fragment. These cDNA products were used for making calibration curve. QPCR was performed using FastStart DNA Master SYBR Green I kit (Roche) in the LightCycler system (Roche). Reaction mixture contained 6 μl prepared cDNA of each sample, 4 μl master mix, 2 μl specific primers (10 μM) and PCR-grade water to a final volume 20 μl. After 10 min pre-incubation at 95°C, PCR conditions were: 10 s denaturation at 95°C, 5 s annealing at 50°C and 10 s extension of primers at 72°C for 30 cycles. The specificity of the PCR reaction was controlled by melting curve analysis (65–95°C, 0.1°C/s) in the LightCycler and by agarose gel electrophoresis. In each experiment, control, sample and serially diluted standard products were detected together. The absolute amount of each gene product could be determined by comparison with the standard curve. Each analysis was done in duplicate and a negative control containing water instead of sample cDNA was included. The transcript level of each sample was normalized with that of GAPDH. Experiment for each gene was performed independently at least three times. The expression level of each sample was expressed as the relative expression compared with that of vector controls.
Table 1.
QPCR primers utilized in this study
| Repair-related genes | Primer sequences (5′→3′) |
|---|---|
| MSH2 | |
| Sense | GCCATTTTGGAGAAAGGACA |
| Antisense | CTCACATGGCACAAAACACC |
| MSH6 | |
| Sense | CCCCACCAGTTGTGACTTCT |
| Antisense | TGTTGGGCTGTCATCAAAAA |
| MLH1 | |
| Sense | TGGGACGAAGAAAAGGAATG |
| Antisense | GATCAGGCAGGTTAGCAAGC |
| PMS2 | |
| Sense | CAGGACATGTCAGCCTCTCA |
| Antisense | GGCTGCTTGATTTTCTCCAG |
| ERCC1 | |
| Sense | ATACCCCTCGACGAGGATGAG |
| Antisense | ACAGTGGGAAGGCTCTGTGTAGA |
| XPA | |
| Sense | GCTGCCCGGCCCTACTC |
| Antisense | TGAAGCCTCCTCCTGTGTCAAT |
| XPB | |
| Sense | CCAGGAAGCGGCACTATGAGG |
| Antisense | GGTCGTCCTTCAGCGGCATTT |
| XPF | |
| Sense | TATCTGGATCCTTTGTGGCACCA |
| Antisense | TTTCTCGACGATATCCTTCCTCG |
| CSA | |
| Sense | TTAAGGTATGACCAAATCCTGCCT |
| Antisense | ACAGCATTTCATGTTTAAGCCAGAT |
| CSB | |
| Sense | TTGAGCTGCAGGGTTTGGGTG |
| Antisense | TGCATCCTCCTCCAGACTGGC |
| BRCA2 | |
| Sense | CGTACACTGCTCAAATCATTC |
| Antisense | GAAATGGAGGTGGACAATCAG |
| Rad51 | |
| Sense | CTTTGGCCCACAACCCATTTC |
| Antisense | ATGGCCTTTCCTTCACCTCCAC |
Cytogenetic analysis
The chromosomal aberrations in EBV DNase transfected cells were determined in the Center for Medical Genetics, Changhua Christian Hospital. Briefly, the cells were treated with colcemid overnight to induce metaphase arrest. The harvested cells were then treated with hypotonic KCl (75 mM) buffer, fixed in a 3 : 1 mixture of methanol and acetic acid, plated onto glass slides for Giemsa staining. Chromosomal aberrations were scored in at least 200 metaphase plates for each cell line. Chromosome aberrations including dicentric chromosomes, chromosome fragments, chromatid gaps, chromosome rings, double minutes and satellite associations were counted (54).
RESULTS
EBV DNase causes formation of MN in various epithelial cell lines
We aligned the amino acid sequences of several herpesvirus ANs: α-herpesvirus HSV-1, β-herpesviruses cytomegalovirus (CMV) and varicella zoster virus (VZV), γ-herpesviruses EBV, KSHV, herpesvirus saimiri (HVS) and rhesus rhadinovirus (RRV) (Supplementary Figure S2a). Using in vitro assays, we have demonstrated that E225 and K227 of motif III are required for cleavage of DNA, while the residue F452, which is in the C-terminus of the EBV DNase but not in a conserved motif, is also critical for enzyme activity (46). To elucidate whether EBV DNase could induce genomic instability, firstly, three different DNase activity-null mutants were constructed: Mutant 1 has an F452A substitution in the C terminus (55), Mutant 2 has the E225A/K227A double substitutions in the catalytic site and Mutant 3 has the E225A/K227A/F452A triple substitutions (Supplementary Figure S2b). All of them were nearly activity-null in nuclease activity assays, compared to the wild type DNase (Supplementary Figure S2c), and their expression was detected by western blotting with DNase specific antibody (Supplementary Figure S2d). Compared to level of DNase expression in EBV reactivated EBV-infected NPC cells (NA), the expression levels of these plasmids were appropriate (Supplementary Figure S3).
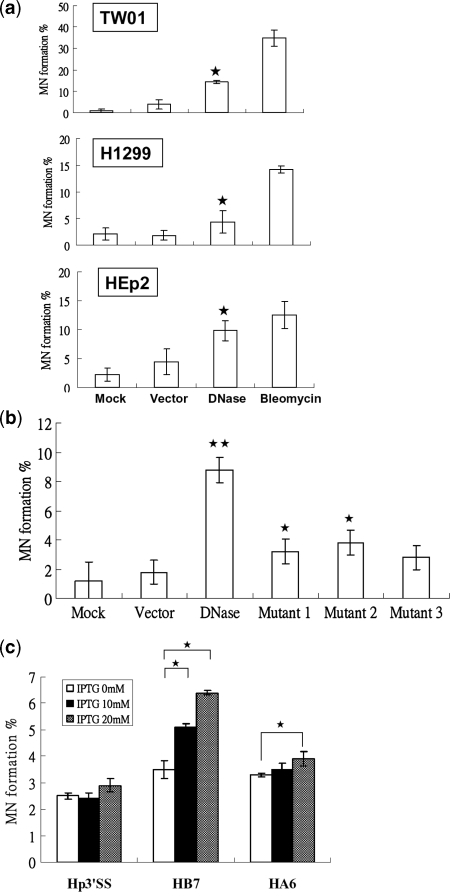
Formation of MN is considered to be indicative of chromosomal damage and is used widely in testing chemicals which may induce cytogenetic damage (56–58). To investigate whether EBV DNase has the ability to cause genomic instability, increased MN formation was detected in various epithelial cells. We carried out transient transfection of the EBV DNase-expressing plasmid into the human epithelial cell lines, TW01, H1299 and HEp2, to determine whether EBV DNase per se could induce the formation of MN. Transfection efficiency of vector and DNase plasmids in TW01, H1299 and HEp2 cell lines were determined (Supplementary Figure S4, vector: 72–82%; DNase: 55–61%; Mutant 1: 66–78%). As shown in Figure 1a and Supplementary Table 1a, EBV DNase induced formation of MN in higher percentages of TW01, H1299 and HEp2 cells than mock-transfected and vector controls. We then transfected vector, wild type DNase and three mutants plasmids into TW01 cells to determine whether nuclease activity is important for MN formation. In contrast to the wild type DNase, mutants retained some ability to induce MN formation (Figure 1b, Supplementary Table S1b). To monitor further the effect of EBV DNase on human epithelial cells, inducible expression of the enzyme was established in HEp2 cells by transfection with vector (p3′SS), wild type DNase (pIn-DNase), or DNase with a point mutation causing loss of enzyme activity (pIn-F452A). Hygromycin-resistant clones emerged 3-4 weeks later and stable transfectants, Hp3′SS, HB7, and HA6 cells, respectively, were established and expanded for further analysis. In HB7, the formation of MN increased following IPTG induction (Figure 1c, Supplementary Table S1c), but there was no difference in MN formation before and after IPTG induction of Hp3′SS. HA6 cells did not differ in the formation of MN after induction with 10 mM IPTG, but, interestingly, showed a little increase in MN formation after induction with 20 mM IPTG (Figure 1c, Supplementary Table S1c). These results indicated that the nuclease activity of EBV DNase was important for MN formation, although other effects could not be ruled out. Because increased formation of MN is considered to be an indicator of genomic instability, these results suggested that EBV DNase may contribute to genomic instability.
Figure 1.
EBV DNase induces formation of MN in epithelial cells. (a) TW01, H1299 and HEp2 cells were mock transfected, transfected with vector control and DNase-expressing plasmid, and treated with bleomycin (1 µM) as positive control. Twenty-four hours post-transfection, cells were harvested and were stained by Hoechst for counting micronuclei. (b) TW01 cells were mock transfected or transfected with vector, DNase, Mutant 1(F452A), Mutant 2 (E225A/K227A) and Mutant 3 (E225A/K227A/F452A), respectively. 24 h post-transfection, cells were harvested and were stained by Hoechst for counting micronuclei. (c) Hp3′SS (vector control), HB7 (DNase-inducible clone), and HA6 (activity-null mutant F452A-inducible clone) were treated with various doses of IPTG. After 48 h induction, cells were stained by Hoechst for counting micronuclei. The counting numbers of MN positive-cells were represented in Supplementary Table S1. In all results, the values are a mean ±SD from at least three separate experiments. Asterisk denotes P < 0.05. Two asterisks denote P < 0.01.
