Abstract
Homing endonucleases have become valuable tools for genome engineering. Their sequence recognition repertoires can be expanded by modifying their specificities or by creating chimeric proteins through domain swapping between two subdomains of different homing endonucleases. Here, we show that these two approaches can be combined to create engineered meganucleases with new specificities. We demonstrate the modularity of the chimeric DmoCre meganuclease previously described, by successfully assembling mutants with locally altered specificities affecting both I-DmoI and I-CreI subdomains in order to create active meganucleases with altered specificities. Moreover these new engineered DmoCre variants appear highly specific and present a low toxicity level, similar to I-SceI, and can induce efficient homologous recombination events in mammalian cells. The DmoCre based meganucleases can therefore offer new possibilities for various genome engineering applications.
INTRODUCTION
The last 15 years have seen the emergence of new reagents enabling precise genome engineering (1–6). Among these new tools are the engineered homing endonucleases, also known as meganucleases, recognizing long DNA sequences (12–45 bp). In nature, these proteins are encoded by mobile introns or inteins, which propagate by cleaving the cognate allele from which the mobile element is absent, thereby stimulating the duplication of the mobile element into the recipient locus by a mechanism of DSB-induced homologous recombination. Mimicking this strategy to manipulate genes by targeted recombination has long been used in many laboratories and has proven to be robust and efficient (7,8). However, until recently, meganuclease-induced recombination was limited to the repertoire of natural meganucleases available which considerably impaired its potential applications. Indeed, the cognate DNA target of natural meganucleases is not necessarily present in a given genome and as a prerequisite prior to tinkering with a gene of interest, it has to be introduced into the desired locus.
Meganucleases are divided into five different families, from which the LAGLIDADG proteins, named after the short consensus LAGLIDADG sequence, constitute the largest and probably the best characterized family. The catalytic site is located within the center of the relatively compact molecule which confers a tight connection between cleavage and DNA-binding activities. Several protein structures including I-CreI, I-MsoI, I-AniI or I-SceI have been solved in complex with their DNA target (9–12) and could be engineered to change their specificity (13–16). Recently, we described a new protein engineering strategy using the modular architecture of the LAGLIDADG proteins. We showed that first, by altering the specificity locally (17) and then by adopting a combinatorial strategy (18), it was possible to redesign the I-CreI meganuclease specificity towards a chosen DNA sequence (18,19). Ultimately, I-CreI derived meganucleases could be used successfully to induce efficient targeted recombination in endogenous genes (2). Thus, these new tools offer the possibility to manipulate a genome in a very precise and safe manner.
Monomeric meganucleases could represent the best reagent for gene targeting applications as they alleviate the problems associated with the delivery and coordinated expression of two molecules needed with dimeric proteins such as I-CreI. The generation of active single-chain meganucleases has been previously reported, for example with I-CreI and I-MsoI, two homodimeric proteins that were successfully monomerized (20,21). In addition, domain swapping between the two subdomains of I-DmoI and between I-DmoI and I-CreI was also shown (20,22,23). Recently, Grizot et al. (2) showed that a single-chain I-CreI variant, targeting the human endogenous RAG1 locus, was potentially safer than the original heterodimeric molecule as it abolished the formation of the homodimer by-products, generated by the co-expression of the two monomers.
In this article, we applied a combinatorial approach to engineer the chimeric DmoCre protein and successfully modified its specificity. The protein activity was first increased to eventually match the activity of natural meganucleases like I-CreI or I-SceI in mammalian cells, while maintaining a high specificity and low toxicity. Taking advantage of the thousands of I-CreI mutants available in our data base, we then showed that I-CreI mutants could be introduced by domain shuffling into the DmoCre molecule. In parallel, variants with altered specificity within the I-DmoI moiety of the molecule were produced and assembled with I-CreI variants in order to create entirely new DmoCre variants. Finally, we show that engineered DmoCre meganucleases show very high specificities, similar to the parental DmoCre scaffold or the natural I-SceI meganuclease, as no toxic effect could be detected in a cell survival assay or in a γ-H2AX foci formation assay. Thus, the DmoCre meganuclease variants appear to represent a new molecular tool in the genome engineering toolbox.
MATERIALS AND METHODS
Construction of target clones
The different 22-bp DNA targets that were used in the yeast screening assay were cloned as follows. Oligonucleotides containing the target site (Proligo) were amplified by PCR to generate double-stranded target DNA. The PCR products were then inserted into reporter vectors with the Gateway protocol (Invitrogen): the yeast vector pFL39-ADH-LACURAZ and the mammalian vector pcDNA3.1-LAACZ, both previously described and containing an I-SceI target site as a control. Yeast reporter vectors were used to transform Saccharomyces cerevisiae strain FYBL2-7B (MATα, ura3Δ851, trp1Δ63, leu2Δ1, lys2Δ202).
Meganuclease production
The ORFs encoding the various meganucleases were amplified by PCR and inserted into the 2 micron-based replicative vector pCLS542, harboring the LEU2 gene. Saccharomyces cerevisiae strain FYC2-6A (MATα, trp1Δ63, leu2Δ1, his3Δ200) was then transformed with the vector using a high-efficiency lithium acetate transformation protocol. For expression in mammalian cells, the various meganuclease genes were also inserted into the mammalian vector pcDNA3.1. The genes were expressed under the control of a CMV promoter. Some of the proteins were also cloned with the addition of an HA (YPYDVPDYA) epitope into the C-terminus of the proteins which does not affect protein expression levels or activities.
Incorporation of I-CreI specific mutations into the DmoCre scaffold
In order to generate DmoCre derived coding sequences that contain mutations in the I-CreI moiety of the protein, a PCR reaction was carried out that amplifies the region between residues 13–148 for each of the I-CreI derived cutters. The different PCR fragments were then pooled. The yeast expression vector for the DmoCre protein was then digested with NgoMIV and MluI removing a fragment covering residues 111–238 of the DmoCre protein. Finally, 25 ng of the PCR pool and 75 ng of the digested vector DNA were used to transform the yeast S. cerevisiae strain FYC2-6A (MATα, trp1Δ63, leu2Δ1, his3Δ200) using a high-efficiency LiAc transformation protocol. An intact DmoCre coding sequence containing the mutations characteristic of the different I-CreI mutants was generated by in vivo homologous recombination in yeast.
Construction of DmoCre mutant libraries
DmoCre mutant libraries randomized at positions 29, 33 and 35 (D10C1lib library) or 75, 76 and 77 (D4C1lib) or 41, 75 and 77 (D4bisC1lib) were constructed using NNK degenerate codons resulting in a theoretical protein diversity of 8000 (203) as previously described (17). All libraries were generated by in vivo homologous recombination in yeast into the pCLS542 vector.
