Abstract
In Saccharomyces cerevisiae, TFIID and SAGA principally mediate transcription of constitutive housekeeping genes and stress-inducible genes, respectively, by delivering TBP to the core promoter. Both are multi-protein complexes composed of 15 and 20 subunits, respectively, five of which are common and which may constitute a core sub-module in each complex. Although genome-wide gene expression studies have been conducted extensively in several TFIID and/or SAGA mutants, there are only a limited number of studies investigating genome-wide localization of the components of these two complexes. Specifically, there are no previous reports on localization of a complete set of Tafs and the effects of taf mutations on localization. Here, we examine the localization profiles of a complete set of Tafs, Gcn5, Bur6/Ncb2, Sua7, Tfa2, Tfg1, Tfb3 and Rpb1, on chromosomes III, IV and V by chromatin immunoprecipitation (ChIP)-chip analysis in wild-type and taf1-T657K mutant strains. In addition, we conducted conventional and sequential ChIP analysis of several ribosomal protein genes (RPGs) and non-RPGs. Intriguingly, the results revealed a novel relationship between TFIIB and NC2, simultaneous co-localization of SAGA and TFIID on RPG promoters, specific effects of taf1 mutation on Taf2 occupancy, and an indirect evidence for the existence of different TFIID conformations.
INTRODUCTION
In eukaryotes, the general transcription factor (GTF) TFIID plays a central role in transcription of protein-coding (class II) genes by RNA polymerase II (pol II) together with other GTFs (i.e. TFIIA, TFIIB, TFIIE, TFIIF and TFIIH), as well as with a series of cofactors including Mediator, NC2, Mot1 and several chromatin modifying complexes (1–4). TFIID is comprised of the TATA box-binding protein (TBP) and 14 TBP-associated factors (TAFs), 5 of which are shared by a distinct histone acetyltransferase complex, SAGA (Spt-Ada-Gcn5 acetyltransferase) (5). These two closely related multi-protein complexes activate transcription by delivering TBP to the core promoters of their own target genes (6–11). In general, TFIID and SAGA mediate transcription of constitutive housekeeping and stress-inducible genes that are mostly driven by TATA-less and TATA-containing promoters, respectively (12–14). Despite such apparent divergence of their target genes, they are functionally redundant at many promoters (15), although not at RNR3, where TFIID and SAGA play different and non-redundant roles in transcription (16).
Gcn5, a catalytic subunit of SAGA, is generally recruited to active genes, including RPL2B, which is one of 138 ribosomal protein genes (RPGs) (17). This is somewhat paradoxical because the expression of RPGs is strongly dependent on TFIID but only modestly dependent on SAGA (9,12,13). However, genome-wide ChIP (chromatin immunoprecipitation)-chip analysis combined with computer modeling has revealed that RPGs exhibit the highest occupancy levels of Spt3, the TBP-delivering subunit of SAGA (18–20). Furthermore, it has also been shown that heat stress can induce transcription of many genes through recruitment of both TFIID and SAGA (18). These observations indicate that TFIID and SAGA may bind to the same set of promoters, even if their requirements for transcription differ depending on the structural properties of the target promoters, such as whether the TATA element is present or not. Nevertheless, it had remained unclear whether TFIID and SAGA bind to these promoters concurrently or alternately. We used sequential ChIP analysis to test whether these two complexes co-localize on the same promoter concurrently.
Genome-wide expression studies have been performed in many taf mutants (10,15,21–24) and have revealed that >80% of class II genes require at least one of 13 essential Tafs (Taf1-Taf13), and that each Taf is required for a distinct subset of genes, ranging from 3% (Taf2) to 59–61% (Taf9) (10). Intriguingly, yeast promoters can be classified into three classes: those that depend on all (or almost all) Tafs, those that depend on only a subset of Tafs, and those that do not require any Tafs (10). These observations indicate that each Taf plays a different role in mediating transcription for different classes of promoters. In metazoans, there are multiple TFIIDs that either lack specific Tafs or contain tissue-specific Tafs (4,25–27). More strikingly, TAF7 dissociates from TFIID at the promoter upon transcriptional initiation (28). Therefore, the aforementioned different roles of Tafs in different classes of promoters may be influenced by the effects of variable composition of TFIIDs on DNA, although yeast TFIID seems to exist in a unified form when purified from cell extracts (29).
It is well established that TFIID undergoes conformational alterations in vitro in response to binding to DNA or to specific proteins such as activators or TFIIA (30–35). However, due to technical limitations, it has remained unclear whether TFIID undergoes conformational alterations when it is bound to different promoters in vivo. Specifically, in vivo footprinting techniques can be used to detect a region that is bound by certain factors, but cannot detect which factor it is.
To address the questions of whether all Tafs bind equally to the same set of promoters and whether TFIID undergoes conformational alterations in vivo, we conducted genome-wide ChIP-chip analysis of chromosomes III, IV and V. To our knowledge, this is the first systematic study examining genome-wide localization of a complete set of Tafs (18,19,36–39). We also examined the effects of a taf1-T657K mutation on Taf occupancy as well as on other related factors, including Gcn5, Bur6/Ncb2, Sua7, Tfa2, Tfg1, Tfb3 and Rpb1, in order to test whether this particular taf1 mutation affects the localization of these factors similarly. Notably, we found a specific effect of this mutation on Taf2 occupancy that may reflect the spatial relationship between Taf1 and Taf2 within TFIID (27,40). We also conducted conventional ChIP analysis to further confirm the presence of different TFIID conformations on different promoters.
MATERIALS AND METHODS
Yeast strains
Standard techniques were used for yeast growth and transformation (41). Yeast strains used in this study are listed in Supplementary Table S1. All strains were derived from Y13.2 or Y22.1, which carry a deletion of the chromosomal TAF1 coding region and the wild type TAF1 gene in a URA3-based low-copy-number vector (pYN1) (42). YTK2741 (43) and YTK3780 were generated from Y22.1 by replacing pYN1 with pM1169 (HA-tagged wild type TAF1/pRS314) (44) and pM1747 (HA-tagged taf1-T657K/pRS314), respectively. The latter plasmid was constructed by site-directed mutagenesis of pM1169 using the primer TK178 (45). The oligonucleotides used in this study are listed in Supplementary Table S2.
The primer pairs TK6352/6353, TK6354/6355, TK6356/6357, TK6358/6359, TK6360/6361, TK6362/6363, TK6364/6365, TK6366/6367, TK6368/6369, TK6370/6371, TK6372/6373, TK6374/6375, TK6376/6377 and TK6378/6379, each of which contains 40 bp of additional nucleotides that are homologous to the region immediately upstream or downstream of the stop codons for TAF2, TAF3, TAF4, TAF5, TAF6, TAF7, TAF8, TAF9, TAF10, TAF11, TAF12, TAF13, TAF14 and GCN5, respectively, were used for PCR to amplify the DNA fragments containing the kanMX6 module (46) and PK epitope tag (47) using pM4376 as a template. Subsequently, 5′- and 3′-flanking regions (ca. 500 bp each) of the stop codons of these genes were amplified by PCR using genomic DNA as a template and two sets of primer pairs: [TK6905/6906 (5′-), TK6907/6908 (3′-)], [TK6909/6910, TK6911/6912], [TK6913/6914, TK6915/6916], [TK6917/6918, TK6919/6920], [TK6921/6922, TK6923/6924], [TK6925/6926, TK6927/6928], [TK6929/6930, TK6931/6932], [TK6933/6934, TK6935/6936], [TK6937/6938, TK6939/6940], [TK6745/6746, TK6747/6748], [TK6749/6750, TK6751/6752], [TK6941/6942, TK6943/6944], [TK6945/6946, TK6947/6948] and [TK6949/6950, TK6951/6952], respectively. For each gene, the DNA fragment obtained from the 1st PCR reaction was fused to the two DNA fragments (5′ and 3′) obtained from the 2nd PCR reaction by two-step PCR (48). The extended PCR products carrying long flanking regions were transformed into Y13.2 to generate YTK6760, YTK6761, YTK6762, YTK6763, YTK6764, YTK6765, YTK6766, YTK6767, YTK6768, YTK6769, YTK6770, YTK6771, YTK6772 and YTK6773, with each containing three repeats of the PK epitope tag at the carboxy-terminal ends of Taf2, Taf3, Taf4, Taf5, Taf6, Taf7, Taf8, Taf9, Taf10, Taf11, Taf12, Taf13, Taf14 and Gcn5, respectively.