EBV DNase induces DNA strand breaks in epithelial cells
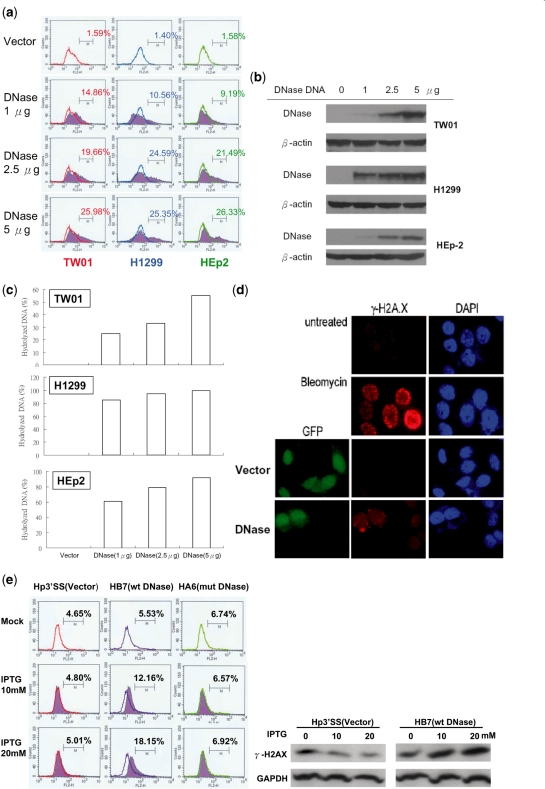
Several factors have been shown to contribute to cellular genomic instability (19,25). Induction of DNA damage and repression of repair of damaged DNA are considered to play the most important roles in genomic instability (21,59,60). Although EBV DNase has been shown to cleave DNA in vitro (8,9,11), it is not clear whether the enzyme causes DNA strand breaks when expressed in cells. Therefore, the ability of EBV DNase to damage DNA was determined in epithelial cells. Since phosphorylated histone H2AX (γH2AX) is a sensitive marker of the presence of DNA DSBs (61), we determined by flow cytometry the amount of γH2AX in epithelial cells transfected with various amounts of DNase. As shown in Figure 2a, EBV DNase caused phosphorylation of H2AX in various human epithelial cell lines in a dose-dependent manner. Protein expression and nuclease activities also were detected (Figure 2b and c). The phosphorylation of H2AX in HEp2 cells transfected with the EBV DNase plasmid also was shown by immunofluorescence staining. The cells which expressed EBV DNase were found to coincide with γH2AX-positive cells, indicating that EBV DNase caused γH2AX formation in human epithelial cells (Figure 2d). Furthermore, in the inducible lines, the phosphorylation of H2AX increased following IPTG induction of HB7 cells but there was no difference in Hp3′SS cells before and after IPTG induction, nor was there a significant increase in IPTG induced HA6 cells, which were performed by flow cytometry and western analysis (Figure 2e). DNase expression was examined and DNase protein expression increased in a dose-dependent manner following IPTG induction (Figure 2f). The nuclease activities in HB7 cells increased following IPTG addition (Figure 2g). These results indicated that EBV DNase can induce phosphorylation of H2AX and support the hypothesis that EBV DNase induces DNA strand breaks in epithelial cells.
Figure 2.
EBV DNase increases γH2AX formation in epithelial cells. (a) TW01, H1299 and HEp2 cells were transfected with various doses of EBV DNase plasmid. Vector was added to adjust for equal DNA amount in each sample. Twenty four hours post-transfection, cells were collected and incubated with γH2AX antibody for cytometry analysis. In histogram, M is defined as the portion of γH2AX-positive cells. (b) Cell lysates were collected to examine protein expression by western blotting. (c) Cell lysates of various clones were collected and assayed for nuclease activity. Nuclease activity is defined as the percentage of hydrolyzed DNA which was digested by EBV DNase. (d) HEp2 cells were mock treated, treated with bleomycin (1 µM) for 24 h or transfected with vector or DNase-expressing plasmid. Twenty four hours post-transfection, cells were examined for phosphorylation of H2AX (red fluorescence), and cell nuclei were visualized with DAPI staining. Transfectants with vector and DNase were monitored with green fluorescence (GFP-positive). (e) Hp3′SS (vector control), HB7 (DNase-inducible clone), and HA6 (activity-null mutant F452A-inducible clone) were treated with various doses of IPTG. After 48 h induction, cells were harvested and incubated with γH2AX antibody for cytometry analysis. In histogram, M is defined as the portion of γH2AX-positive cells. The protein expression of γH2AX in Hp3′SS and HB7 cells was also detected by western analysis (right panel). (f) Cell lysates were collected for western blotting to examine EBV DNase expression. (g) Cell lysates were collected and assayed for nuclease activity. Nuclease activity is defined as the percentage of hydrolyzed DNA which was digested by EBV DNase.
EBV DNase activity is important for damage to DNA
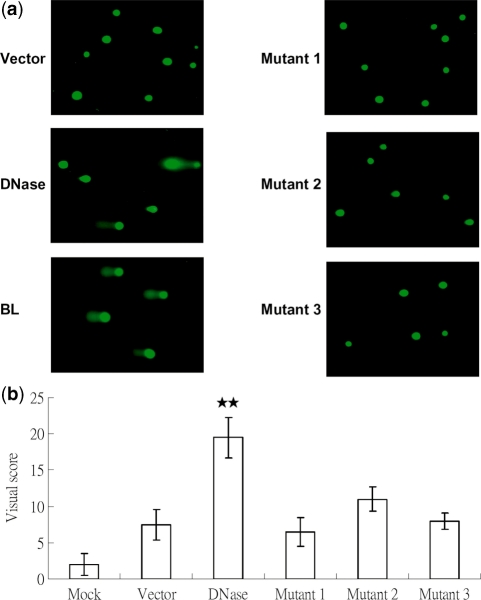
In order to confirm that nuclease activity is critical for damaging DNA, an alternative to γH2AX formation was used for detection. The comet assay (or single cell gel electrophoresis assay) is a sensitive, widely-used method to detect DNA damage (48,62). Under alkaline conditions, the comet assay can detect effectively single and double strand breaks in DNA (62). The principle and detailed methodology are described in ‘Materials and Methods’ section. For the comet assay, wild-type DNase and three activity-null mutants were transfected into TW01 cells. After 24 h, cells which were transfected with the wild-type displayed robust comet tails compared to the control, but the three mutants did not (Figure 3a). This result is consistent with the detection of γH2AX formation, implying that EBV DNase could induce DNA strand breaks in human epithelial cells. We also selected 100 cells randomly to quantify their comet centre intensity and tail lengths. After referring to standard images (Supplementary Figure S1), the wild-type DNase had the highest comet score. This meant that wild type DNase caused the heaviest DNA damage. None of the mutants had significant effects by visual quantification, compared to the vector control (Figure 3b). The results of the comet assays suggested that the nuclease activity of EBV DNase is important for damage to DNA in human epithelial cells.
Figure 3.
The nuclease activity of EBV DNase is important for the effect of DNA damage. (a) TW01 cells were transfected with vector, DNase, Mutant 1, Mutant 2 and Mutant 3, respectively. After 24 h transfection, cells were collected for detection of DNA damage. DNA damage was analyzed by comet assay, which is as described in ‘Materials and Methods’ section. (b) The results of comet assay were quantitated by visual scoring. Visual scores were presented as classes 0–4 which increase with the level of genomic DNA damage (Supplementary Figure S1). All data presented represent the means and the standard deviations of at least three independent experiments. Two asterisks denote P < 0.01.