Mating of meganuclease expressing clones and screening in yeast
A colony gridder (QpixII, Genetix) was used for the mating of yeast strains. Mutants were gridded on nylon filters placed on YPGlycerol plates, using high gridding density (∼20 spots/cm2). A second gridding process was performed on the same filters for spotting of a second layer consisting of reporter-harboring yeast strains for each target. Membranes were placed on solid agar containing YPGlycerol rich medium, and incubated overnight at 30°C, to allow mating. The filters were then transferred onto synthetic medium, lacking leucine and tryptophan, with glucose (2%) as the carbon source (and with G418 for coexpression experiments) and incubated for 5 days at 30°C, to select for diploids carrying the expression and target vectors. Finally, filters were transferred onto YPGalactose rich medium for 2 days at 37°C to induce the expression of the meganuclease. Filters were then placed on solid agarose medium with 0.02% X-Gal in 0.5 M sodium phosphate buffer, pH 7.0, 0.1% SDS, 6% dimethyl formamide (DMF), 7 mM β-mercaptoethanol, 1% agarose and incubated at 37°C, to monitor β-galactosidase activity. Filters were scanned and each spot was quantified using the median values of the pixels constituting the spot. We attribute the arbitrary values 0 and 1 to respectively white and dark pixels. β-Galactosidase activity is directly associated with the efficiency of homologous recombination (17).
Activity improvement
Error-prone PCR was used to introduce random mutations in a pool of chosen mutants. Libraries were generated by error-prone PCR using Mn2+ at a concentration of 0.3 mM (24,25). To maintain a relatively low rate of mutagenesis, the concentrations of dCTP and dTTP were not increased. Under these conditions, the analysis of the resulting protein sequences showed that half of the molecules were not mutated, while the other half of the mutants carried 1.4 mutations on average. In addition, 60% of the amino acid was mutated at least once in the library.
Extrachromosomal assay in CHO-K1 cells
CHO-K1 cells were transfected with the meganuclease expression vectors and the reporter plasmid, in the presence of Polyfect transfection reagent in accordance with the manufacturer’s protocol (Qiagen). The culture medium was removed 72 h after transfection and 150 µl of lysis/detection buffer was added for β-galactosidase liquid assay [typically, for 1 l of buffer, we used 100 ml of lysis buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% Triton ×100, 0.1 mg/ml BSA, protease inhibitors), 10ml of Mg 100 × buffer (MgCl2 100 mM, 2-mercaptoethanol 35%), 110 ml of an 8 mg/ml solution of ONPG and 780 ml of 0.1 M sodium phosphate pH 7.5]. After incubation at 37°C, we measured optical density at 420 nm. The entire process was performed on 96-well plate format using an automated Velocity11 BioCel platform. Conditions have been adapted so that the cell number and transfection efficiency are constant in every wells through out the plates. To insure repeatability and reproducibility of the experiments, control plates are included in all plates. Correlation between intra- and inter-plates controls allows us to validate the assay.
Targeting of a chromosomal reporter gene in CHO-K1 cells
The introduction of different meganuclease sites in the same chromosomal context has already been described (19). Briefly, a transgene containing an I-SceI cleavage site has been stably expressed in CHO-K1 cells in single copy. In order to create a cell line containing the meganuclease target site, the cells were cotransfected with an I-SceI expression vector and a repair plasmid containing a LacZ expression cassette interrupted by the meganuclease site. In a second step, the CHO-K1 cell lines harboring the reporter system were seeded at a density of 2 × 105 cells per 10 cm dish in complete medium [Kaighn’s modified F-12 medium (F12-K), supplemented with 2 mM l-glutamine, penicillin (100 IU/ml), streptomycin (100 µg/ml), amphotericin B (Fongizone) (0.25 µg/ml) (Invitrogen-Life Science) and 10% FBS (Sigma-Aldrich Chimie)]. The next day, cells were cotransfected in the presence of Polyfect transfection reagent (Qiagen) with 2 µg of LacZ repair matrix vector and various amounts of meganuclease expression vector. After 72 h of incubation at 37°C, β-galactosidase activity was measured using the Beta-Glo® Assay System (Promega). The signal value is plotted against a standard curve established by measuring the luminescent signal emitted by a mixed LacZ+/LacZ− cell population. The frequency of LacZ positive cells is expressed as a percentage and is calculated using the standard curve and corrected for the transfection efficiency.
Cell survival assay
The CHO-K1 cell line was used to seed plates at a density of 2 × 105 cells per 10 cm dish. The next day, various amounts of meganuclease expression vectors and a constant amount of GFP-encoding plasmids were used to transfect the cells. GFP levels were monitored on days 1 and 6 after transfection, by flow cytometry (Guava EasyCyte, Guava Technologies). Cell survival is expressed as a percentage and was calculated as a ratio: (meganuclease-transfected cell expressing GFP on day 6/control transfected cell expressing GFP on day 6) corrected for the transfection efficiency determined on day 1.
γ-H2AX immunocytochemistry
For γ-H2AX immunocytochemistry, CHO-K1 cells were transfected with a mixture containing 1 µg of plasmid encoding a HA-tagged meganuclease and with DNA levels totaling up to 4.5 µg with empty vector, in the presence of Polyfect reagent (Qiagen). Cells were fixed 48 h after transfection, by incubation with 2% of paraformaldehyde for 20 min and permeabilized by incubation for 5 min at room temperature in 0.5% Triton. Cells were washed and incubated for 1 h in 0.3% Triton buffer supplemented with 10% normal goat serum (NGS) and 3% BSA, to block non-specific staining. Cells were then incubated for 1 h at RT with anti-γ-H2AX (Upstate: 1/10000) and anti-HA (Santa Cruz:1/700) antibodies diluted in Triton 0.3% in PBS supplemented with 3% BSA and 10% NGS and then for 1 h with Alexa Fluor 546 goat anti-mouse (Invitrogen-Molecular Probes: 1/1000) and Alexa Fluor 488 goat anti-rabbit secondary antibodies (Invitrogen-Molecular Probes 1/1000) diluted in 0.3% Triton in PBS supplemented with 3% BSA and 10% NGS. Coverslips were incubated with 1 µg/ml 4′,6-diamino-2′-phenylindole (DAPI, Sigma) mounted and the γ-H2AX foci were visualized in transfected cells (HA-positive) by fluorescence microscopy. The inactive I-CreI V2(G19S)/V3(G19S) heterodimer was used as a negative control at a dose of 1 µg for each expression plasmid.