The 5′- and 3′-flanking regions (ca. 500 bp each) of the stop codons of SPT15, SUA7, TFA2, TFG1, TFB3, BUR6 and NCB2 that had been amplified by PCR using genomic DNA as a template and two sets of primer pairs: [TK7772/7773 (5′-), TK7774/7775 (3′-)], [TK7112/9359 (5′-), TK7114/7115 (3′-)], [TK7478/7117, TK7118/7119], [TK7120/7121, TK7122/7123], [TK7124/7125, TK7126/7479], [TK7884/7885, TK7886/7887] and [TK7888/7889, TK7890/7891], respectively, were fused individually with the DNA fragments containing kanMX6 and a PK tag that had been amplified by PCR using pM4376 as a template and the primer pair TK6382/6383. Subsequently, as described above, the extended PCR products carrying long flanking regions were transformed into Y13.2 to generate YTK6783, YTK6779, YTK6780, YTK6781, YTK6782, YTK6784 and YTK6785, containing three repeats of a PK tag at the carboxy-terminal end of TBP, Sua7, Tfa2, Tfg1, Tfb3, Bur6 and Ncb2, respectively.
YTK6818 to YTK6831 and YTK6837 to YTK6843 were generated from YTK6760 to 6773 and YTK6779 to YTK6785, respectively, by replacing pYN1 with pM1169. Similarly, YTK6845 to YTK6858 and YTK6864 to YTK6870 were generated from YTK6760 to 6773 and YTK6779 to YTK6785, respectively, by replacing pYN1 with pM1747.
The expression of HA or PK epitope-tagged proteins in these strains was confirmed by immunoblotting analysis (Supplementary Figure S1). None of these strains showed growth defects at 25 or 37°C on YPD plates (Supplementary Figure S2).
Chromatin immunoprecipitation, sequential ChIP and ChIP-chip analysis
Cells were grown to log phase in YPD media at 25°C, and then each culture was shifted to 37°C, and incubation was continued for 2 h. ChIP analysis was conducted using monoclonal antibodies against the HA (F7, #sc-7392, Santa Cruz Biotechnology Inc.), PK (SV5-Pk1, #MCA1360, AbD Serotec), and CTD epitopes (8WG16, #MMS-126R, Covance) according to the methods described by Katou et al. (49) with minor modifications. Namely, in this study, the chromatin fraction was sheared by sonication using a bioruptor (UCD-250; Tosho Denki, Yokohama, Japan) (30 min, 30 s ON and 30 s OFF at the ‘High’ [250 W] position) instead of Branson Sonifier 2508 (Danbury, CT, USA) (49).
Quantitative PCR was performed using an iCycler iQ real-time detection system (Bio-Rad) using SYBR Green I (Molecular Probes) as a fluorescent dye for detection of DNA. All PCRs were carried out under the same conditions: 3 min at 95°C followed by 40 cycles of 30 s at 94°C, 30 s at 45°C and 60 s at 72°C. For each PCR reaction, titrations of known amounts of DNA were used as a standard to calculate the ratio of precipitated DNA to input DNA (immunoprecipitation efficiency). The PCR primer pairs used to amplify the respective genes were: RPS11A, TK9493-TK9494; RPS17B, TK9497-TK9498; RPL27B, TK9501-TK9502; RPL31A, TK9487-TK9488; RPL35A, TK9614-TK9615; RPL35B, TK9483-TK9484; RPL10, TK5413-TK5414 (50); PGK1, TK4234-TK4235; HTB1, TK10475-TK10476; YCL049C, TK10334-TK10335; DLD3, TK8413-TK8414; and the transcribed region of POL1, TK3506-TK3507 (51).
For sequential ChIP analysis, the first immunoprecipitation was performed as described above (49) except that beads were only washed twice with lysis buffer [50 mM Hepes–KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 1% Triton-X100, 0.1% Na-deoxycholate, 350 ng/ml bestatin, 2 mM benzamidine, 400 ng/ml pepstatin A, 500 ng/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride] to minimize the loss of precipitate. The eluates (50 µl), which were prepared by incubation at 65°C for 10 min in elution buffer [50 mM Tris–HCl (pH 8.0), 10 mM EDTA, 1% SDS] were then incubated with beads attached to the appropriate antibodies for the second immunoprecipitation in 1 ml of lysis buffer (49). Washes, elution and crosslink reversal were carried out as described for ChIP (49).
High-density oligonucleotide microarrays covering every 300 bp region of Saccharomyces cerevisiae chromosomes III–V and part of chromosome VI by 16 or 11 sets of unique 25 nt probes (SC3456a 52005F, P/N 520015) were purchased from Affymetrix and used in this study for ChIP-chip analysis. ChIP DNA was prepared as described above. Subsequent steps including DNA amplification, hybridization to microarrays and data analysis were all carried out as described by Katou et al. (49).
RESULTS
TFIIB and NC2 appear to co-regulate a subset of class II gene promoters
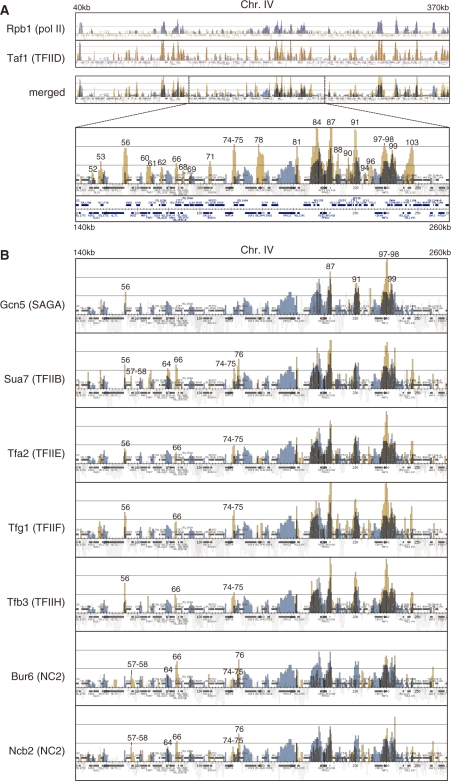
To investigate whether TFIID (Taf1), SAGA (Gcn5), NC2 (Bur6/Ncb2) and several other GTFs, including TFIIB (Sua7), TFIIE (Tfa2), TFIIF (Tfg1) and TFIIH (Tfb3), bind to the same or different sets of class II gene promoters, we compared localization profiles of these transcription factors on chromosomes III, IV and V by conducting ChIP-chip analysis. A complete set of the data is graphically represented in Supplementary Figures S5–S10 and summarized in Supplementary Table S3. The results obtained for a portion of chromosome IV (40–370 kb or 140–260 kb) are shown in Figure 1. First, we sought to confirm whether the experimental system used in this study could detect specific occupancy sites of these transcription factors. A merged figure showing the localization profiles of both Taf1 and Rpb1, which are shown in different colors, demonstrates that these two factors have overlapping, but not equivalent, localization on the yeast genome (Figure 1A). In general, their localization profiles are consistent with the conventional view that TFIID binds specifically to the promoter region, whereas pol II binds to the promoter, coding region and/or 3′-end of class II genes. For example, the RPO21 promoter is bound by both Taf1 and Rpb1 (corresponding to signal #81 in an enlarged image of the merged figure in Figure 1A), whereas only the latter factor localizes to the entire ORF region of this gene. Furthermore, strong Taf1 and Rpb1 signals were observed for the promoters of three ribosomal protein genes, RPL35B (#84), RPL41B (#87) and RPP1B (#91), all of which are known to be highly expressed TFIID-dependent genes. These observations indicate that the signals shown in our ChIP-chip analysis represent specific occupancy sites of these transcription factors.