EBV DNase inhibits repair of chemical-elicited and UV-induced DNA damage
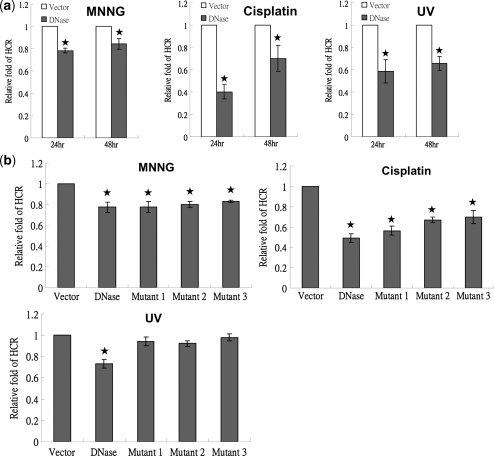
DNA repair is essential for maintenance of genome integrity and inefficient or defective repair leads indirectly to genomic instability (21). We asked next whether EBV DNase has the potential to affect DNA repair. We carried out HCR assay to investigate the effect of DNase on DNA repair (63). The HCR assay can easily measure the capacity for DNA repair quantitatively for various types of DNA damage (64,65). In the HCR assay, either the empty vector or plasmid expressing DNase, control plasmid pRL-CMV, and either a damaged or untreated luciferase reporter plasmid (pCMV-Luc) were cotransfected into TW01 epithelial cells. Because DNA-damaging chemicals and UV are the most common causes of damage used experimentally (63), first, we assayed the effect of DNase on repair of chemical-induced damage. Reporter plasmids were treated with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) or cisplatin for 3 h, purified on columns and cotransfected with DNase or vector plasmid and the internal control pRL-CMV. Surprisingly, compared to the vector control, the cells transfected with DNase had an efficiency of repair of the MNNG-damaged reporter plasmids reduced to 0.78 at 24 h and 0.84 at 48 h (P < 0.05, t-test) in the TW01 cells (Figure 4a, left). Similarly, DNase reduced cisplatin-elicited repair at 24 and 48 h (Figure 4a, middle). The effect of DNase on UV-induced repair was investigated to verify this phenomenon. Reporter plasmids were irradiated with UV before transfection and, 24 and 48 h post-transfection, the cells were harvested and examined by HCR assay. The repair efficiency of the cells transfected with EBV DNase for the UV-damaged reporter plasmids was reduced to 0.58 at 24 h and 0.65 at 48 h (P < 0.05, t-test) compared to the vector control (Figure 4a, right). Furthermore, similar results were demonstrated in the DNase-inducible lines. After 24 and 48 h of mock treatment or IPTG induction, repair of MNNG-, cisplatin- and UV-damaged DNA was repressed significantly in HB7 cells compared to that in Hp3′SS cells (data not shown). These results showed that EBV DNase caused a distinct repression of repair of DNA damage induced by various damaging agents, suggesting that EBV DNase also uses an alternative pathway to induce genomic instability.
Figure 4.
EBV DNase represses repair of chemical- and UV-damaged DNA. (a) TW01 cells were cotransfected with 2 μg/well of empty vector or a construct encoding DNase, together with a damaged or undamaged pCMV-luciferase reporter construct (pCMV-Luc, 0.1 μg/well) and Renilla reporter construct (0.1 μg/well). In the left panel, pCMV-Luc was pre-treated with MNNG (100 μM) for 3 h. In the middle panel, pCMV-Luc was pre-treated with cisplatin (10 μg/μl) for 3 h. In the right panel, pCMV-Luc was exposed to UV (2000 J/m2). Twenty-four hours and fourty-eight hours after cotransfection, luciferase activity and Renilla intensity were determined. Relative fold of HCR represents DNA repair activity, calculated as described in ‘Materials and Methods’ section. (b) TW01 cells were transfected with 2 μg/well of vector, DNase, Mutant 1, Mutant 2 or Mutant 3, together with an MNNG-treatment (100 μM) for 3 h, or cisplatin-treated (10 μg/μl), or UV-exposed (2000 J/m2), or undamaged pCMV-luciferase reporter construct (pCMV-Luc) (0.1 μg/well) and Renilla reporter construct (0.1 μg/well), respectively. Forty-eight hours after cotransfection, HCR assays were determined. Relative fold of HCR represents DNA repair activity, calculated as described in ‘Materials and Methods’ section. All data presented represent the means and the standard deviations of at least three independent experiments. Asterisk denotes P < 0.05.
Furthermore, we determined whether the nuclease activity of EBV DNase is important for repression of DNA repair. Wild type and three mutants of EBV DNase were transfected into TW01 cells and the HCR assay was used to measure the repair efficiencies of chemical- and UV-induced damage. As shown in Figure 4b, compared to wild type DNase, all three activity-null mutants were able to inhibit repair of MNNG- and cisplatin-elicited DNA damage, showing that the nuclease activity of EBV DNase was not important for repression of chemical-elicited repair. In contrast, surprisingly, the ability to inhibit UV damage-induced repair was lost in all three mutants (Figure 4b). These results indicated that, besides interference of DNA repair by nuclease activity, another mechanism for repression may exist.
EBV DNase represses transcription and translation of repair-related genes
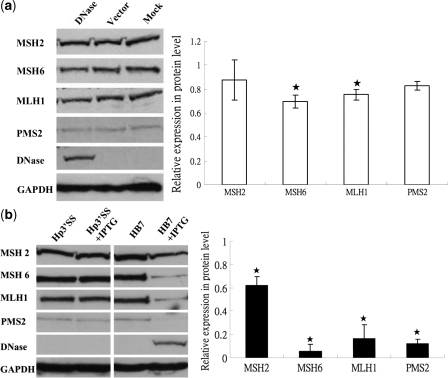
For extrinsic nucleases, the cellular repair pathway is activated by DNA damage which is sensed by cellular repair systems (66). Chang et al. reported that the APE suppressed MMR activity by repressing expression of MSH6, which is one of the MMR genes (35). In addition, MNNG and cisplatin treatment have been shown to induce expression of MMR genes (67,68) and MMR plays important roles in the repair of MNNG- and cisplatin-generated adducts (67,69), although other repair systems are also important in this process (67,70). To determine whether EBV DNase affects the protein expression levels of MMR repair genes, western blotting was used to detect expression of four of them, MSH2, MSH6, MLH1 and PMS2, in the parental, vector and DNase-transfected TW01 cells. The results indicated a decrease in the levels of MSH6 and MLH1 expression in the DNase-transfected cells compared to the vector control (P < 0.05), but no decrease was detected in MSH2 and PMS2 (Figure 5a). Although the decreased expressions of MSH6 and MLH1 were not very profound, it may be due to the transfection efficiency was only around 50–60% (Supplementary Figure 4). To investigate whether EBV DNase also can suppress expression of the repair genes in inducible lines, Hp3′SS and HB7 were treated with IPTG or not. Forty-eight hours after induction, cells were harvested and the protein expression of MMR genes was determined by western blot analysis. Surprisingly, all four proteins from HB7 cells were expressed at significantly decreased levels compared to those from the Hp3′SS vector controls (Figure 5b). Similarly, in DNase-expressing H1299 cells, MSH2 was decreased slightly and MSH6 was suppressed markedly, compared to vector-expressing cells (data not shown).
Figure 5.
EBV DNase decreases protein expressions of repair-related genes in epithelial cells. (a) In the left panel, 48 h post-transfection, the protein extracts of DNase and the empty vector transfected TW01 cells, and the parental TW01 cells (Mock) were examined by western blot using MSH2, MSH6, MLH1 and PMS2 antibodies. The detection of GAPDH was used as the loading control. In the right panel, the expression levels were quantified by densitometry. After standardizing with loading control, they were expressed as the relative expression compared with the vector controls. The values are a mean ± SD from three separate experiments. Asterisk denotes P < 0.05. (b) In the left panel, after 48 h IPTG (20 mM) induction or mock-treated, the protein extracts of Hp3′SS and HB7 were examined by western blot using MSH2, MSH6, MLH1 and PMS2 antibodies. The detection of GAPDH was used as the loading control. In the right panel, the expression levels were quantified. After standardization with loading control, they were expressed as the relative expression compared to those from the Hp3′SS vector controls. The values are a mean ±SD from three separate experiments. Asterisk denotes P < 0.05.
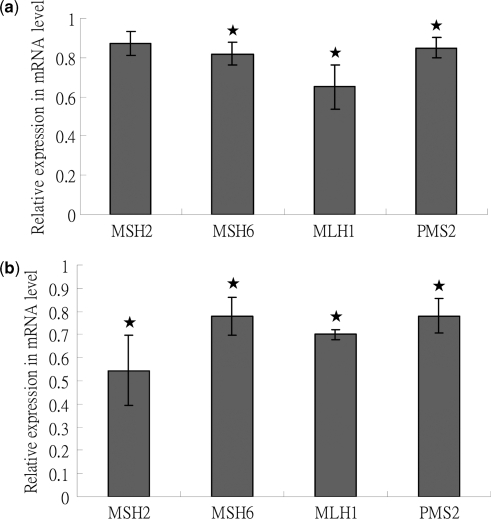
To investigate this phenomenon further, we measured the concentrations of MSH2, MSH6, MLH1 and PMS2 mRNAs from DNase-expressing TW01 cells by QPCR to investigate whether the transcription of MMR-related genes was repressed. As shown in Figure 6a, the transcripts of MSH6, MLH1 and PMS2 were decreased to nearly 0.8-fold of vector control (P < 0.05, t-test). This result was consistent with the results of protein expression. Concentrations of MMR mRNA were also quantified in Hp3’SS and HB7 cells after 48 h induction with IPTG or not. MSH2 expression was decreased the most (0.55-fold of vector control, P < 0.05, t-test) while expression of MSH6, MLH1 and PMS2 decreased to 0.7–0.78-fold compared with the vector control (P < 0.05, t-test, Figure 6b). The mRNA concentrations of four genes in the IPTG-untreated Hp3′SS and HB7 were similar to IPTG-treated Hp3′SS cells (data not shown). The decreasing levels of RNA expression were not correlated with protein expression in HB7 cells, although repression of protein expression also had occurred. These unexpected findings may result from specific differences between TW01 and HEp2 cells. To address the effect of DNase activity on the decreased mRNA of these repair genes, we further detected the mRNA expressions of these genes in Mutants 1, 2 and 3-transfected TW01 cells. The mRNA levels of repair genes in Mutants 1, 2 and 3-transfected TW01 cells were similar to that of wild type DNase-transfected cells (data not shown). This result suggested that DNase activity may not be involved in decreased expression of repair genes.