RESULTS
Activity improvement and characterization of the hybrid DmoCre protein
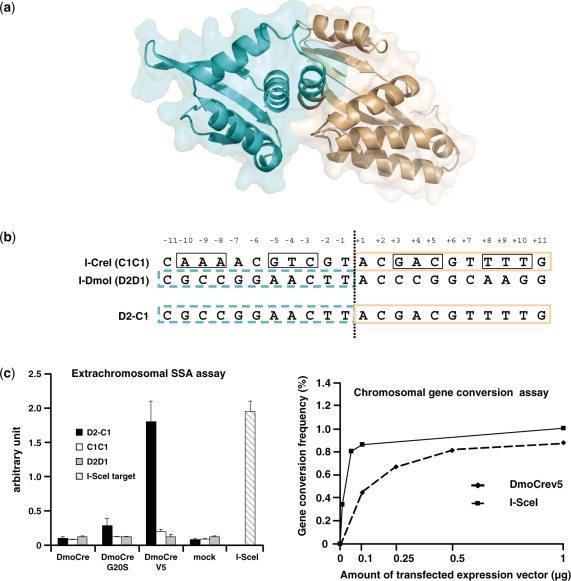
We previously described the engineering of a chimeric DmoCre meganuclease (20) resulting from the fusion of the N-terminal domain of I-DmoI with one I-CreI monomer (Figure 1a) able to cleave the combined D2-C1 target (Figure 1b). Due to the hyperthermophilic origin of I-DmoI, the DmoCre protein was functional at a high temperature on the D2-C1 target but displayed a modest in vivo activity in living cells (data not shown). In order to improve the cleavage activity of the DmoCre protein, two rounds of random mutagenesis using error-prone PCR were performed on the I-DmoI half of the DmoCre coding sequence. Following the first round of optimization, we identified a variant carrying the G20S mutation that showed maximum cleavage activity toward the D2-C1 target using our previously described yeast assay (20). However, as shown in Figure 1c, this mutant displayed a very low level of activity using an extrachromosomal single-strand annealing (SSA) assay in CHO-K1 cells (17). A second round of mutagenesis was thus performed on the DmoCre G20S variant. Two thousand two hundred mutants were screened for high level of activity in CHO-K1 cells. The protein with the highest activity (Figure 1c), called DmoCreV5, displayed three mutations located in the LAGLIDADG helix of the I-DmoI moiety: L15Q, I19D and G20S (the protein sequence is provided in the Supplementary Material). In order to estimate the full potential of the newly generated DmoCreV5 meganuclease, the protein activity was assessed using a chromosomal gene targeting assay that has already been described (19). Briefly, a CHO-K1 strain carrying a single copy of the lacZ gene interrupted by the D2-C1 target was produced. Subsequently, cells were cotransfected with increasing amounts of plasmid coding for the DmoCreV5 protein and a fixed quantity of plasmid carrying a truncated LacZ gene as a repair matrix. Cleavage of the D2-C1 target by the DmoCreV5 endonuclease results in the restoration of the LacZ gene by gene conversion and thus production of active β-galactosidase. The frequency of LacZ positive cells was measured 72 h post-transfection. Figure 1c shows that the new DmoCreV5 protein can induce gene correction in up to 0.8% of transfected cells and has an activity profile very similar to I-SceI.
Figure 1.
The DmoCre protein. (a) Representation of the chimeric protein DmoCre. The N-terminal domain of I-DmoI (residues 1–109) is depicted in blue and the I-CreI monomer (residues 13–163) in brown. (b) Nomenclature used for various of DNA targets. C1C1 is the palindromic target derived from the I-CreI target. The 10NNN ( ±8, ±9, ±10 nt) and 5NNN ( ±3, ±4, ±5 nt) regions of the target are boxed. D2D1 is the I-DmoI target. D2-C1 is the target cleaved by the hybrid DmoCre protein. (c) In vivo activity.of optimized DmoCre. DmoCre variants with improved activity were first characterized by an extrachromosomal assay in CHO-K1 cells. Cells were transfected with meganuclease or empty vector and the resulting cleavage was monitored against the three targets described in (b). Activity of I-SceI against its natural S1234 target is shown as a positive control. Activity of the meganuclease DmoCreV5 was also monitored in a chromosomal gene conversion assay in CHO-K1 cells. CHO-K1 cells containing a single chromosomal copy of the lacZ gene interrupted by the DNA target site were co-transfected with increasing amounts of the meganuclease expression vector and a fixed quantity of the lacZ repair plasmid. Activity of the DmoCreV5 and I-SceI meganucleases was monitored using two different cell lines that differ only by the meganuclease target site (D2-C1 for DmoCre and S1234 for I-SceI).
Insertion of I-CreI derived mutants into the DmoCre scaffold
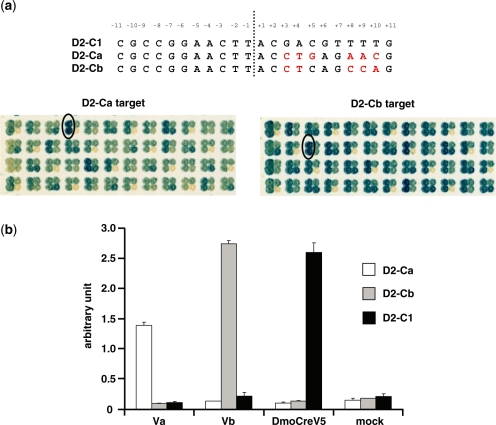
In addition to its monomeric architecture, the DmoCre scaffold offers the possibility to use I-CreI derived mutants that have already been obtained and characterized (17–19). Therefore, we then tested whether the I-DmoI and I-CreI moities could be modified independently. In order to answer this question, we used a subset of I-CreI mutants with new substrate specificities towards either nucleotides located at positions ±10, ±9, ±8 (10NNN) or at positions ±5, ±4, ±3 (5NNN) of the palindromic I-CreI target C1C1 (Figure 1b) or I-CreI variants generated by a combinatorial process and able to cleave 10NNN-5NNN hybrid targets (18). These I-CreI derived mutants were incorporated into the DmoCreV5 coding sequence by in vivo cloning in yeast (see ‘Material and methods’ section) and activity of the resulting DmoCre variants was monitored in yeast against the corresponding target. Table 1 summarizes the data. The cleavage activity is expressed as an arbitrary value between 0 and 1. In all cases, active DmoCre variants could be obtained, showing the relative independence of the two domains of the protein. Although most of the resulting DmoCre mutants showed a cleavage activity similar to the corresponding I-CreI mutants, the introduction of the I-CreI mutants targeting the Ca or the Cb DNA sequences within the DmoCreV5 protein resulted in a relatively low success rate for the combinatorial process (respectively 17 and 24%) and in a slight loss of cleavage activity of the active variants. We then optimized these proteins in order to increase their cleavage activities on D2-Ca or D2-Cb targets. For each target, a corresponding pool of three active DmoCre mutants (Table 2, #1, #2, #3 and #4, #5, #6) was used as a template in error-prone PCR, covering the entire length of the molecule. Two thousand two hundred and thirty two (2232) clones generated by in vivo cloning in yeast were then screened against the target of interest. For both D2-Ca and D2-Cb targets, highly active proteins could thus be obtained (Figure 2a).
Table 1.