Figure 1.
Localization of general transcription factors (GTFs) and RNA polymerase II (pol II) on chromosomes III, IV and V. (A) Comparison of the localization of Rpb1 (a subunit of pol II, top panel) and Taf1 (a subunit of TFIID, second panel) by combining these two images into one (third panel) denoted as ‘merged’ at left. The strain YTK2741 expressing HA-tagged Taf1 was grown in YPD (yeast extract, peptone, dextrose) medium to mid-log phase at 25°C. Two hours after the temperature shift to 37°C, the cross-linked chromatin was prepared and precipitated with anti-CTD (Rpb1) or anti-HA (Taf1) monoclonal antibodies, and then analyzed by GeneChip (SC3456a 52005F, P/N 520015; Affymetrix). The blue and orange vertical bars represent the significant occupancy by Rpb1 and Taf1 at the region from 40 000 to 370 000 on chromosome IV, respectively. The light gray bars underneath the colored bars (e.g. blue) indicate the signals derived from other-colored factors (e.g. orange) that are not enriched significantly in the immunoprecipitated fraction (49). The horizontal small squares with a different color in each panel indicate the ORFs. The bottom panel represents the enlarged image corresponding to the region from 140 000 to 260 000 of the third ‘merged’ panel. In these ‘merged’ images, the thick colored vertical bars are shown in light colors to increase their transparency. Note that the ORFs are also drawn below the bottom panel as they were partly overlapped by the vertical bars in their original positions. The numbers above the occupancy signals correspond to those in Supplementary Table S3. (B) Comparison of the localization of Gcn5 (a subunit of SAGA, top panel), Sua7 (TFIIB, second panel), Tfa2 (a subunit of TFIIE, third panel), Tfg1 (a subunit of TFIIF, fourth panel), Tfb3 (a subunit of TFIIH, fifth panel), Bur6 (a subunit of NC2, sixth panel) and Ncb2 (another subunit of NC2, bottom panel), with that of Rpb1 by generating merged images of the same region (from 140 000 to 260 000) of chromosome IV as described for Taf1 in (A). The strains YTK6831 (Gcn5), YTK6837 (Sua7), YTK6838 (Tfa2), YTK6839 (Tfg1), YTK6840 (Tfb3), YTK6842 (Bur6) and YTK6843 (Ncb2) were cultured and cross-linked as described in (A). ChIP analysis was conducted as described in (A) except that anti-PK monoclonal antibody was used to precipitate these transcription factors. Note that the localization profile of Rpb1 represented as ‘blue’ signals in each panel is the same as described in A, which was obtained from YTK2741. The complete data sets including Tafs are represented in Supplementary Figures S5–7.
Next, we compared the localization profile of Taf1 with those of Gcn5, Sua7, Tfa2, Tfg1, Tfb3, Bur6 and Ncb2 (Figure 1B). The latter factors appear to bind to only a subset of the promoters that are bound by Taf1. For example, significant Gcn5 occupancy was detected at the RPL41B (#87), RPP1B (#91), CDC48 (#97) - HNT1 (#98) and SNA4 (#99) promoters, but not at other promoters, including NRP1 (#60), RPN5 (#74) - NOP14 (#75), COP1 (#78), RPO21 (#81), CDC53 (#88) and CYK3 (#103). In contrast, the CDC9 (#62) promoter is bound by only Taf1. We found that Taf1 occupancy sites on the promoter regions on chromosomes III, IV and V were consistently more numerous than occupancy sites for other factors (Figure 5A).
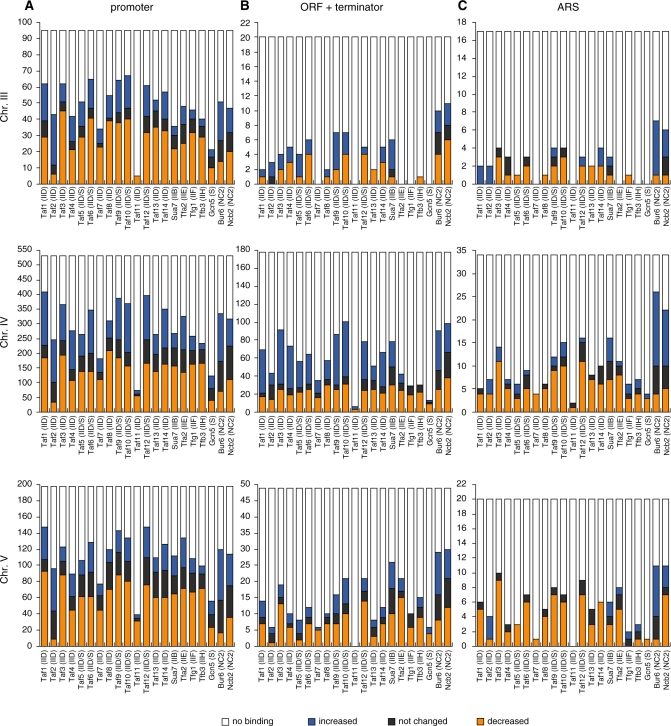
Figure 5.
Effects of taf1-T657K mutation on the number of occupancy sites for GTFs, Tafs, Gcn5 and NC2 on chromosomes III, IV and V. The number (y-axis) of occupancy sites for each factor that is shown below the x-axis (letters inside parentheses are abbreviations that specify originated GTF or complexes; e.g. IIB/D/E/F/H and S refer to TFIIB/D/E/F/H and SAGA, respectively) was counted separately for the promoter regions of class II genes (A), open reading frames (ORF) and the terminator regions of class II genes (B), and autonomously replicating sequences (ARS) (C). The results are summarized for each chromosome: III (top panels), IV (middle panels), and V (bottom panels). In all panels, the occupancy sites are categorized into four groups that are labeled with different colors. Orange, blue and black bars indicate the sites where the occupancy level of a given factor was weakened, strengthened, or not changed, respectively, in taf1-T657K strains when compared with wild-type strains, while white bars indicate the sites that are not bound by a given factor but are bound by one of the other factors shown below the x-axis. These data were originally derived from Supplementary Figures S8–10, and the counting details are summarized in Supplementary Table S3.
Interestingly, the localization profiles of NC2 (Bur6/Ncb2) and GTFs seem to vary depending on the promoter. For example, the STE7 (#66) and RPN5 (#74) - NOP14 (#75) promoters are bound by both GTFs (Taf1, Sua7, Tfa2, Tfg1 and Tfb3) and NC2 (Bur6/Ncb2), whereas the UGA3 (#56) promoter is bound by GTFs, but not by NC2. Notably, only Sua7 and NC2 localize to the YDL160C-A (#64) and LDB17 (#76) promoters with apparently different occupancy ratios for Sua7/NC2. This indicates that TFIIB and NC2 may co-regulate a subset of promoters at some step after the incorporation of TFIIB but prior to completion of the pre-initiation complex assembly. This is unexpected since these two factors are competitive in association with the TBP-DNA complex in vitro (52,53). However, a recent in vivo study demonstrated that NC2 can regulate TFIIB binding positively or negatively in a promoter-specific manner (54). In addition, a wide variation in TFIIB/NC2 ratios from one gene to another has also been observed in human cells (55). Therefore, we believe that co-regulation by TFIIB and NC2 may be more prevalent than previously thought. This view is supported by hierarchical clustering analysis showing that the localization profiles of Bur6 and Ncb2 cluster most closely with that of Sua7 (Supplementary Figure S4).