Figure 6.
EBV DNase decreases mRNA amounts of MMR-related genes in epithelial cells. (a) Forty-eight hours post-transfection, total RNA of DNase and the empty vector transfected TW01 cells, and the parental TW01 cells were extracted. After RT, cDNA were examined for the mRNA expression of MSH2, MSH6, MLH1 and PMS2. The detection of GAPDH was used as the control. The expression level of each sample was standardized with that of GAPDH and then expressed as the relative expression compared to the vector controls. (b) After 48 h IPTG (20 mM) induction, total RNA of Hp3′SS and HB7 cells were extracted. After RT, cDNA were examined for the mRNA expression of MSH2, MSH6, MLH1 and PMS2. The detection of GAPDH was used as the control. The expression level of each sample was standardized with that of GAPDH and expressed as the relative expression compared to the decreased levels of vector controls. In all of results, the values are a mean ±SD from at least three separate experiments. Asterisk denotes P < 0.05.
The repression of expression of MMR-related genes at the mRNA and protein levels prompted us to ask whether expression of other repair-related genes also was affected by EBV DNase. To evaluate this possibility, several repair-related genes were selected to investigate their mRNA expression in the mock-treated and IPTG-treated Hp3′SS and HB7 cells. As shown in Table 2, as well as the MMR-related genes, almost all of the selected genes were decreased. Among the NER-related genes, ERCC1 was decreased significantly compared to the control (0.38-fold of control, P < 0.01, t-test) and XPF was decreased 0.8-fold compared to the vector control (P < 0.05, t-test). However, XPA and XPB were not significantly down-regulated. Among the BER-related genes, CSA and CSB were reduced 0.68- and 0.77-fold compared to the vector controls (P < 0.05, t-test). Among the HR-related genes, expression of BRCA2 and Rad51 also was down-regulated 0.8- and 0.58-fold compared to the vector controls (P < 0.05, t-test). These results revealed that EBV DNase can suppress the expression of various repair-related genes and thus diminish cellular repair activities, however, it could not down-regulate all of the NER- related genes, which may imply certain target specificity.
Table 2.
Relative expression of repair-related genes in EBV DNase-expressing cells
| Repair-related genes | Decrease folds of mRNA (mean±SD) |
|---|---|
| MMR | |
| MSH2 | 0.546±0.152* |
| MSH6 | 0.779±0.083* |
| MLH1 | 0.700±0.022* |
| PMS2 | 0.781±0.075* |
| NER | |
| ERCC1 | 0.384+0.360** |
| XPA | 0.992+0.083 |
| XPB | 0.934±0.054 |
| XPF | 0.796+0.045* |
| BER/TCR | |
| CSA | 0.678±0.102* |
| CSB | 0.765±0.045* |
| HR | |
| BRCA2 | 0.793±0.030* |
| Rad51 | 0.577±0.027* |
The values are a mean ±SD from at least three separate experiments.
*P<0.05.
**P<0.01.
The function of host-shut off of EBV DNase may contribute in repression of DNA repair
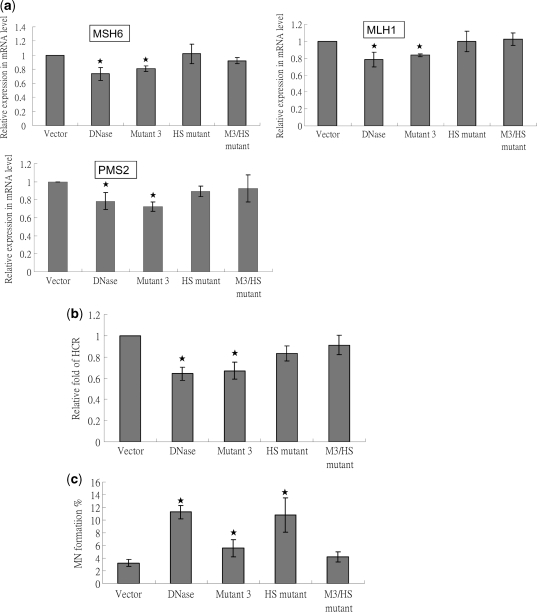
Recently, EBV DNase and its homolog KSHV SOX were demonstrated to shut-off universal host mRNA in expressing cells (71,72), and thus affect cellular functions. We then addressed whether the function of host shut-off of EBV DNase plays an important role in repression of DNA repair. Several groups had defined the important host shut-off residues of gammaherpesviral DNases (73,74). Based on these data, we cloned two host shut-off-null mutants, which were designated as HS mutant (K231M) and M3/HS mutant (E225A/K227A/F452A/K231M), to perform further experiments (Supplementary Figure 2a and b). After confirming by DNA sequencing, DNase activity and protein expression of these mutants were detected (Supplementary Figure 2c and d). The results revealed that HS mutant had DNase activity but M3/HS mutant did not (Supplementary Figure 2c). In addition, mRNAs of several repair genes were detected. As shown in Figure 7a, compared to wild type and Mutant 3, HS and M3/HS mutants had lost the ability to down-regulate repair genes MSH6, MLH1 and PMS2. This loss of host shut-off ability was compatible with previous studies by others (72,74) and obviously affected the repression of cellular repair activity (Figure 7b).
Figure 7.
Host shut-off mutants do not induce repair repression but the mutants with DNase activity induce MN formation in epithelial cells. (a) Total RNAs of vector, DNase and mutants transfected TW01 cells were extracted 48 h post-transfection. After RT, cDNA were examined for the mRNA expression of MSH6, MLH1 and PMS2 with GAPDH as the internal control. Then the relative expression compared to the vector control was presented. The values are a mean ±SD from at least three separate experiments. (b) TW01 cells were cotransfected with 2 μg/well of empty vector or a construct encoding DNase, together with a damaged or undamaged pCMV-luciferase reporter construct (pCMV-Luc, 0.1 μg/well) and Renilla reporter construct (0.1 μg/well). pCMV-Luc was pre-treated with cisplatin (10 μg/μl) for 3 h. Luciferase activity and Renilla intensity were determined 48 h after cotransfection. Relative fold of HCR represents DNA repair activity, calculated as described in ‘Materials and Methods’ section. (c) TW01 cells were transfected with vector control, DNase and mutants-expressing plasmids. Cells were harvested and were stained by Hoechst for counting micronuclei 24 h post-transfection. The values are a mean ±SD from at least three separate experiments. In all of results, asterisk denotes P < 0.05.
Finally, we tried to define the importance of host shut-off in DNase-induced genomic instability. As shown in Figure 7c, mutants which defected in DNase activity also significantly decreased the ability of MN induction (Mutant 3 and M3/HS mutant). However, since Mutant 3 had a little increase and M3/HS mutant had no significant effect compared to vector control suggesting host shut-off has a little contribution to MN formation. These results provided the strong evidence that, in EBV DNase-inducing genomic instability, DNA damage plays a major role and repair repression is a cooperative and auxiliary role.
In conclusion, these results indicate host shut-off may contribute partly in the induction of genomic instability, although its effect is relatively weaker.
EBV DNase increases chromosomal aberrations
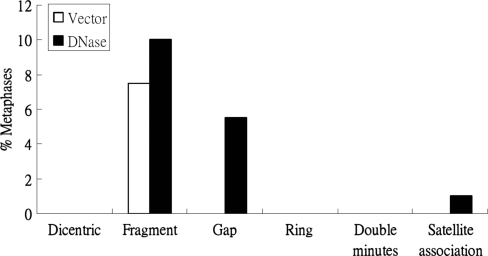
Because, based on MN formation, EBV DNase could induce genomic instability, we investigated whether EBV DNase had the ability to cause chromosomal aberration (CA). To this end, TW01 cells transfected EBV DNase or vector for 24 h were subjected to karyotype analysis. CA, including dicentric chromosomes, chromosome fragments, chromatid gaps, chromosome rings, double minutes and satellite associations were scored in 200 metaphase plates of each vector and DNase transfected TW01 cells. As shown in Figure 8, the cells transfected with DNase had higher CA than the vector control in terms of chromosome fragments, gaps and satellite associations. The results of chromosome analysis showed that EBV DNase had the ability to enhance chromosome aberrations.