Cleavage activity of DmoCre variants with altered specificity within the I-CreI moiety
| Targets derived from C1221 that are cleaved by I-CreI mutantsa | Number and strength of the I-CreI mutants | Targets cleaved by DmoCre mutantsa, number of positive DmoCre mutants and maximal cleavage activity |
|---|---|---|
| CAAAACTTGGTACCAAGTTTTG | 16 0.11 < AU < 0.30 | CGCCGGAACTTACCAAGTTTTG 16 AUmax = 0.56 |
| CAAAACGAGGTACCTCGTTTTG | 36 0.2 < AU < 1.0 | CGCCGGAACTTACCTCGTTTTG 32 AUmax = 0.92 |
| CCCAACGTCGTACGACGTTGGG | 14 0.15 < AU < 0.70 | CGCCGGAACTTACGACGTTGGG 12 AUmax = 0.92 |
| CTGGCTGCTGTACAGCAGCCAG | 26 0.61 < AU < 0.94 | CGCCGGAACTTACAGCAGCCAG 19 AUmax = 0.89 |
| CGTTCTCAGGTACCTGAGAACG (Ca) | 33 0.4 < AU < 1.0 | CGCCGGAACTTACCTGAGAACG (D2-Ca) 8 AUmax = 0.40; (0.91)b |
| CTGGCTGAGGTACCTCAGCCAG (Cb) | 35 0.2 < AU < 0.74 | CGCCGGAACTTACCTCAGCCAG ( D2-Cb) 6 AUmax = 0.48; (0.97)b |
AU: arbitrary unit.
aBases bold underlined are bases for which the I-CreI specificity has been modified.
bCleavage activity after protein optimization.
Table 2.
Sequences of engineered DmoCre variants
| D | R | B | F | N | A | K | N | Y | Q | S | Q | Y | R | R | D | I | V | I | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Target | Mutant | 75 | 77 | 93 | 101 | 102 | 108 | 125 | 127 | 130 | 135 | 137 | 141 | 163 | 165 | 167 | 172 | 174 | 202 | 229 |
| D2-Ca | #1 | K | A | A | Y | S | Y | K | ||||||||||||
| #2 | R | C | A | Y | S | Y | K | |||||||||||||
| #3 | R | C | A | Y | S | R | ||||||||||||||
| Va | R | C | A | H | Y | S | Y | K | ||||||||||||
| D2-Cb | #4 | Q | S | R | R | T | S | Y | ||||||||||||
| #5 | N | S | R | R | Y | S | Q | V | ||||||||||||
| #6 | N | S | R | R | A | N | N | |||||||||||||
| Vb | Q | S | R | R | A | N | Q | V | ||||||||||||
| D(4AGT)-C1 | S1 | R | V | |||||||||||||||||
| D(4AGT)-Ca | M 1 | R | V | R | C | A | H | Y | S | Y | K | |||||||||
| M 2 | R | N | S | R | C | A | H | Y | S | Y | K | |||||||||
| M 3 | R | A | R | C | A | H | Y | S | Y | K | ||||||||||
| OM 1 | R | N | L | R | C | A | H | Y | S | Y | K | V | ||||||||
| OM 2 | R | N | S | E | R | C | A | H | Y | S | Y | K | ||||||||
| OM 3 | R | N | S | R | C | A | H | Y | S | Y | K | A | ||||||||
| D(4AGT)-Cb | P 1 | R | A | Q | S | R | R | A | N | Q | V | |||||||||
| P 2 | Y | V | Q | S | R | R | A | N | Q | V | ||||||||||
| P 3 | T | C | Q | S | R | R | A | N | Q | V | ||||||||||
| OP 1 | T | C | S | Q | S | R | R | A | N | Q | V | V | ||||||||
| OP 2 | R | A | Q | S | R | R | A | N | Q | V | V |
Figure 2.
In vivo activity of DmoCre variants with altered specificity in the I-CreI moiety. (a) Yeast secondary screening of optimized DmoCre mutants toward respectively the D2-Ca and D2-Cb targets. The optimized mutants displayed on the two yeast filters had already been selected based on a cleavage activity equivalent or greater than the initial activity level during a primary screening. For each target, one optimized mutant is located in the two left dots on each four-dot yeast cluster. The bottom right dot represents internal experimental controls (negative or positive) while the upper right dot is one of the DmoCre mutants that have been used as a template for the optimization process (respectively mutant #2 for the D2-Ca target and mutant #6 for the D2-Cb target, Table 2). The Va and Vb mutants have been circled. For both targets, the nucleotides in the C1 part of the D2-C1 target for which the specificity has been modified are in red. (b) Extrachromosomal assay in CHO-K1 cells. Cells were transfected with different meganucleases or an empty vector and the cleavage against the D2-Ca, D2-Cb and D2-C1 targets was monitored.
Two improved meganucleases, Va and Vb (Table 2), specific to D2-Ca and D2-Cb respectively, were selected for further activity analyses in mammalian cells. The ORF’s were further subcloned into a mammalian expression vector and their activity was determined in our extrachromosomal SSA assay in CHO-K1 cells. Figure 2b shows the activity of the original DmoCreV5 meganuclease and the two selected DmoCre variants tested against all three targets (D2-C1, D2-Ca and D2-Cb). Each meganuclease showed specific cleavage of its cognate target. Interestingly, the analysis of the mutant sequences revealed that the mutations introduced during error-prone PCR affected only the I-CreI moiety of the molecule (Table 2). While one additional mutated position (Y163H) was found in the Va variant cleaving the D2-Ca target, the improved mutant Vb, specific to the D2-Cb target, appears to result from a shuffling of the mutations present on the three initial DmoCre variants during the error-prone PCR step.
Functionally engineered meganucleases with new specifities toward the D2 portion of the D2-C1 target
The I-CreI domain of the DmoCre scaffold can be easily shuffled with I-CreI derived mutants already available. However, the engineering of the protein–DNA interface of the Dmo part of the molecule would considerably expand its potential application. Therefore we tried to locally alter the DNA specificity of the I-DmoI moiety of the molecule. The structure of I-DmoI and E-DreI, a meganuclease very similar to our DmoCre protein, have been resolved alone or in complex with their DNA target (23,26,27). The E-DreI protein is slightly different from the DmoCre protein, but these differences do not involve residues in direct contact with the DNA or in close proximity. Moreover, the I-DmoI subdomain, common to the E-DreI and I-DmoI proteins, has been described as having the same protein–DNA interaction pattern (27). Thus the analysis of the I-DmoI structure brought us to divide the D2 part of the D2-C1 target into three triplets: 10NNN (bases at positions −10, −9 and −8), 7NNN (bases at position −7, −6 and −5) and 4NNN (bases at positions −4, −3 and −2) (Figure 3b). Analysis of the E-DreI structure leads to similar hypothesis. For each degenerated triplet, the corresponding 64 DNA targets were produced [D(10NNN)-C1, D(7NNN)-C1 and D(4NNN)-C1] and several DmoCre mutant libraries were generated in yeast.
Figure 3.