Finally, significant differences were not observed in localization of Tfa2, Tfg1 and Tfb3. In fact, hierarchical clustering analysis showed that their localization profiles are clustered very closely (Supplementary Figure S4). These observations support the view that TFIIF does not travel along with elongating pol II in living cells (19,56).
TFIID containing Taf1-T657K is defective in TFIIB recruitment and in later steps at several ribosomal protein gene promoters
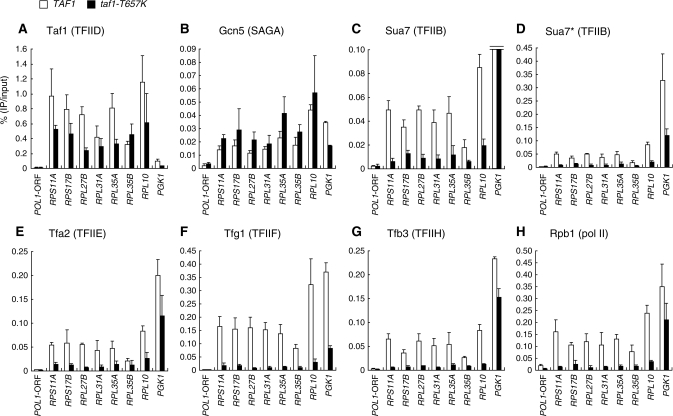
In taf1-T657K mutant cells, nearly 40% of the genes in the genome were affected positively or negatively two hours after the temperature shift from 25°C to 37°C (Ohtsuki et al., manuscript in preparation). Taf1-T657K mutant protein was still stably expressed under these conditions (45) and appeared to form a normal TFIID complex (45) (Supplementary Figure S12). Thus, it remained unclear whether the binding step itself or subsequent steps after TFIID binding are affected by this mutation. To address this question, we conducted ChIP analysis to compare the occupancy levels of Taf1, Gcn5, Sua7, Tfa2, Tfg1, Tfb3 and Rpb1 at several TFIID-dependent ribosomal protein gene (RPG) promoters: RPS11A, RPS17B, RPL27B, RPL31A, RPL35A, RPL35B and RPL10, and SAGA-dependent PGK1 promoter (13) in wild-type and taf1-T657K strains (Figure 2). Consistent with the transcription levels (data not shown) (45), Rpb1 occupancy at the RPG promoters was greatly decreased in the taf1-T657K strain, whereas its occupancy at the PGK1 promoter was only slightly decreased (Figure 2H). Significantly, the taf1-T657K mutation only partially affected Taf1 occupancy at RPG promoters (Figure 2A) but affected the occupancy by Sua7 (Figure 2C and D), Tfa2 (Figure 2E), Tfg1 (Figure 2F) and Tfb3 (Figure 2G) more severely, i.e. to a similar extent as for Rpb1 (Figure 2H). These observations indicate that TFIID containing Taf1-T657K, even if it remains bound to the promoter at higher temperatures, cannot effectively recruit TFIIB and thereby other GTFs. In contrast, Gcn5 occupancy at the RPG promoters appears to be increased in the taf1-T657K strain (Figure 2B), suggesting that the SAGA recruitment occurs independently of Taf1/TFIID and may be competitive with TFIID binding at these promoters. This is consistent with a previous notion that SAGA and Bdf1, a TFIID-associated factor, are competitive in promoter binding (57). It is also notable that significantly less TFIIB binds to the RPG promoters than to the PGK1 promoter (Figure 2D), although they are equally active promoters at the highest levels (24). This is in stark contrast with Taf1 occupancy (Figure 2A), supporting the view that TFIID- and SAGA-dependent promoters are regulated by different mechanisms.
Figure 2.
Effects of the taf1-T657K mutation on the localization of GTFs and pol II at a subset of class II gene promoters. Strains expressing HA-tagged TAF1 (YTK2741) or taf1-T657K (YTK3780) alone or in combination with PK-tagged GCN5 (YTK6831/YTK6858; this depicts TAF1/taf1-T657K strains, respectively), SUA7 (YTK6837/YTK6864), TFA2 (YTK6838/YTK6865), TFG1 (YTK6839/YTK6866) or TFB3 (YTK6840/YTK6867) were cultured and cross-linked as described in Figure 1. The cross-linked chromatin was prepared and precipitated with anti-CTD (Rpb1, H), anti-HA (Taf1, A) or anti-PK (Gcn5, B; Sua7, C–D; Tfa2, E; Tfg1, F; Tfb3, G) monoclonal antibodies. After the cross-link reversal, quantitative PCR was carried out in triplicate to determine the recovery ratio of DNA corresponding to the promoters of several genes or POL1 ORF (negative control) as indicated at the bottom of each panel. The average values from three independent experiments with standard deviations of the ratios of precipitated DNA to the inputs are shown as white (TAF1) or black (taf1-T657K) bars in each panel. YTK2741 and YTK3780 strains were used to assess occupancy levels of Taf1 and Rpb1. Note that the data for Sua7 are shown in two panels (C and D) with different scales on the vertical axis as occupancy levels varied greatly between PGK1 and ribosomal protein genes.
TFIID and SAGA co-localize to RPG promoters concurrently
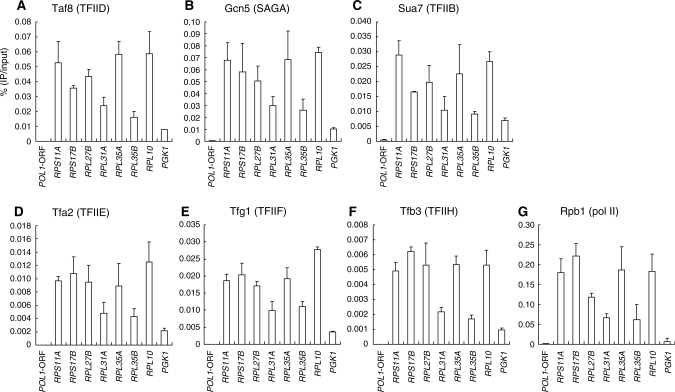
Previous genome-wide expression studies have demonstrated that housekeeping genes such as RPGs are predominantly regulated by TFIID, whereas stress-inducible genes are regulated by SAGA (13,15). However, the genome-wide promoter occupancy by Gcn5 has been reported to be well correlated with gene transcription rates (17), suggesting that SAGA also binds to highly active RPG promoters. In fact, Gcn5 has been shown to bind to the upstream activating sequence (UAS) of RPL2B (17). Consistent with this, our ChIP-chip analysis showed that Gcn5 is localized to nearly all RPG promoters on chromosomes III, IV and V, except for RPL4B (chromosome IV), which appears to be less active (Supplementary Figures S5–7). Therefore, SAGA (or other Gcn5-containing complexes like SLIK/SALSA) (58) might be involved in transcriptional regulation of RPGs even though its impairment does not necessarily lead to reduced RPG expression, probably due to functional redundancy with TFIID (13,15). However, it had remained unclear whether TFIID and SAGA bind to the RPG promoters concurrently or alternately. To address this question, we conducted sequential ChIP analysis using anti-HA (Taf1) antibody for the first immunoprecipitation step and anti-PK (Taf8, Gcn5, Sua7, Tfa2, Tfg1, Tfb3) or anti-Rpb1 antibodies for the second immunoprecipitation step (Figure 3). As expected, promoter DNA for several RPGs and PGK1 can be recovered with the TFIID-specific subunit Taf8 (Figure 3A), several other GTFs (Figure 3C–F) and pol II (Figure 3G) in the second immunoprecipitation step, indicating that they bind to these promoters together with Taf1. Significantly, a similar co-localization profile was observed for Gcn5 (Figure 3B). The signals obtained here appear to be specific because they could not be detected when strains lacking either the HA (Taf1) or PK (Gcn5) tag were used in the same experiments (Supplementary Figure S3). Thus, we conclude that TFIID and SAGA co-localize concurrently to TFIID-dependent RPG promoters. However, we cannot formally exclude the possibility that these promoters are bound by TFIID or SAGA alone in some cells. In fact, the occupancy signal for Gcn5 at the PGK1 promoter was stronger than those at many other RPG promoters in a single-step ChIP experiment (Figure 2B), whereas it was the lowest in a sequential ChIP experiment (Figure 3B). These results imply that in some cells, the PGK1 promoter is bound by Gcn5/SAGA but not by Taf1/TFIID, even though it is bound by both factors in other cells.