Figure 8.
DNase induces chromosomal aberrations in epithelial cells. Chromosomal aberrations, including dicentric chromosomes, chromosome fragments, chromatid gaps, chromosome rings, double minutes and satellite associations were scored in 200 metaphase plates of DNase-expressing cells and vector control. The frequency of chromosomal aberrations was represented as the percentage of scoring metaphase plates.
EBV DNase causes microsatellite instability and genetic mutation
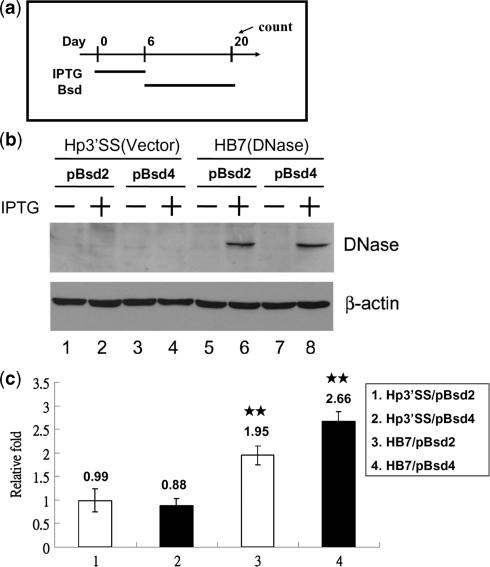
Because EBV DNase can induce DNA strand breaks, inhibit DNA repair and chromosome aberrations, it was assumed to act as a mutagen. We carried out a mutation assay to investigate whether the expression of EBV DNase causes genetic mutation and MSI. Hp3′SS (a vector control) and HB7 (wild-type) cells were transfected with two plasmids, pBsd2-puro and pBsd4-puro. After transfection in HEp2 cells, the transfectants were selected with puromycin (1 µg/ml). Puromycin-resistant clones emerged 2-3 weeks later and were designated Hp3′SS/pBsd2, Hp3′SS/pBsd4, HB7/pBsd2 and HB7/pBsd4. Parental HEp2 cells did not grow in the presence of blasticidin (5 µg/ml) (survival ratio < 10−6). These transfectants were incubated with IPTG to induce DNase expression for 6 days and selected with blasticidin (5 µg/ml) for 14 days (Figure 9a and 9b). If genetic mutations (for pBsd2-puro: TAG changed to ATG and for pBsd4-puro: out-frame shifted to in-frame) did occur, these cells would become blasticidin resistant and grow in the presence of blasticidin. As a consequence, the blasticidin resistant colonies were considered to have genetic mutations. We found that the mutation rates obtained from the DNase-induced cells were increased to 1.95 and 2.66-fold for HB7/pBsd2 and HB7/pBsd4, respectively (P < 0.01) (Figure 9c). These data indicated that expression of EBV DNase is able to cause genetic mutations. Because these mutations also may have resulted from MSI, we propose that EBV DNase may induce DNA mutation, MSI and lead to a mutator phenotype.
Figure 9.
EBV DNase enhances MSI and gene mutation. (a) Schematic representation of treatment with IPTG and blasticidin. Hp3′SS and HB7 cells were transfected with pBsd2-puro or pBsd4-puro, respectively, and then transfectants were selected with puromycin (1 µg/ml). Puromycin-resistant cells were collected and treated with 10 mM of IPTG or not for 6 days. These cells were maintained at a density of 2 × 103 or 1 × 104 cells per 100- mm culture dishes in the presence of blasticidin (5 µg/ml). After 14 days of incubation, the cells were stained with 0.05% p-iodonitrotetrazolium violet dye and then colonies larger than 1 mm were counted. (b) Hp3′SS or HB7 cells were transfected with pBsd2-puro or pBsd4-puro, and puromycin-resistant cells were mock treated (lanes 1, 3, 5 and 7) or treated with IPTG (lanes 2, 4, 6 and 8) for 24 h. Cell lysates were analyzed by western blotting with antibodies against EBV DNase and β-actin. (c) The frequency of gene mutation was calculated by dividing the number of blasticidin-resistant colonies by the number of cells seeded. The relative fold changes indicate the frequency of gene mutation in the presence of IPTG, calculated by dividing by that in the absence of IPTG. The values are a mean ±SD from three separate experiments. Two asterisks denote P < 0.01.
DISCUSSION
In this study, EBV DNase, a member of viral ANs, was transfected into human epithelial cells, TW01, H1299 and HEp2. The results indicate that EBV DNase could induce genomic instability in various human epithelial cells (Figure 1a). This phenomenon was further corroborated with the results obtained from DNase inducible cells (Figure 1b). DNA damage and repression of DNA repair may contribute to the induction of genomic instability. Using γH2AX formation and comet assay, we found that EBV DNase is able to induce DNA strand breaks in transient transfection and inducible DNase expression system (Figures 2 and 3). By HCR, EBV DNase was able to repress repair of DNA damage by both chemical treatments (MNNG and cisplatin) and UV irradiation (Figure 4). Through western blot and QPCR, expressions of repair-related genes were reduced in transient expression and inducible DNase expressing cells (Figures 5 and 6). We further performed QPCR to quantitate relative expression of repair-related genes in EBV DNase expressing cells and found many of genes involved in MMR, NER, BER/TCR and HR were reduced in their expressions (Table 2). These results provide a possible mechanism for viruses carrying ANs in the carcinogenesis of human cancers.
It is conceivable that a protein possesses DNase activity would damage DNA. γH2AX formation is an immediate early step after the occurrence of DNA DSBs (75). In our study, EBV DNase increased phosphorylation of H2AX in a dose-dependent manner (Figure 2a). Furthermore, activity-null mutants were found to be less effective in the induction of DNA damage (Figure 3). These results provided strong evidence that nuclease activity is important for induction of DNA damage.
Repression of DNA repair may result in genomic instability (21). It was surprising for us to find EBV DNase repressed HCR in this study (Figure 4). Although it revealed that inhibition of DNA repair seemed just only play as an auxiliary worker in the process of DNase-induced genomic instability (Figure 7), it would be interesting to see by what mechanism(s) did EBV DNase repress these cellular repair activities? Several lines of evidence suggested that appropriate expression of nucleases increased repair activities and is beneficial to the maintenance of genome integrity (76–78), however, other investigations supported the concept that expression of nucleases would not only repress repair activity but also increase the mutation rate (34,35,79). Furthermore, ectopic expression of restriction endonucleases may increase repair activities because of DNA damage (80). In our experiments, EBV DNase clearly down-regulated chemical- and UV-induced repair (Figure 4). These results led logically to postulate that repair mechanisms were likely impaired in these cells. The damage signal was probably activated, because H2AX was phosphorylated (Figure 2), however, repair activity was suppressed at the same time (Figure 4). It can be speculated repression of DNA repair overwhelmed the damage signaling. The finding that repression of repair resolved slightly from 24 to 48 h may support the possibility that increasing strength of damage signal overcame the DNA repair-repressing effect of EBV DNase (Figure 4). In addition, because down-regulation of repair-related components would decrease repair activity significantly, the result of down-regulation of repair-related proteins by EBV DNase may explain how chemical- and UV-induced repair repression occurred (Figures 5 and 6, Table 2). Interestingly, the nuclease activity of EBV DNase appeared to be important for repair of UV-damaged DNA, but not for repair of MNNG- and cisplatin-induced damage (Figure 4). Because NER was suggested to be involved mostly in repairing UV-damaged DNA, these findings imply that the nuclease activity of EBV DNase interferes with the NER pathway. However, since EBV DNase interfered NER, BER or MMR expressions differently (Table 2), the detailed roles that the nuclease plays in different repair processes require careful clarification.
Another interesting question can be raised. How did EBV DNase repress expression of these repair-related genes? Three possible mechanisms may be considered. First, EBV DNase may eliminate expression of repair-related genes through turnover of mRNA. It has been shown that EBV DNase blocks the synthesis of HLA class I and II by eliminating mRNAs (72). Our results showing that EBV DNase inhibits expression of repair genes support this possibility (Table 2). Furthermore, as shown in Figure 7, compared to wild type, although host shut-off mutant HS increased approximately similar level of MN formation, which suggested that host shut-off may not be important for induction of genomic instability, however, Mutant 3 lost DNase activity but keeping host shut-off function still increased little but significant level of MN formation. Of note, EBV DNase likely did not inhibit all repair gene expression (Table 2). This implies that EBV DNase may act preferentially on mRNA turnover. We also measured by QPCR the amounts of mRNA of the housekeeping genes GAPDH, β-actin and p53 which were normalized with the amount of 18S RNA (Supplementary Figure 5a). Results showed that the amount of GAPDH RNA of IPTG-treated HB7 cells is almost equal to that of mock-treated HB7, while that of β-actin and p53 decreased in the cells under IPTG induction (Supplementary Figure S5a). Western blot indicated that levels of the protein expressions from β-actin and GAPDH were coincident with that of mRNA by QPCR (Supplementary Figure S5b). This is also why we used GAPDH as internal control in the host shut-off experiments. Similarly, an earlier report suggested that the DNase of KSHV affected mRNA turnover (71). Microarray data revealed that a small population of host mRNA escaped elimination by KSHV DNase (81). These results demonstrated that host shut-off may contribute in repair repression auxiliarily (Figure 7). Unfortunately, so far, we do not know which mRNAs are eliminated preferentially by the γ-herpesvirus DNases. Recently, Lee et al. suggested that aberrant herpesvirus –induced polyadenylation correlates with host shut-off function (82). These studies revealed that the precise mechanism(s) of RNA turnover may be quite complex and thus far they remain unclear.