Profiling of the DmoCre protein on DNA targets deriving from D2-C1. (a) The D2 part of the D2-C1 target has been subdivided into three different domains: the − 10, − 9, − 8 triplet (D(10NNN)-C1), the − 7, − 6, − 5 triplet (D(7NNN)-C1) and the − 4, − 3, − 2 triplet (D(4NNN)-C1]. For each DNA triplet, the DmoCreV5 protein has been profiled in yeast against the 64 corresponding degenerated targets. The gray level is proportional to the cleavage intensity obtained with DmoCreV5 in yeast. (b) Hitmaps of different DmoCreV5 mutant libraries that were screened in yeast. The number of identified active proteins is shown below each target and the gray level is proportional to the average signal intensity obtained with these cutters in yeast. Amino acids that were randomized are indicated on top of the hitmaps. See Supplementary Data for statistical analysis.
In order to create a DmoCre library targeting the group of D(10NNN)-C1 targets, the residues Tyr29, Arg33 and Glu35 of the protein were randomized. 2232 clones, representing 28% of the library diversity, were screened against the 64 D(10NNN)-C1 targets. Three hundred and eighty seven active mutants with unique sequences were thereby identified. Figure 3a shows that the DmoCreV5 protein is able to cleave 16 out of the 64 targets, five of which were very faintly cleaved. The protein exhibits a low specificity toward the base at position −10 as it is able to cleave the four D(NCC)-C1 targets. The whole collection of newly produced DmoCre variants allowed cleavage for 50 out of 64 D(10NNN)-C1 targets (Figure 3b). Moreover, with an average of six targets cleaved per variant, these engineered meganucleases appeared to retain a specificity equivalent, if not higher than, the DmoCreV5 protein.
Using the same strategy, the residues Asp75, Thr76 and Arg77 of the protein were randomized in order to generate a DmoCreV5 library targeting the DNA triplet at positions −4, −3 and −2 of D2-C1 (D(4NNN)-C1). The screening of the library (2232 clones) against the 64 D(4NNN)-C1 DNA targets yielded 570 active mutants with unique sequences. Six D(4NNN)-C1 targets were cleaved by the initial DmoCreV5 protein (Figure 3a) while 23 out of the 64 D(4NNN)-C1 DNA triplets were recognized by the new DmoCre variants (Figure 3b). Furthermore these mutants appear highly specific since the average number of tolerated targets is three targets per protein. We noticed that the D(4ANN)-C1 sequences were preferentially targeted by the library (Figure 3b). As an example of this engineering step, a mutant called S1 specific for the D(4AGT)-C1 target and carrying the D75R and R77V mutations was chosen and cloned into a mammalian expression vector for further characterizations. Analysis of the E-DreI and I-DmoI structures in complex with their DNA targets show that the residue Thr41 makes a van der Waals contact with the methyl group of the complementary strand containing a thymidine at position − 4. Therefore, we generated a new library randomized at residues Thr41, Asp75 and Arg77, while keeping the Thr76 fixed to limit the diversity of the library. Screening against the D(4NNN)-C1 targets allowed the isolation of 221 unique mutants which cleaved an average of two targets. In addition to the 35 D(4NNN)-C1 sequences recognized by at least one mutant, the randomization of the residue Thr41 allowed the cleavage of new D(4GNN)-C1 and D(4TNN)-C1 targets. Still, almost no cleavage for the D(4CNN)-C1 targets could be detected.
The analysis of the E-DreI and I-DmoI structures shows that Arg37 and Arg81 interact extensively with the nucleotides at positions −7, −6 and −5 of the D2-C1 target and appear resistant to engineering since mutations in any of the arginines led either to an inactive protein or to a protein with much reduced activity (data not shown). However, the original DmoCreV5 meganuclease tolerates a purine at positions −7 and −6 and does not display strong specificity at position −5. Overall, DmoCre V5 is able to cleave 9 out of the 64 D(7NNN)-C1 DNA targets in our yeast assay (Figure 3a).
Combination of two mutations sets in the I-DmoI subdomain
Hundreds of DmoCre mutants recognizing the D(10NNN)-C1 or D(4NNN)-C1 targets have been obtained. We have previously described for the I-CreI protein, how to combine two mutation sets by a combinatorial approach to generate engineered I-CreI mutants (18). We then decided to apply this approach to the I-DmoI subdomain of DmoCre. The two sets of mutations Tyr29, Arg33, Glu35, and Asp75, Thr76, Arg77, constitute two distinct groups of mutations, which could therefore be combined into the same DmoCre coding sequence. Four D(10NNN)-C1 targets were selected (D(10CAG)-C1, D(10CCA)-C1, D(10CCG)-C1, D(10GCG)-C1), together with 24 corresponding DmoCre variants for each target. The same procedure was performed for three D(4NNN)-C1 targets where three sets of 24 DmoCre mutants recognizing respectively, D(4AGA)-C1, D(4AGT)-C1 and D(4ATA)-C1 DNA sequences, were chosen. Finally, the two groups of mutations were combined by in vivo cloning in yeast. Eight mutant libraries were created in such a manner and screened against their corresponding combined targets. None of the libraries enabled us to detect active proteins (data not show). This result prompted us to adopt an alternative approach in which the two mutation sets would be introduced sequentially. Thus, a pool of eight DmoCre variants cleaving the D(4AGT)-C1 target was selected as a template to generate a mutant library randomized at residues Tyr29 and Arg33. The residue Glu35 was ignored in order to maintain the diversity of the library. The library was then screened against four combined targets [D(10GCG4AGT)-C1, D(10CCG4AGT)-C1 and D(10CAG4AGT)-C1, D(10CCA4AGT)-C1]. No active protein cleaving the first three targets could be detected, but 13 unique proteins were able to cleave the D(10CCA4AGT)-C1 target, albeit with a low efficiency. Four mutants were further selected as templates in an error-prone PCR experiment in order to improve the cleavage activity. Although their endonuclease activities could be improved, these mutants did not reach the level of I-SceI activity in yeast, and no activity could be detected using our extrachromosomal assay in CHO-K1 cells (data not shown).
Combination of mutation sets present in both I-DmoI and I-CreI subdomains
To explore the engineering possibilities of the DmoCre molecule, we assembled mutations affecting the specificity of both the I-DmoI and I-CreI moieties. The improved meganucleases Va and Vb, specific to D2-Ca and D2-Cb were selected and used as templates to generate I-DmoI libraries randomized at residues Tyr29, Arg33, Glu35, or, Asp75, Thr76, Arg77. The four resulting libraries were then screened in yeast against 18 randomly chosen hybrid targets. Table 3 summarizes the results, confirming that the modularity between the I-DmoI and I-CreI subdomains as active DmoCre variants could be obtained against 13 out of the 18 tested targets.
Table 3.