Figure 3.
Co-occupancy by TFIID and SAGA at the promoters of several class II genes. Strains expressing HA-tagged TAF1 (YTK2741) alone or in combination with PK-tagged TAF8 (YTK6824), GCN5 (YTK6831), SUA7 (YTK6837), TFA2 (YTK6838), TFG1 (YTK6839) or TFB3 (YTK6840) were cultured and cross-linked as described in Figure 1. The cross-linked chromatin was prepared and precipitated with anti-HA monoclonal antibody. The eluates were then re-precipitated with anti-CTD (Rpb1, G) or anti-PK (Taf8, A; Gcn5, B; Sua7, C; Tfa2, D; Tfg1, E; Tfb3, F) monoclonal antibodies. After the cross-link reversal, quantitation of the ratio of DNA recovered after the second immuno-precipitation step to the input was performed as described in Figure 2. The data from three independent experiments are presented as mean ± SD.
TFIID may have different conformations and is affected differently by the taf1-T657K mutation at different promoters
In higher eukaryotes, TBP and three TBP-related factors (TRFs) bind to a different subset of promoters (4,59–61). Furthermore, cell-type-specific TAFs may generate several distinct forms of TFIID to mediate tissue-specific gene expression (4,27). In contrast to such complexities in higher eukaryotes, there are no known variants for TBP and Tafs in yeast. In fact, only a single form of TFIID can be immunopurified from cell extracts using antibodies against TBP or Tafs (29). However, it was not clear whether all Tafs bind stably to the same set of promoters or if some Tafs dissociate from TFIID after transcriptional initiation as observed for human TAF7 (28,62). In addition, we wondered whether some Tafs would dissociate from TFIID-dependent promoters at restrictive temperatures in the taf1-T657K strain, despite the fact that considerable amounts of Taf1 remain bound to RPG promoters under these conditions (Figure 2A).
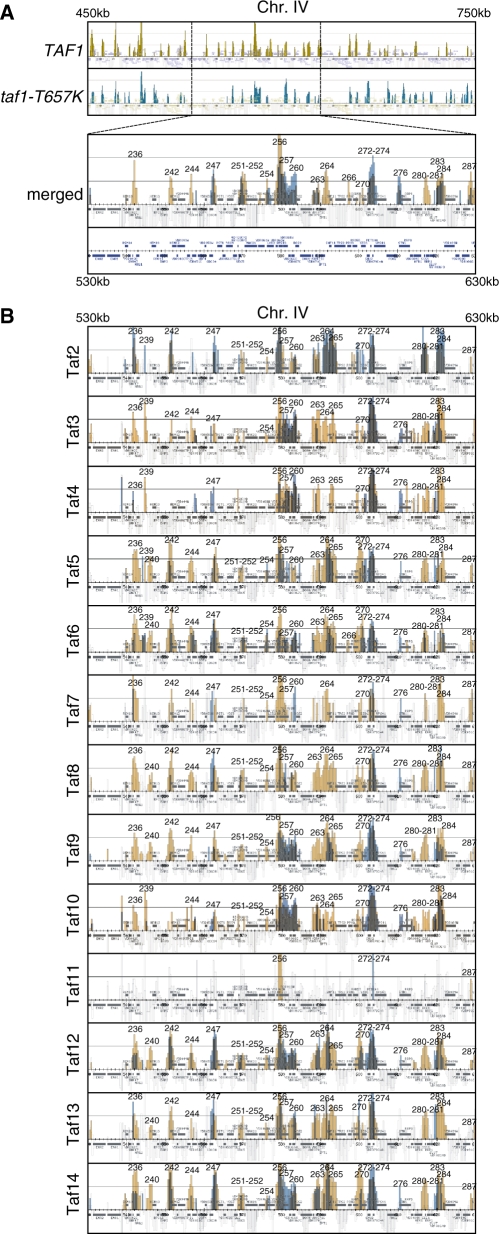
To address these issues, we compared the localization profiles of all Tafs on chromosomes III, IV and V in the wild-type and taf1-T657K strains by ChIP-chip analysis (Figure 4 and Supplementary Figures S8–10). To make the effects of the taf1-T657K mutation easily recognizable and comparable among genes, we merged the localization profiles for each Taf obtained from wild-type (orange) and taf1-T657K (blue) strains in one figure, which are shown in Figure 4A (Taf1) and 4B (Taf2-14). For some reason, the Taf11 occupancy signals were very few in number and quite weak [PK-tagged Taf11 was expressed at a reasonable level (Supplementary Figure S1) and appeared to form a normal TFIID complex (Supplementary Figure S12)]. In previous studies, Taf11 has been shown to bind to promoter DNA in vitro (63) as well as in vivo (10). Thus, we postulate that the PK tag fused to the carboxy-terminus of Taf11 used in this experiment may sterically hinder antibody binding when bound to DNA. This may be related to our previous observation that TAND (Taf1 N-terminal domain) can be transferred to the amino-terminus but not the carboxy-terminus of Taf11 due to incompatibility with other Tafs (43).
Figure 4.
Effects of the taf1-T657K mutation on the localization of Tafs on chromosomes III, IV and V. (A) Comparison of Taf1 localization in wild-type (top panel; significant occupancy is indicated by orange vertical bars) and taf1-T657K (second panel; significant occupancy is indicated by blue vertical bars) strains by combining two images into one (bottom panel, denoted as ‘merged’). Note that the region from 450 000 to 750 000 of chromosome IV is shown in the top two panels, whereas only a part of this region (from 530 000 to 630 000) is selectively shown as an enlarged image in the bottom ‘merged’ panel. The strains YTK2741 and YTK3780 expressing HA-tagged Taf1 and Taf1-T657K, respectively, were cultured and cross-linked as described in Figure 1A. The cross-linked chromatin was prepared and precipitated with anti-HA monoclonal antibody, and then analyzed by GeneChip as described in Figure 1A. The numbers above the occupancy signals correspond to those in Supplementary Table S3. (B) Comparison of the localization of the Tafs (i.e. Taf2-Taf14 as denoted at the left of each panel) in wild-type (orange bars) and taf1-T657K (blue bars) strains by generating merged images of the same region (from 530 000 to 630 000) of chromosome IV as described for Taf1 in A. Each pair of strains expressing one of the PK-tagged TAF genes as well as either HA-tagged TAF1 or taf1-T657K, which is abbreviated as [PK-tagged]TAFX (strain #[HA-tagged TAF1]/strain #[HA-tagged taf1-T657K]), i.e. TAF2 (YTK6818/YTK6845), TAF3 (YTK6819/YTK6846), TAF4 (YTK6820/YTK6847), TAF5 (YTK6821/YTK6848), TAF6 (YTK6822/YTK6849), TAF7 (YTK6823/YTK6850), TAF8 (YTK6824/YTK6851), TAF9 (YTK6825/YTK6852), TAF10 (YTK6826/YTK6853), TAF11 (YTK6827/YTK6854), TAF12 (YTK6828/YTK6855), TAF13 (YTK6829/YTK6856) or TAF14 (YTK6830/YTK6857), was cultured and cross-linked as described in (A). The cross-linked chromatin was prepared and precipitated with anti-PK monoclonal antibody, and then analyzed by GeneChip as described in (A). The complete data sets including Gcn5, NC2, pol II and other GTFs are represented in Supplementary Figures S8–10.