Secondly, overexpression of APE alone reduced MMR function markedly and increased MSI through repression of MSH6 expression (35). Ectopic expression of E. coli exonuclease III repressed certain repair activities in the breast cancer cells (83). These reports suggested that unbalanced expression of nucleases may repress expression of repair proteins, disturb cellular repair pathways, and increase genomic instability and genetic mutation. On this basis, it is possible that unbalanced expression of EBV DNase may cause down-regulation of repair proteins, disturb repair systems, and eventually repress repair activity.
Finally, we cannot rule out the possibility that EBV DNase may destroy the genes that are responsible for repair. However, if this is the case, it would be difficult to explain how activity-null mutants repressed chemical-elicited repairs (Figure 4b). On other hand, the finding that the luciferase-reporter genes were also detectable by PCR provided more indirect evidence to suggest that EBV DNase may not destroy large numbers of repair-related genes (data not shown). This interesting question is worth further investigation.
EBV, KSHV, hepatitis B virus (HBV), HCV, HTLV and HPV infections are considered to be associated with the development of various human cancers (84). All these viral-associated human malignancies possess significant genomic instabilities and chromosomal aberrations. Genetic changes in these malignancies have been linked explicitly to the virus infections and expression of viral proteins. HTLV Tax has been suggested to inhibit BER by interfering with DNA polymerase β (85) and thereby induce increase of MN formation (86). HBV HBx diminished NER by interacting with UV-damaged binding protein (87) and could induce MN formation (88). HBV preS also has been shown to cause oxidative DNA damage by generating endoplasmic reticulum stress (89). HCV has been shown to induce DNA damage or mutation through expression of the core protein or nonstructural protein 3 (27,49,90). In addition, HPV E6/E7 has been shown to induce numerical chromosome instability (91,92) and E6 was suggested to inhibit NER and BER by E6-mediated interfering with DNA repair proteins (93). In the herpesviruses, the KSHV latent protein LANA plays an important role in induction of chromosomal instability (94). Moreover, regarding the EBV latent proteins, LMP1 was reported to induce genomic instability through repression of NER (47,95), EBNA 1 induces genomic instability via induction of reactive oxygen species (96) and EBNA2 and EBNA3C disrupt the mitotic checkpoint and causes chromosomal instability (97,98). Among these viral gene products, most are involved in the generation of genomic instability by induction of DNA damage indirectly, repression of DNA repair, and disturbance of DNA replication and cytokinesis. However, we find that EBV DNase is unique in that it may contribute both through direct induction of DNA damage and indirect repression of DNA repair leading to increased chromosomal alteration (Figure 8), gene mutation and MSI (Figure 9). Interestingly, it was found that lytic proteins were expressed immediately but transiently in virus primary infection (99,100).This phenomenon implies that EBV lytic proteins may have chance to ‘hit’ or ‘affect’ cellular DNA in the temporary expression and then play an important role in the subsequent carcinogenesis. Our results provide a new paradigm of a viral protein which may contribute to the carcinogenesis of human malignancies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council (grant numbers NSC89-2320-B002-198, NSC90-2320-B002-135, NSC94-2321-B002-023, NSC95-2320-B002-057 and NSC96-2320-B002-072). Funding for open access charge: National Institute of Cancer Research, National Health Research Institutes, Taiwan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are indebted to Dr T. J. Harrison (UCL Medical School, London, U.K.), Dr S. F. Lin (National Institute of Cancer Research, NHRI, Taiwan) and Dr Y. Chang (National Institute of Cancer Research, NHRI, Taiwan) for critical reviews and comments on the paper.
REFERENCES
- 1.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Rohrmann GF. Characterization of a baculovirus alkaline nuclease. J. Virol. 2000;74:6401–6407. doi: 10.1128/jvi.74.14.6401-6407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter IM, Stow ND. Virus particles produced by the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12 contain abnormal genomes. J. Gen. Virol. 2004;85:583–591. doi: 10.1099/vir.0.19657-0. [DOI] [PubMed] [Google Scholar]
- 5.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 6.Feederle R, Bannert H, Lips H, Muller-Lantzsch N, Delecluse HJ. The Epstein-Barr virus alkaline exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 2009;83:4952–4962. doi: 10.1128/JVI.00170-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang CX, Decaussin G, de Turenne Tessier M, Daillie J, Ooka T. Identification of an Epstein-Barr virus-specific desoxyribonuclease gene using complementary DNA. Nucleic Acids Res. 1987;15:2707–2717. doi: 10.1093/nar/15.6.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baylis SA, Purifoy DJ, Littler E. The characterization of the EBV alkaline deoxyribonuclease cloned and expressed in E. coli. Nucleic Acids Res. 1989;17:7609–7622. doi: 10.1093/nar/17.19.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stolzenberg MC, Ooka T. Purification and properties of Epstein-Barr virus DNase expressed in Escherichia coli. J. Virol. 1990;64:96–104. doi: 10.1128/jvi.64.1.96-104.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen MR, Hsu TY, Chen JY, Yang CS. Molecular characterization of a cDNA clone encoding the Epstein-Barr virus (EBV) DNase. J. Virol. Methods. 1990;29:127–141. doi: 10.1016/0166-0934(90)90107-q. [DOI] [PubMed] [Google Scholar]
- 11.Lin SF, Lin SW, Hsu TY, Liu MY, Chen JY, Yang CS. Functional analysis of the amino terminus of Epstein-Barr virus deoxyribonuclease. Virology. 1994;199:223–227. doi: 10.1006/viro.1994.1115. [DOI] [PubMed] [Google Scholar]
- 12.Cheng YC, Chen JY, Glaser R, Henle W. Frequency and levels of antibodies to Epstein-Barr virus-specific DNase are elevated in patients with nasopharyngeal carcinoma. Proc. Natl Acad. Sci. USA. 1980;77:6162–6165. doi: 10.1073/pnas.77.10.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JY, Hwang LY, Beasley RP, Chien CS, Yang CS. Antibody response to Epstein-Barr-virus-specific DNase in 13 patients with nasopharyngeal carcinoma in Taiwan: a retrospective study. J. Med. Virol. 1985;16:99–105. doi: 10.1002/jmv.1890160202. [DOI] [PubMed] [Google Scholar]
- 14.Sbih-Lammali F, Berger F, Busson P, Ooka T. Expression of the DNase encoded by the BGLF5 gene of Epstein-Barr virus in nasopharyngeal carcinoma epithelial cells. Virology. 1996;222:64–74. doi: 10.1006/viro.1996.0398. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 16.Michor F, Iwasa Y, Lengauer C, Nowak MA. Dynamics of colorectal cancer. Semin. Cancer Biol. 2005;15:484–493. doi: 10.1016/j.semcancer.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 18.Huang DP, Lo KW, Choi PH, Ng AY, Tsao SY, Yiu GK, Lee JC. Loss of heterozygosity on the short arm of chromosome 3 in nasopharyngeal carcinoma. Cancer Genet. Cytogenet. 1991;54:91–99. doi: 10.1016/0165-4608(91)90035-s. [DOI] [PubMed] [Google Scholar]
- 19.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 20.Nowak MA, Komarova NL, Sengupta A, Jallepalli PV, Shih Ie M, Vogelstein B, Lengauer C. The role of chromosomal instability in tumor initiation. Proc. Natl Acad. Sci. USA. 2002;99:16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon K, Kopras E. Genetic alterations and DNA repair in human carcinogenesis. Semin. Cancer Biol. 2004;14:441–448. doi: 10.1016/j.semcancer.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 23.Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 2004;104:14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- 24.Gollin SM. Mechanisms leading to chromosomal instability. Semin. Cancer Biol. 2005;15:33–42. doi: 10.1016/j.semcancer.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Jefford CE, Irminger-Finger I. Mechanisms of chromosome instability in cancers. Crit. Rev. Oncol. Hematol. 2006;59:1–14. doi: 10.1016/j.critrevonc.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Tsukasaki K. Genetic instability of adult T-cell leukemia/lymphoma by comparative genomic hybridization analysis. J. Clin. Immunol. 2002;22:57–63. doi: 10.1023/a:1014471500757. [DOI] [PubMed] [Google Scholar]