Cleavage activity in yeast of DmoCre variants with altered specificity in both I-DmoI and I-CreI moieties
| Library template (target specificity) | Randomized positions | Target tested | Maximal cleavage activity observed in yeast (AU value) |
|---|---|---|---|
| Vb (D2-Cb) | 75, 76, 77 | D(4AGT)-Cb | 0.91 |
| D(4ATA)-Cb | 0.57 | ||
| D(4AGA)-Cb | 0.66 | ||
| D(4AGG)-Cb | 0.50 | ||
| 29, 33, 35 | D(10CTC)-Cb | 0.19 | |
| D(10TAG)-Cb | 0 | ||
| D(10GCG)-Cb | 0.64 | ||
| D(10ACG)-Cb | 0.64 | ||
| D(10CCA)-Cb | 0.41 | ||
| Va (D2-Ca) | 75, 76, 77 | D(4AGT)-Ca | 0.87 |
| D(4ATA)-Ca | 0 | ||
| D(4AGA)-Ca | 0.34 | ||
| D(4AGG)-Ca | 0.37 | ||
| 29, 33, 35 | D(10CTC)-Ca | 0 | |
| D(10TAG)-Ca | 0 | ||
| D(10GCG)-Ca | 0.43 | ||
| D(10ACG)-Ca | 0 | ||
| D(10CCA)-Ca | 0.2 |
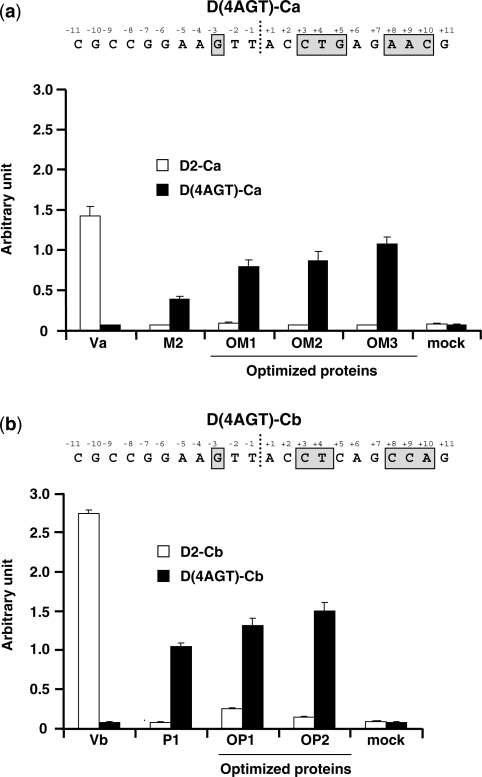
DmoCre variants, able to cleave the D(4AGT)-Cb and D(4AGT)-Ca targets (Figure 4a and b), were further selected for in vivo activity characterization. For each target, a pool of three mutants, M1 to M3 and P1 to P3 (Table 2), showing good activity on their respective targets in yeast, were selected and their activities further improved. Upon random mutagenesis, new optimized variants, OM1 to OM3, and OP1, OP2, displaying high cleavage activities against D4(AGT)-Ca or D(4AGT)-Cb, respectively, were chosen and cloned into a mammalian expression vector in order to perform an extrachromosomal recombination assay in CHO-K1 cells. Figure 4a and 4b show that all the DmoCre variants tested were active in CHO-K1 cells. However, the engineering step that shifts the protein specificity from the D2-Ca or D2-Cb target to D(4AGT)-Ca or D(4AGT)-Cb, respectively, also reduced, to a certain extent, the mutant cleavage activity which could be partially recovered after our optimization step. Importantly, the newly obtained DmoCre mutants are specific for their targets as they display no cleavage activity towards the D2-Ca or D2-Cb targets (Figure 4a and b). Analysis of mutant sequences confirmed that activity improvements resulted mainly in the addition of mutations in residues located in the I-CreI moiety of the DmoCre molecule or in the region linking the two subdomains (residues 93–108) (Table 2). In agreement with previous data (28), the C-terminal part of the I-CreI protein seems to play a major role in DmoCre activity since mutations affecting this domain (amino acids 202 and 229) can increase cleavage efficacy in living cells. The residues Val202 and Ile229 belong to the protein hydrophobic core. In both cases, the mutations consist in the replacement of one hydrophobic residue with another hydrophobic residue having a smaller side chain. Such a mutation could slightly increase the protein flexibility, which could better accommodate its DNA target.
Figure 4.
Activity of DmoCre mutants with altered specificity in both I-DmoI and I-CreI subdomains. (a) Extrachromosomal assay in CHO-K1 cells. Cells were transfected with different meganucleases or an empty vector and the cleavage against the two D2-Ca and D(4AGT)-Ca (targets was measured). The nucleotides in the D(4AGT)-Ca target for which the specificity has been modified have been gray boxed. (b) Extrachromosomal assay in CHO-K1 cells. Cells were transfected with different meganucleases or an empty vector (mock) and the cleavage against the two D2-Cb and D(4AGT)-Cb targets was measured. The nucleotides in the D(4AGT)-Cb target for which the specificity has been modified have been gray boxed.
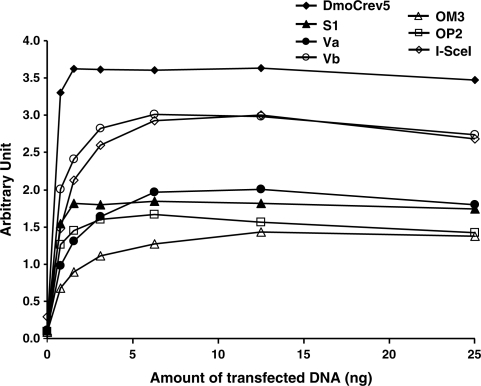
Finally, a set of DmoCre variants representative of each engineering step (Va, Vb, S1, OM3 and OP2) (Figure 5) were tested in a dose-response study (from 0.5 to 25 ng of meganuclease coding vectors) using an extrachromosomal assay in CHO-K1 (19). We compared the cleavage activities of several engineered DmoCre variants specific to D2-Cb, D2Ca, D(4AGT)-C1, D(4AGT)-Ca and D(4AGT)-Cb respectively, as well as the parental hybrid DmoCreV5 molecule and the natural meganuclease I-SceI against their cognate targets. Figure 5 shows that all of the mutants are active in mammalian cells, however, the DmoCre variants show less activity, compared to the initial DmoCreV5 protein.
Figure 5.
In vivo cleavage activity of different engineered DmoCre mutants monitored in an extrachromosomal SSA assay. CHO-K1 cells were transfected with increasing amounts of meganuclease expression vectors (from 0 to 25 ng). Activity of the DmoCreV5 and I-SceI proteins and the S1, Va, Vb, OM3 and OP2 DmoCre mutants was monitored against their respective cognate targets, D2-C1, S1234, D(AGT)-C1, D2-Ca, D2-Cb, D(4AGT)-Ca and D(4AGT)-Cb targets.