All Tafs, including Taf11, bound to the promoter of TFIID-dependent RPS13 (signal #256) while only a subset of Tafs, including Taf5, 6, 12 and 14, significantly bound to the promoter of SAGA-dependent SED1 (#270) (13) (Figure 4). Taf5, 6, 9, 10 and 12 are common components shared by TFIID and SAGA. Taf14 is a component of TFIID, TFIIF, RSC, SWI/SNF, INO80, NuA3 and Mediator (64). Thus, the Taf signals at the SED1 (#270) promoter mainly represent the occupancy by SAGA and certain other complexes rather than TFIID. However, these signals were greatly reduced in the taf1-T657K strain, suggesting that TFIID might be also involved in transcriptional regulation of this gene. Consistent with this, occupancy signals of TFIID-specific Tafs such as Taf7/8 were also detected at this promoter (#270) and were reduced in the taf1-T657K strain. On the other hand, Taf signals were all partially reduced at the RPS13 (#256) promoter (e.g. Taf1, consistent with the results for other RPGs in Figure 2A) or completely abolished (e.g. Taf7/11). Collectively, these observations indicate that the ChIP-chip analysis reveals specific localization of Tafs as well as localization changes caused by the taf1-T657K mutation.
Taf1 and Taf7 appeared to bind to the IPT1/SNF11 promoter (#264) much more strongly than to the neighboring TPS2 promoter (#265), whereas other Tafs bound to these two promoters at similar levels (Figure 4). This may imply that Taf1 and Taf7 dissociate from TFIID or that they are positioned at a less cross-linkable distance from DNA specifically on the latter promoter. Remarkably, the localization profiles of these two Tafs are clustered more closely with each other than with those of other Tafs (Supplementary Figure S4). Similar selective occupancy by Tafs was observed at the BAP3/HEM12 (#242) promoter where very low amounts of Taf4 and Taf10 are localized (Figure 4). This is in contrast with the AFR1 (#280) or SSS1 (#281) promoters, where all Tafs are localized fairly equally (Figure 4). Collectively, these observations indicate that TFIID may exist in different conformations in which cross-linking efficiency is different for each Taf, depending on the promoter structure.
Intriguingly, the taf1-T657K mutation can either decrease or increase Taf occupancy. For example, occupancy by all Tafs was increased at the SHU2/PET100 (#272) or TFB5/VPS41 (#274) promoters, whereas the occupancy by only Taf1, 3, 9, 10, 12 and 14 was significantly increased in the region encompassing the YDR065W (#257), RTR2 (#259), and OCA6/DOS2 (#260) promoters (Figure 4). These results indicate that Taf1 can inhibit binding of either the entire TFIID complex or of only a subset of Tafs at different promoters.
Finally, we found that the effects of the taf1-T657K mutation on the localization profile of Taf2 differ significantly from its effects on other Tafs. For example, this mutation did not decrease Taf2 occupancy at the RPS13 (#256) promoter (Figure 4). Furthermore, it increased Taf2 occupancy at the SLU7 (#283) or YDR089W (#284) promoters even though it decreased occupancy by other Tafs at the same promoters. We consistently found that Taf2 occupancy was increased rather than decreased at most promoters on chromosomes III, IV and V in this mutant (Figure 5). This specific localization profile for Taf2 may be related to a previous finding that only a small portion of the entire genome (∼3%) is affected in taf2 mutants (10) and the physical properties of Taf2 (27,40,65,66) as discussed below.
Taf2 occupancy is affected differently than other Tafs by the taf1-T657K mutation
We roughly counted the number of occupancy signals for each Taf (Taf1-14), GTF (Sua7, Tfa2, Tfg1, Tfb3), SAGA (Gcn5) and NC2 (Bur6, Ncb2) on chromosomes III, IV and V that were strengthened (blue), weakened (orange) or not changed (black) in the taf1-T657K strain (Figure 5) compared to the wild-type strain by manually assessing the color of each signal in the merged figures (Supplementary Figures S8–10). The effects of the taf1-T657K mutation on the occupancy by these factors were originally scored using five categories: ++ (significantly increased), + (marginally increased), ± (not changed), – (marginally decreased) or −− (significantly decreased) (Supplementary Table S3). However, since the differences between the ++ and + groups (and the −− and − groups) were not statistically significant, the groups were combined into three classifications, i.e. ‘increased’, ‘not changed’ or ‘decreased’ (Figure 5).
As expected, most of the occupancy signals for these factors were observed in the promoter regions (Figure 5A). However, a smaller but still significant number of occupancy signals were also found within the ORF and terminator regions (Figure 5B) and in the ARS (autonomously replicating sequence) (Figure 5C). This may be consistent with a recent view that the whole-yeast genome may be expressed more extensively than previously appreciated (67–69). Importantly, the localization profiles for these factors on the three chromosomes appear to be similar to each other, indicating that manual counting can be used to assess the effects of taf1-T657K mutation in a reliable manner.
Several intriguing observations can be made from this figure (Figure 5). The occupancy signals for Taf11 were the fewest in number, and most of these were decreased by the taf1-T657K mutation (Figure 5A). In contrast, the occupancy signals for Gcn5 were also few in number, but were only decreased by approximately half (or less) by this mutation (Figure 5A). Most interestingly, out of the 14 Tafs, the taf1-T657K mutation differentially affected Taf2 occupancy; the number of ‘increased’ signals was much larger than the number of ‘decreased’ signals for Taf2, while the other Tafs typically had more ‘decreased’ signals, or an equivalent number of ‘increased’ and ‘decreased’ signals (Figure 5A). In other words, the taf1-T657K mutation increased rather than decreased Taf2 occupancy at many promoters. This is in stark contrast with, for example, Taf7, which showed relatively few occupancy signals, but most of these were decreased by the taf1-T657K mutation (Figure 5A). Although the basis for these specific changes in Taf2 localization remains unknown, it may be somehow linked to the physical properties of Taf2, which is closely associated with Taf1 as well as being the most easily dissociable component of TFIID (27,40,65,66).
A similar number of occupancy signals were observed for the Tafs (except Taf11), GTFs and NC2 at promoter sites (Figure 5A). However, the number of NC2 occupancy signals relative to those of other factors appeared to be higher in the ORF, terminator and ARS regions (Figure 5B and C), implying that NC2 may have a novel function that is independent of Tafs and GTFs. Furthermore, it is also intriguing that the taf1-T657K mutation affected occupancy by the two NC2 subunits differently. Specifically, this mutation tends to increase the occupancy by Bur6 more than Ncb2 (or decrease the occupancy by Ncb2 more than Bur6) at many sites. Consistent with this, recent findings have shown that the two NC2 subunits may carry out distinct functions in yeast (37,70), Drosophila (71) and mammalian (72) cells. In mammals, NC2β (but not NC2α) and the POLE3-POLE4 heterodimer were shown to be components of the ATAC (Ada2a-containing)-type histone acetyltransferase (HAT) complex (72). The POLE3-POLE4 heterodimer is also known to be a component of DNA polymerase ε (73), which is primarily responsible for leading-strand synthesis (74), suggesting a functional link between NC2 and ARS. However, it is currently unknown whether a similar HAT complex exists in yeast cells.