- 27.Machida K, Cheng KT, Sung VM, Shimodaira S, Lindsay KL, Levine AM, Lai MY, Lai MM. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl Acad. Sci. USA. 2004;101:4262–4267. doi: 10.1073/pnas.0303971101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadaja M, Sumerina A, Verst T, Ojarand M, Ustav E, Ustav M. Genomic instability of the host cell induced by the human papillomavirus replication machinery. EMBO J. 2007;26:2180–2191. doi: 10.1038/sj.emboj.7601665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan H, Zhou F, Gao SJ. Kaposi's; sarcoma-associated herpesvirus induction of chromosome instability in primary human endothelial cells. Cancer Res. 2004;64:4064–4068. doi: 10.1158/0008-5472.CAN-04-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gualandi G, Giselico L, Carloni M, Palitti F, Mosesso P, Alfonsi AM. Enhancement of genetic instability in human B cells by Epstein-Barr virus latent infection. Mutagenesis. 2001;16:203–208. doi: 10.1093/mutage/16.3.203. [DOI] [PubMed] [Google Scholar]
- 31.Kamranvar SA, Gruhne B, Szeles A, Masucci MG. Epstein-Barr virus promotes genomic instability in Burkitt's; lymphoma. Oncogene. 2007;26:5115–5123. doi: 10.1038/sj.onc.1210324. [DOI] [PubMed] [Google Scholar]
- 32.Fang CY, Lee CH, Wu CC, Chang YT, Yu SL, Chou SP, Huang PT, Chen CL, Hou JW, Chang Y. Recurrent chemical reactivations of EBV promotes genome instability and enhances tumor progression of nasopharyngeal carcinoma cells. Int. J. Cancer. 2009;124:2016–2025. doi: 10.1002/ijc.24179. [DOI] [PubMed] [Google Scholar]
- 33.Russo CA, Weber TK, Volpe CM, Stoler DL, Petrelli NJ, Rodriguez-Bigas M, Burhans WC, Anderson GR. An anoxia inducible endonuclease and enhanced DNA breakage as contributors to genomic instability in cancer. Cancer Res. 1995;55:1122–1128. [PubMed] [Google Scholar]
- 34.Gladdy RA, Taylor MD, Williams CJ, Grandal I, Karaskova J, Squire JA, Rutka JT, Guidos CJ, Danska JS. The RAG-1/2 endonuclease causes genomic instability and controls CNS complications of lymphoblastic leukemia in p53/Prkdc-deficient mice. Cancer Cell. 2003;3:37–50. doi: 10.1016/s1535-6108(02)00236-2. [DOI] [PubMed] [Google Scholar]
- 35.Chang IY, Kim SH, Cho HJ, Lee DY, Kim MH, Chung MH, You HJ. Human AP endonuclease suppresses DNA mismatch repair activity leading to microsatellite instability. Nucleic Acids Res. 2005;33:5073–5081. doi: 10.1093/nar/gki829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fautrel A, Andrieux L, Musso O, Boudjema K, Guillouzo A, Langouet S. Overexpression of the two nucleotide excision repair genes ERCC1 and XPC in human hepatocellular carcinoma. J. Hepatol. 2005;43:288–293. doi: 10.1016/j.jhep.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryant PE, Riches AC. Oncogenic transformation of murine C3H 10T1/2 cells resulting from DNA double-strand breaks induced by a restriction endonuclease. Br. J. Cancer. 1989;60:852–854. doi: 10.1038/bjc.1989.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryant PE, Johnston PJ. Restriction-endonuclease-induced DNA double-strand breaks and chromosomal aberrations in mammalian cells. Mutat. Res. 1993;299:289–296. doi: 10.1016/0165-1218(93)90105-m. [DOI] [PubMed] [Google Scholar]
- 40.Singh B, Bryant PE. Induction of mutations at the thymidine kinase locus in CHO cells by restriction endonucleases. Mutagenesis. 1991;6:219–223. doi: 10.1093/mutage/6.3.219. [DOI] [PubMed] [Google Scholar]
- 41.Chenet-Monte C, Mohammad F, Celluzzi CM, Schaffer PA, Farber FE. Herpes simplex virus gene products involved in the induction of chromosomal aberrations. Virus Res. 1986;6:245–260. doi: 10.1016/0168-1702(86)90073-0. [DOI] [PubMed] [Google Scholar]
- 42.Lin CT, Wong CI, Chan WY, Tzung KW, Ho JK, Hsu MM, Chuang SM. Establishment and characterization of two nasopharyngeal carcinoma cell lines. Lab Invest. 1990;62:713–724. [PubMed] [Google Scholar]
- 43.Kim MS, Li SL, Bertolami CN, Cherrick HM, Park NH. State of p53, Rb and DCC tumor suppressor genes in human oral cancer cell lines. Anticancer Res. 1993;13:1405–1413. [PubMed] [Google Scholar]
- 44.Toolan HW. Transplantable human neoplasms maintained in cortisone-treated laboratory animals: H.S. No. 1; H.Ep. No. 1; H.Ep. No. 2; H.Ep. No. 3; and H.Emb.Rh. No. 1. Cancer Res. 1954;14:660–666. [PubMed] [Google Scholar]
- 45.Tsai CH, Liu MT, Chen MR, Lu J, Yang HL, Chen JY, Yang CS. Characterization of monoclonal antibodies to the Zta and DNase proteins of Epstein-Barr virus. J. Biomed. Sci. 1997;4:69–77. doi: 10.1007/BF02255596. [DOI] [PubMed] [Google Scholar]
- 46.Liu MT, Hu HP, Hsu TY, Chen JY. Site-directed mutagenesis in a conserved motif of Epstein-Barr virus DNase that is homologous to the catalytic centre of type II restriction endonucleases. J. Gen. Virol. 2003;84:677–686. doi: 10.1099/vir.0.18739-0. [DOI] [PubMed] [Google Scholar]
- 47.Liu MT, Chen YR, Chen SC, Hu CY, Lin CS, Chang YT, Wang WB, Chen JY. Epstein-Barr virus latent membrane protein 1 induces micronucleus formation, represses DNA repair and enhances sensitivity to DNA-damaging agents in human epithelial cells. Oncogene. 2004;23:2531–2539. doi: 10.1038/sj.onc.1207375. [DOI] [PubMed] [Google Scholar]
- 48.Collins AR. The comet assay for DNA damage and repair: principles, applications and limitations. Mol. Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 49.Naganuma A, Dansako H, Nakamura T, Nozaki A, Kato N. Promotion of microsatellite instability by hepatitis C virus core protein in human non-neoplastic hepatocyte cells. Cancer Res. 2004;64:1307–1314. doi: 10.1158/0008-5472.can-03-2992. [DOI] [PubMed] [Google Scholar]
- 50.McGurk CJ, Cummings M, Koberle B, Hartley JA, Oliver RT, Masters JR. Regulation of DNA repair gene expression in human cancer cell lines. J. Cell Biochem. 2006;97:1121–36. doi: 10.1002/jcb.20711. [DOI] [PubMed] [Google Scholar]
- 51.Cheng L, Guan Y, Legerski RJ, Einspahr J, Bangert J, Alberts DS, Wei Q. Expression in normal human tissues of five nucleotide excision repair genes measured simultaneously by multiplex reverse transcription-polymerase chain reaction. Cancer Epidemiol. Biomarkers Prev. 1999;8:801–7. [PubMed] [Google Scholar]
- 52.Vogel U, Dybdahl M, Frentz G, Nexo BA. DNA repair capacity: inconsistency between effect of over-expression of five NER genes and the correlation to mRNA levels in primary lymphocytes. Mutat. Res. 2000;461:197–210. doi: 10.1016/s0921-8777(00)00051-3. [DOI] [PubMed] [Google Scholar]
- 53.Yuan R, Fan S, Wang JA, Meng Q, Ma Y, Schreiber D, Goldberg ID, Rosen EM. Coordinate alterations in the expression of BRCA1, BRCA2, p300, and Rad51 in response to genotoxic and other stresses in human prostate cancer cells. Prostate. 1999;40:37–49. doi: 10.1002/(sici)1097-0045(19990615)40:1<37::aid-pros5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 54.Bale SS. Cytological effects of kepone on Chinese hamster cells. J. Hered. 1983;74:123–124. doi: 10.1093/oxfordjournals.jhered.a109737. [DOI] [PubMed] [Google Scholar]
- 55.Liu MT, Hsu TY, Lin SF, Seow SV, Liu MY, Chen JY, Yang CS. Distinct regions of EBV DNase are required for nuclease and DNA binding activities. Virology. 1998;242:6–13. doi: 10.1006/viro.1997.8974. [DOI] [PubMed] [Google Scholar]
- 56.Heddle JA. A rapid in vivo test for chromosomal damage. Mutat Res. 1973;18:187–190. doi: 10.1016/0027-5107(73)90035-3. [DOI] [PubMed] [Google Scholar]
- 57.Parry JM, Parry EM. Comparisons of tests for aneuploidy. Mutat. Res. 1987;181:267–287. doi: 10.1016/0027-5107(87)90104-7. [DOI] [PubMed] [Google Scholar]
- 58.Sternes KL, Vig BK. Micronuclei, kinetochores and hypoploidy: tests with some agents. Mutagenesis. 1989;4:425–431. doi: 10.1093/mutage/4.6.425. [DOI] [PubMed] [Google Scholar]