The engineered DmoCre mutants display a minimal toxicity
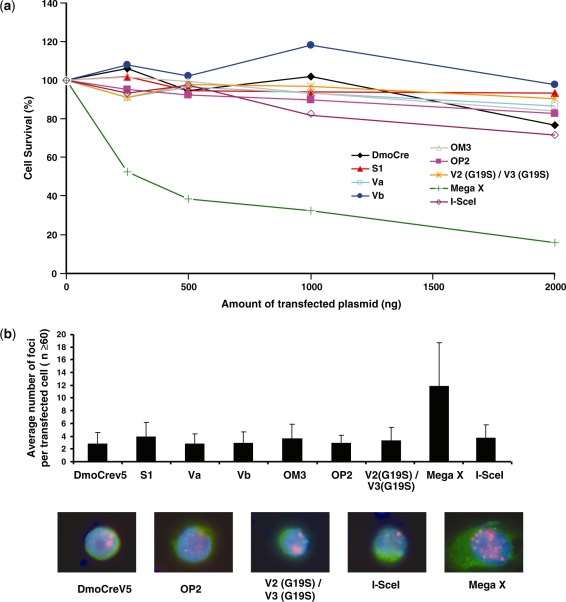
The specificity/toxicity ratio is a crucial parameter in DSB-induced recombination technology. Thus, the five representative engineered meganucleases described above (Va, Vb, S1, OM3 and OP2), along with the I-SceI and DmoCreV5 meganucleases, were evaluated for their potential toxicity using a cell survival assay in CHO-K1 cells (2). Two meganucleases derived from I-CreI were added as control. The V2(G19S)/V3 (G19S) heterodimer is an inactive meganuclease (2) while MegaX (4) is a meganuclease with relaxed specificity. MegaX is a heterodimeric meganuclease constituted by two I-CreI mutants, which display a large cleavage profile in a yeast screening assay performed on several thousands of DNA targets (data not shown). Figure 6a shows that the five engineered DmoCre mutants, as well as the initial DmoCreV5 protein, do not display any visible toxicity and present the same profile as I-SceI. As expected, the inactive meganuclease shows a flat pattern, while expression of the non-specific MegaX induces significant toxicity.
Figure 6.
Evaluation of the potential toxicity of the DmoCre meganucleases. (a) Toxicity of the DmoCre engineered meganucleases, as monitored by a cell survival assay. Increasing amounts of meganuclease expression vector and a constant amount of plasmid encoding GFP were used to cotransfect CHO-K1 cells. Cell survival is expressed as a percentage of cells expressing GFP 6 days after transfection as described in the ‘Materials and methods’ section. (b) The potential genotoxicity of the engineered DmoCre meganucleases was monitored by visualizing the formation of γ-H2AX foci at DNA double-strand breaks. CHO-K1 cells were transfected with 1 µg of a plasmid coding for different meganucleases carrying a C-terminal HA epitope. For each condition, the average number of γ-H2AX foci per transfected cell (HA positive) was determined. Pictures below are representative images of cells transfected with the DmoCreV5, OP2, V2 (G19S)/V3 (G19S), I-SceI and Mega X meganucleases: Green, HA labeling; red, γ-H2AX foci and blue, DAPI staining. The inactive V2 (G19S)/V3 (G19S) I-CreI heterodimer is shown as a control for non-toxicity, while Mega X is a meganuclease derived from I-CreI with relaxed specificity.
Toxicity might be related to the specificity profile of the endonuclease, which may generate off-site cleavage events. The characterization of the engineered DmoCre mutants was completed by monitoring the γ-H2AX foci content in CHO-K1 cells, expressing the endonucleases, since the γ-H2AX focus formation is one the first responses of the cell to DNA DSBs. CHO-K1 cells were transfected with 1 µg of the expression vector for the different meganucleases carrying a C-terminal HA epitope. At this high dose, no toxicity was revealed by the cell survival assay. Expression of DmoCreV5, as well as the five representative engineered DmoCre mutants, induces on average, approximately two to three γ-H2AX foci per transfected cells, similar to the background level (expression of the inactive V2 (G19S)/V3 (G19S) meganuclease) and to the number of foci induced by I-SceI (Figure 6b). In contrast, the expression of the non-specific MegaX that displayed toxicity in the cell survival assay induces an average number of 12 γ-H2AX foci per transfected cell. Altogether, this data indicates that the modification of the protein specificity has not been achieved at the expense of toxicity.
DISCUSSION
Double-strand break induced recombination technology has proven to be an efficient genome engineering strategy. Moreover, recent advances in DNA-binding protein engineering enabled scientists to alter the specificity of such proteins in order to precisely target a chosen locus in the genome (29,30). However, its application in molecular medicine requires highly specific reagents, capable to cleave a unique sequence in the genome. In this regard, homing endonucleases and more specifically, monomeric meganucleases present certain advantages. The tight coupling of the binding site to cleavage activity ensures a high degree of specificity and minimal off-site target cleavage events. Moreover, in contrast to zinc-finger nucleases or heterodimeric meganucleases that need the coordinate expression of two proteins, monomeric molecules simplify problems related to the delivery and expression of the protein.
The fusion of protein domains is an important strategy in both molecular evolution and protein engineering. In this report we have explored the modularity properties of a previously described chimeric meganuclease, DmoCre (20). We show that the specificity of this monomeric meganuclease could be diversified, and highly active engineered DmoCre variants could be obtained while keeping the non-toxic character of the original scaffold.
Production of hybrid meganucleases created by the fusion of domains from two distinct LAGLIDADG homing endonucleases is a powerful means to enrich the collection of DNA sequences that can be targeted by such endonucleases. In this sense, the DmoCre protein appears to be a promising scaffold since numerous I-CreI monomers with altered specificities have already been obtained (17,18) and could, at least theoretically, be inserted in this scaffold. However, the DmoCre protein initially exhibited only residual activity at 37°C because of the thermophilic origin of I-DmoI and therefore was not an appropriate tool for most of the genome engineering applications. Mesophilic I-DmoI variants have already been described (31). However, the mutations isolated in the I-DmoI context did not have any impact on the DmoCre activity level (data not shown). Therefore, an engineering step was first performed in order to obtain a DmoCre variant active at 37°C. Two successive rounds of random mutagenesis allowed us to isolate a mesophilic DmoCre enzyme, presenting high cleavage activity in yeast and CHO cells. The newly generated DmoCreV5 protein has three mutations in comparison with the initial DmoCre protein: L15Q, I19D and G20S. All three mutated residues are located in the LAGLIDADG helix of the I-DmoI moiety. Structure superimposition reveals that the G20S mutation is the equivalent of the G19S mutation that has been isolated and described on I-CreI heterodimers (2). This mutation decreases or even abolishes the activity of homodimers and increases activity of the heterodimer when present in only one monomer. In accordance with these data, the G20S mutation present in the DmoCre protein strongly enhances its activity. Interestingly, the introduction of the glycine to serine substitution in the LAGLIDADG helix of the I-CreI moiety had no effect on the activity of DmoCre (our unpublished data). The Ile19 residue of I-DmoI superimposes with Asp18 of I-CreI. In the I-CreI homodimer structure, Asp18 of one monomer makes a hydrogen bond with Tyr12 of the second monomer. Thus, it is possible that the I19D mutation creates a new interaction between the I-DmoI and I-CreI moieties, conferring a greater stability to the DmoCreV5 hybrid protein. The impact of the L15Q mutation is more uncertain. Inspection of the E-DreI structure indicates that the residue Leu15 resides in a hydrophobic pocket of the protein. Replacement of a hydrophobic leucine residue by a polar glutamine residue in such an environment should have a potentially adverse effect. However, this mutation is significant, since the engineering of a DmoCreV5 mutant carrying only the I19D and G20S mutations, results in a meganuclease slightly less active in our extrachromosomal assay in CHO-K1 cells. Biochemical and biophysical characterization of this mutant could be very valuable; however, the protein proved to be insoluble in bacteria and could therefore not be produced in sufficient amounts to allow in vitro analyses.