Finally, most of the GTF occupancy signals were decreased by the taf1-T657K mutation as observed for several RPGs (Figure 2), indicating that Taf1 may play a stimulatory role in PIC formation at many sites. Conversely, Taf2, whose occupancy was increased at many sites in the taf1-T657K strain, may regulate PIC formation in a negative manner.
The occupancy profiles of the 14 Tafs are similar but not identical when compared at several different class II gene promoters
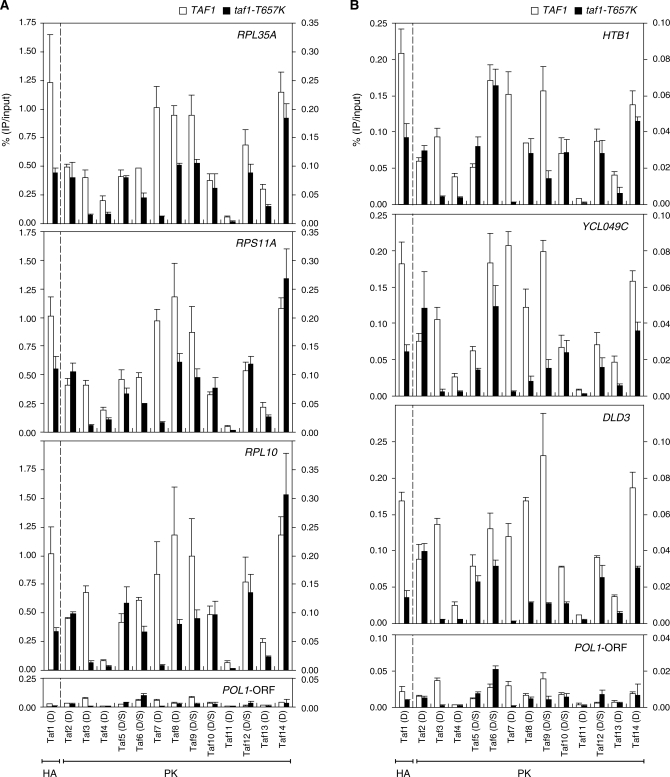
The ChIP-chip analysis described above is suitable for comparisons of relative occupancy by one particular Taf among genes in a genome-wide manner but not for comparison of the absolute occupancy levels of a set of Tafs at a single locus. Thus, we next sought to compare the occupancy profiles of a complete set of Tafs at several class II gene promoters, including the three RPGs (RPL35A, RPS11A and RPL10) (Figure 6A) examined in Figure 2 as well as three other randomly selected non-RPGs (HTB1, YCL049C and DLD3) (Figure 6B), by conventional ChIP analysis.
Figure 6.
Effects of taf1-T657K mutation on the occupancy levels of Tafs at a subset of class II gene promoters. (A) Comparison of Taf occupancy at the promoters of three ribosomal protein genes, RPL35A (top panel), RPS11A (second panel) and RPL10 (third panel), or POL1-ORF (bottom panel, negative control) in wild-type (indicated by white bars) and taf1-T657K (indicated by black bars) strains. ChIP analysis was conducted as described in Figure 2 using the same set of strains described in Figure 4. Quantitation of DNA that was recovered after the immuno-precipitation was performed as described in Figure 2. The data from three independent experiments are presented as mean ± SD. Note that the left and right y-axes are scaled differently to better display the data for Taf1 (obtained using an anti-HA monoclonal antibody) and the other Tafs (obtained using an anti-PK monoclonal antibody) in the same panel. Thus, each panel is divided into two parts by a broken vertical line. (B) Comparison of Taf occupancy at the promoters of the other three class II genes, i.e. HTB1 (top panel), YCL049C (second panel) and DLD3 (third panel), or POL1-ORF (bottom panel, negative control) in wild-type (white bars) and taf1-T657K (black bars) strains. ChIP and DNA quantitation were performed as described in (A). Note that the data for POL1-ORF are the same as those in (A) but shown here on a different scale for comparison with the data for HTB1, YCL049C and DLD3.
We found that the relative occupancy ratios of the 14 Tafs to the three RPGs were quite similar (Figure 6A). Consistent with the results shown in Figure 4, Taf11 occupancy was the weakest among the 14 Tafs. Taf4 and Taf13 occupancy was stronger than that of Taf11 but weaker than that of the other Tafs. Taf7, 8, 9 and 14 occupancies were the strongest, whereas Taf2, 3, 5, 6, 10 and 12 occupancies were intermediate. Note that the occupancy level of Taf1 cannot be compared directly with those of other Tafs since Taf1 was tagged with a different epitope (HA instead of PK). Furthermore, the effects of the taf1-T657K mutation on the occupancy by the 14 Tafs appeared to be similar when compared among the three RPG promoters. Specifically, this mutation affected the occupancy by Taf3 and Taf7 most severely, whereas it affected occupancy by Taf1, 4, 6, 8, 9 and 13 less strongly, but still significantly. In contrast, this mutation had less of an effect on the occupancy by Taf2, 5, 10, 12 and 14, indicating that part of the structure of TFIID may remain bound to these RPG promoters in taf1-T657K cells at restrictive temperatures.
Intriguingly, the relative occupancy ratios of the 14 Tafs to the three non-RPG promoters was similar but not identical to those observed for the three RPG promoters (Figure 6B). For example, the Taf6 occupancy level was much lower than that of Taf8 at the RPG promoters (Figure 6A), but this occupancy ratio was reversed at the HTB1 and YCL049C promoters (Figure 6B). Furthermore, the taf1-T657K mutation affected the occupancy by Taf10 and Taf14 specifically at the DLD3 and YCL049C/DLD3 promoters, respectively. Conversely, taf1-T657K affected less the occupancy by Taf6 and Taf8 specifically at the HTB1 promoter. These observations indicate that TFIID may have an identical or similar conformation when bound to the RPG promoters, and different and presumably specific conformations when bound to the HTB1, YCL049C and DLD3 promoters. This is consistent with an aforementioned view that TFIID may have different conformations depending on the promoter structure (Figure 4). Notably, only Taf2 occupancy was not decreased by the taf1-T657K mutation at these six promoters, indicating that this Taf may play a specific role in TFIID function.
DISCUSSION
In this study, we have made several novel observations by examining the localization profiles of Tafs (TFIID and/or SAGA), Sua7 (TFIIB), Tfa2 (TFIIE), Tfg1 (TFIIF), Tfb3 (TFIIH), Gcn5 (SAGA) and Bur6/Ncb2 (NC2) on chromosomes III, IV and V in wild-type and taf1-T657K Saccharomyces cerevisiae strains.