- 59.Coleman WB, Tsongalis GJ. Multiple mechanisms account for genomic instability and molecular mutation in neoplastic transformation. Clin. Chem. 1995;41:644–657. [PubMed] [Google Scholar]
- 60.Kastan MB. Genetic instability and tumorigenesis: introduction. Curr. Top Microbiol. Immunol. 1997;221:1–4. doi: 10.1007/978-3-642-60505-5_1. [DOI] [PubMed] [Google Scholar]
- 61.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 62.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 63.Johnson JM, Latimer JJ. Analysis of DNA repair using transfection-based host cell reactivation. Methods Mol. Biol. 2005;291:321–335. doi: 10.1385/1-59259-840-4:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cenni B, Kim HK, Bubley GJ, Aebi S, Fink D, Teicher BA, Howell SB, Christen RD. Loss of DNA mismatch repair facilitates reactivation of a reporter plasmid damaged by cisplatin. Br. J. Cancer. 1999;80:699–704. doi: 10.1038/sj.bjc.6690412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, Wei Q, Shi Q, Guo Z, Qiao Y, Spitz MR. A modified host-cell reactivation assay to measure repair of alkylating DNA damage for assessing risk of lung adenocarcinoma. Carcinogenesis. 2007;28:1430–1436. doi: 10.1093/carcin/bgm029. [DOI] [PubMed] [Google Scholar]
- 66.Thacker J. The study of responses to ‘model’ DNA breaks induced by restriction endonucleases in cells and cell-free systems: achievements and difficulties. Int. J. Radiat. Biol. 1994;66:591–596. doi: 10.1080/09553009414551671. [DOI] [PubMed] [Google Scholar]
- 67.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin. Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 68.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA–repair mechanisms and medical significance. DNA Repair. 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 71.Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell. 2004;13:713–723. doi: 10.1016/s1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 72.Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, Ganem D, Middeldorp J, Wiertz EJ, Ressing ME. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl Acad. Sci. USA. 2007;104:3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Glaunsinger B, Chavez L, Ganem D. The exonuclease and host shutoff functions of the SOX protein of Kaposi's; sarcoma-associated herpesvirus are genetically separable. J. Virol. 2005;79:7396–7401. doi: 10.1128/JVI.79.12.7396-7401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuo J, Thomas W, van Leeuwen D, Middeldorp JM, Wiertz EJ, Ressing ME, Rowe M. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J. Virol. 2008;82:2385–2393. doi: 10.1128/JVI.01946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim PM, Allen C, Wagener BM, Shen Z, Nickoloff JA. Overexpression of human RAD51 and RAD52 reduces double-strand break-induced homologous recombination in mammalian cells. Nucleic Acids Res. 2001;29:4352–4360. doi: 10.1093/nar/29.21.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferry KV, Hamilton TC, Johnson SW. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells: role of ERCC1-XPF. Biochem. Pharmacol. 2000;60:1305–1313. doi: 10.1016/s0006-2952(00)00441-x. [DOI] [PubMed] [Google Scholar]
- 78.Muotri AR, Marchetto MC, Suzuki MF, Okazaki K, Lotfi CF, Brumatti G, Amarante-Mendes GP, Menck CF. Low amounts of the DNA repair XPA protein are sufficient to recover UV-resistance. Carcinogenesis. 2002;23:1039–1046. doi: 10.1093/carcin/23.6.1039. [DOI] [PubMed] [Google Scholar]
- 79.Gibson SL, Narayanan L, Hegan DC, Buermeyer AB, Liskay RM, Glazer PM. Overexpression of the DNA mismatch repair factor, PMS2, confers hypermutability and DNA damage tolerance. Cancer Lett. 2006;244:195–202. doi: 10.1016/j.canlet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 80.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chandriani S, Ganem D. Host transcript accumulation during lytic KSHV infection reveals several classes of host responses. PLoS ONE. 2007;2:1–10. doi: 10.1371/journal.pone.0000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee YJ, Glaunsinger BA. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 2009;7:e1000107. doi: 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shokolenko IN, Alexeyev MF, Robertson FM, LeDoux SP, Wilson GL. The expression of exonuclease III from E. coli in mitochondria of breast cancer cells diminishes mitochondrial DNA repair capacity and cell survival after oxidative stress. DNA Repair. 2003;2:471–482. doi: 10.1016/s1568-7864(03)00019-3. [DOI] [PubMed] [Google Scholar]
- 84.Weinberg RA. The Biology of Cancer. 1 edn. New York: Garland Science; 2007. [Google Scholar]
- 85.Jeang KT, Widen SG, Semmes O.Jt, Wilson SH. HTLV-I trans-activator protein, tax, is a trans-repressor of the human beta-polymerase gene. Science. 1990;247:1082–1084. doi: 10.1126/science.2309119. [DOI] [PubMed] [Google Scholar]
- 86.Majone F, Semmes OJ, Jeang KT. Induction of micronuclei by HTLV-I Tax: a cellular assay for function. Virology. 1993;193:456–459. doi: 10.1006/viro.1993.1145. [DOI] [PubMed] [Google Scholar]
- 87.Lee TH, Elledge SJ, Butel JS. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J. Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Livezey KW, Negorev D, Simon D. Increased chromosomal alterations and micronuclei formation in human hepatoma HepG2 cells transfected with the hepatitis B virus HBX gene. Mutat. Res. 2002;505:63–74. doi: 10.1016/s0027-5107(02)00140-9. [DOI] [PubMed] [Google Scholar]
- 89.Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023–2032. doi: 10.1093/carcin/bgh207. [DOI] [PubMed] [Google Scholar]
- 90.Machida K, Cheng KT, Sung VM, Lee KJ, Levine AM, Lai MM. Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes. J. Virol. 2004;78:8835–8843. doi: 10.1128/JVI.78.16.8835-8843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Macoska JA, Beheshti B, Rhim JS, Hukku B, Lehr J, Pienta KJ, Squire JA. Genetic characterization of immortalized human prostate epithelial cell cultures. Evidence for structural rearrangements of chromosome 8 and i(8q) chromosome formation in primary tumor-derived cells. Cancer Genet. Cytogenet. 2000;120:50–57. doi: 10.1016/s0165-4608(99)00248-4. [DOI] [PubMed] [Google Scholar]
- 92.Duensing S, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002;62:7075–7082. [PubMed] [Google Scholar]
- 93.Iftner T, Elbel M, Schopp B, Hiller T, Loizou JI, Caldecott KW, Stubenrauch F. Interference of papillomavirus E6 protein with single-strand break repair by interaction with XRCC1. EMBO J. 2002;21:4741–4748. doi: 10.1093/emboj/cdf443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Si H, Robertson ES. Kaposi's; sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 2006;80:697–709. doi: 10.1128/JVI.80.2.697-709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen YR, Liu MT, Chang YT, Wu CC, Hu CY, Chen JY. Epstein-Barr virus latent membrane protein 1 represses DNA repair through the PI3K/Akt/FOXO3a pathway in human epithelial cells. J. Virol. 2008;82:8124–8137. doi: 10.1128/JVI.00430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc. Natl Acad. Sci. USA. 2009;106:2313–2318. doi: 10.1073/pnas.0810619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pan SH, Tai CC, Lin CS, Hsu WB, Chou SF, Lai CC, Chen JY, Tien HF, Lee FY, Wang WB. Epstein-Barr virus nuclear antigen 2 disrupts mitotic checkpoint and causes chromosomal instability. Carcinogenesis. 2009;30:366–375. doi: 10.1093/carcin/bgn291. [DOI] [PubMed] [Google Scholar]
- 98.Gruhne B, Sompallae R, Masucci MG. Three Epstein-Barr virus latency proteins independently promote genomic instability by inducing DNA damage, inhibiting DNA repair and inactivating cell cycle checkpoints. Oncogene. 2009;28:3997–4008. doi: 10.1038/onc.2009.258. [DOI] [PubMed] [Google Scholar]
- 99.Altmann M, Hammerschmidt W. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 2005;3:2148–2157. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wen W, Iwakiri D, Yamamoto K, Maruo S, Kanda T, Takada K. Epstein-Barr virus BZLF1 gene, a switch from latency to lytic infection, is expressed as an immediate-early gene after primary infection of B lymphocytes. J. Virol. 2007;81:1037–42. doi: 10.1128/JVI.01416-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.