To widen the number of DNA sequences that could be targeted by DmoCre derivatives, the DmoCreV5 protein was further engineered to modify its specificity. First, the inter-domain modularity was investigated. We showed that the assembly of the I-Dmo moiety with I-CreI variants, previously isolated (17,18) was very effective and results in a highly active meganuclease. Next, we tackled the engineering of the I-DmoI moiety. A close inspection of the protein–DNA interactions of the I-DmoI and E-DreI meganucleases (23,27) allowed us to identify three potential subdomains necessary for protein–DNA interactions. The sequence DNA corresponding to D2 of the D2-C1 DNA target of DmoCre was separated into three DNA triplets (D(10NNN)-C1, D(7NNN)-C1 or D(4NNN)-C1). We obtained locally engineered DmoCre variants that showed altered specificities toward the D(10NNN)-C1 or D(4NNN)-C1 DNA triplets, whereas no altered specificities toward the D7NNN-C1 DNA triplet could be isolated by mutating the two arginine residues at positions 37 and 81, which interact with this central region. The I-DmoI structure shows that these two amino acids are in an extended conformation to establish six hydrogen bonds with bases at positions − 7 to − 5. Any mutation will thus result in amino acids with a shorter side chain that would be unable to interact with the DNA. Moreover, the strong interaction of Arg37 and Arg81 with the DNA could also be responsible for the failure in combining the two sets of mutations associated with the D(10NNN)-C1 and D(4NNN)-C1 targets respectively, and seems to be a stumbling block of the I-DmoI intradomain modularity. Therefore one can envision that a DmoCre variant with a modified specificity towards the D(7NNN)-C1 target might improve this engineering approach.
In conclusion, we have generated a DmoCre variant able to induce efficient gene conversion events in mammalian cells. Moreover, target site diversification could be achieved since we successfully produced active, engineered DmoCre mutants, with combined altered specificities for both the I-DmoI and I-CreI moieties., In addition to the obvious advantage of alleviating the need of coordinate expression of two proteins to form active heterodimers, our newly generated and engineered versions of DmoCre, as the monomeric I-SceI or engineered single-chain I-CreI meganucleases (2), appear to present minimal toxicity in mammalian cells and therefore qualify as new tools for targeted gene modification in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The EU Sixth Framework Programme for Research (contract no: LSHG-CT-2006-037226, MEGATOOLS).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank F. Daboussi for her technical help, and A. Holmes for valuable discussion and critical reading of the manuscript.
REFERENCES
- 1.Cannata F, Brunet E, Perrouault L, Roig V, Ait-Si-Ali S, Asseline U, Concordet JP, Giovannangeli C. Proc. Natl Acad. Sci. USA. 2008;105:9576–9581. doi: 10.1073/pnas.0710433105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. Nucleic Acids Res. 2009;37:5405–19. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redondo P, Prieto J, Munoz IG, Alibes A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 5.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 7.Rouet P, Smih F, Jasin M. Mol. Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choulika A, Perrin A, Dujon B, Nicolas JF. Mol. Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevalier BS, Monnat R.J., III, Stoddard BL. Nat. Struct. Biol. 2001;8:312–316. doi: 10.1038/86181. [DOI] [PubMed] [Google Scholar]
- 10.Chevalier B, Turmel M, Lemieux C, Monnat R.J., III, Stoddard BL. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 11.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moure CM, Gimble FS, Quiocho FA. J. Mol. Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 13.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat R.J., Jr, Stoddard BL, Baker D. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McConnell Smith A, Takeuchi R, Pellenz S, Davis L, Maizels N, Monnat R.J., Jr, Stoddard BL. Proc. Natl Acad. Sci. USA. 2009;106:5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu Y, Tenney K, Li H, Gimble FS. J. Mol. Biol. 2008;382:188–202. doi: 10.1016/j.jmb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R., Jr, Stoddard BL, Seligman LM. J. Mol. Biol. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 17.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 18.Smith J, Grizot S, Arnould S, Duclert A, Epinat J.-C, Prieto J, Redondo P, Blanco FJ, Bravo J, Montoya G, et al. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 20.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Pellenz S, Ulge U, Stoddard BL, Monnat R.J., Jr Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva GH, Belfort M, Wende W, Pingoud A. J. Mol. Biol. 2006;361:744–754. doi: 10.1016/j.jmb.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 23.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 24.Cadwell RC, Joyce GF. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 25.Cadwell RC, Joyce GF. PCR Methods Appl. 1994;3:S136–140. doi: 10.1101/gr.3.6.s136. [DOI] [PubMed] [Google Scholar]
- 26.Silva GH, Dalgaard JZ, Belfort M, van Roey P. J. Mol. Biol. 1999;286:1123–1136. doi: 10.1006/jmbi.1998.2519. [DOI] [PubMed] [Google Scholar]
- 27.Marcaida MJ, Prieto J, Redondo P, Nadra AD, Alibes A, Serrano L, Grizot S, Duchateau P, Paques F, Blanco FJ, et al. Proc. Natl Acad. Sci. USA. 2008;105:16888–16893. doi: 10.1073/pnas.0804795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prieto J, Redondo P, Padro D, Arnould S, Epinat JC, Paques F, Blanco FJ, Montoya G. Nucleic Acids Res. 2007;35:3262–3271. doi: 10.1093/nar/gkm183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paques F, Duchateau P. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 30.Galetto R, Duchateau P, Paques F. Expert Opin. Biol. Ther. 2009 doi: 10.1517/14712590903213669. [DOI] [PubMed] [Google Scholar]
- 31.Prieto J, Epinat JC, Redondo P, Ramos E, Padro D, Cedrone F, Montoya G, Paques F, Blanco FJ. J. Biol. Chem. 2008;283:4364–4374. doi: 10.1074/jbc.M706323200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.