First, we found that the localization profiles of Sua7 (TFIIB) and Bur6/Ncb2 (NC2) are closely related to each other. Specifically, we found that some promoters were bound by these two factors but not by other GTFs. In principle, the method employed here can be used to analyze the relative occupancy by a given factor on a chromosomal scale such that if a subset of signals were extraordinary strong, they should interfere with other weaker signals. Accordingly, TBP occupancy to class II promoters was obscured in our analysis owing to much stronger signals for the class III promoters (Supplementary Figures S8–10 and Supplementary Table S3) (37,75–77). Thus, it remains unclear whether TFIIB/NC2-bound promoters are also bound by TBP, which can be ascribed to the less significant signals of the latter factor at class II promoters, e.g. as observed for the YDL160C-A (#64) and LDB17 (#76) promoters on chromosome IV (Supplementary Figure 9). From a structural viewpoint, TFIIB and NC2 cannot bind to the TBP-TATA complex simultaneously (53). However, recent studies have shown that an NC2–TBP–DNA complex can be formed even in the absence of TATA (78), and that NC2 can mobilize TBP on TATA-containing DNA (79). These observations indicate the possibility that the NC2–TBP–DNA complex can assume conformations that are different from the crystallographically characterized one (53) and may allow simultaneous binding of TFIIB and NC2. It is also possible that TBP may not bind to TFIIB/NC2-bound promoters, considering that TBP and NC2 are involved in TATA- and DPE-mediated transcription, respectively, in an antagonistic manner in Drosophila cells (80). Certainly, we cannot exclude the more likely possibility that two distinct complexes, i.e. NC2–TBP–DNA and TFIIB–TBP–DNA, exist separately on TFIIB/NC2-bound promoters, representing two different intermediate states during transcriptional activation. Even so, it is still remarkable in the sense that these promoters are regulated by a novel mechanism occurring after the incorporation of TFIIB but before that of other GTFs. In the case of LDB17 (#76) promoter (Figure 1), only Taf3 occupancy signals were significantly detected among the 14 Tafs (Supplementary Figure S9), suggesting that a specific form of TFIID may bind to such promoters. In fact, TAF3 forms a novel complex with TRF3 that mediates transcription of muscle-specific genes in mammalian cells (81). Furthermore, the localization profiles of TFIIB and NC2 are clustered most closely among those of more than 20 transcription factors in yeast (Supplementary Figure S4) and mammalian cells (38). Therefore, TFIIB and NC2 may co-regulate a subset of genes through a novel mechanism in eukaryotic cells.
Second, we found that the taf1-T657K mutation decreased the occupancy levels of several GTFs and pol II, but not of SAGA, in at least a subset of RPGs (Figure 2). Previous studies have shown that mutations in most tafs, except taf3 and taf5, decrease TBP occupancy at the RPG promoters (10,12,82). Consistent with this, we found that the taf1-T657K mutation also decreased TBP occupancy at a subset of RPG promoters (Supplementary Figure S11), confirming a previous view that Tafs are required for TBP recruitment but not vice versa (10,12,82). Notably, although most taf mutations except taf11 and taf13 significantly decrease Taf1 occupancy at the RPG promoters (10), the taf1-T657K mutation decreased its own occupancy only partially (Figure 2A). Thus, this particular taf1 mutation may affect both pre- and post-Taf(s) recruitment steps, including the recruitment of other GTFs.
Intriguingly, the occupancy levels of TBP and TFIIB appear to be much weaker at RPG promoters than at the PGK1 promoter, even in wild-type cells (our unpublished observations) (Figure 2). This suggests that TBP and TFIIB are located at a less cross-linkable distance from DNA on the TATA-less promoters. More importantly, we have shown for the first time that SAGA binds to RPG promoters independent of TFIID. This is consistent with a mechanism proposed previously in which activators recruit SAGA to DNA via direct binding to a Tra1 subunit (83) and also with the idea that SAGA functions upstream of TFIID in RNR3 transcription (16,84). In the latter case, however, SAGA binding itself could not be examined in taf1 mutants due to weak signals in ChIP experiments (16).
To our knowledge, this is the first report to show co-occupancy by TFIID and SAGA at a single locus on DNA (Figure 3). In general, yeast genes can be classified as either TFIID- or SAGA-dominated (13). Currently, only RNR3 is known to be a gene for which TFIID and SAGA play non-redundant roles in transcription (16). Namely, TFIID mediates chromatin remodeling via the action of Rap1 and a Swi/Snf complex, whereas SAGA delivers TBP to the core promoter of this gene (16). Hence, our findings that TFIID and SAGA are co-localized concurrently to TFIID-dominated RPG promoters (13) (Figure 3) indicate that SAGA may also play specific (albeit minor) roles in transcription even from TFIID-dominated promoters.
Third, this is the first investigation of the genome-wide localization of a complete set of Tafs, as previous studies have examined the localization of only a subset of Tafs such as Taf1 (18,36,37,39), Taf1/5/6/9 (19) or Taf1/4/7/10/12 (38). Most remarkably, our extensive analysis revealed that the taf1-T657K mutation affects Taf2 occupancy quite specifically; that is, it increased Taf2 occupancy at many promoters even though it decreased the occupancy by other Tafs (Figures 4 and 5). This specific effect may be partly due to the steric proximity of Taf1 and Taf2 within TFIID (40). It appears that TFIID is composed of two sub-complexes: a module containing a single copy of Taf1, Taf2, Taf7 and TBP and a crescent-shaped core complex containing more than two copies of other Tafs (27). Intriguingly, the dissociation of Taf2 from TFIID that occurs easily does not affect the integrity of TFIID but alters its conformation (40). Thus, it is possible that Taf1 may regulate this dissociation step of Taf2 to achieve the appropriate conformation of TFIID on a given promoter. Another intriguing observation is the variable binding of some Tafs: e.g. Taf1 and Taf7 appear to bind to the IPT1/SNF11 (#264) promoter much more strongly than to the neighboring TPS2 (#265) promoter, whereas other Tafs bind to these two promoters at similar levels as described above (Figure 4). It is likely that such variable binding may be due to different conformations of TFIID that are induced by binding to each promoter. A recent electron microscopy study showed that the structure of TFIID is not drastically altered upon binding to activators such as Sp1, c-Jun and p53 (34). However, it is still possible that simultaneous binding of activators and promoter DNA could induce more drastic structural changes in TFIID because DNA functions not only as a docking site but also as a ligand (85,86).
The localization profiles of the TFIID-specific Tafs, such as Taf1 and Taf7 or Taf11 and Taf13, are clustered very closely (Supplementary Figure 4). Notably, Taf7 interacts with Taf1 to regulate its histone acetyltransferase activity (28), whereas the Taf11–Taf13 heterodimer plays a specialized role in TBP recruitment (10). Therefore, the localization profiles obtained here likely reflect physical and/or functional relationships between Tafs and may be useful in predicting unknown features of TFIID.
Lastly, the three RPGs (i.e. RPL35A, RPS11A and RPL10) showed similar relative occupancy profiles for the 14 Tafs, while three randomly selected non-RPGs (HTB1, YCL049C and DLD3) showed different and gene-specific occupancy profiles (Figure 6). Importantly, the effects of the taf1-T657K mutation on the relative occupancy profiles of the 14 Tafs were more similar among the RPGs than among non-RPGs (Figure 6). These results indicate that TFIID has a similar conformation on the RPG promoters but also that it may vary on other promoters. Most of the RPG promoters are TATA-less and are activated by Rap1 (12,82,87). Hence, the relative occupancy profiles for the complete set of Tafs and the effects of taf mutations on these could provide a molecular index to predict variability in the structure of TFIID depending on the promoter context.
In summary, this study has revealed a novel relationship between TFIIB and NC2, the simultaneous co-localization of SAGA and TFIID on RPG promoters, the specific effects of taf1 mutation on Taf2 occupancy, and indirect evidence indicative of different conformations of TFIID. The effects of the taf1-T657K mutation on the recruitment of other Tafs are limited (Figure 6) when compared with those of other taf mutations (10). Thus, future studies examining the effects of different taf alleles on the genome-wide localization of a complete set of Tafs, GTFs and other factors could provide more detailed views of TFIID- and/or SAGA-mediated transcription in eukaryotic cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science; the Ministry of Education, Culture, Sports, Science and Technology of Japan; the CREST program of the Japan Science and Technology Corporation, and the Mitsubishi Foundation. Funding for open access charge: Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr H. Iwasaki and other members of our laboratory for advice and helpful discussions. We also thank Drs Y. Katou and T. Itoh for their help with genome-wide ChIP analysis.
REFERENCES
- 1.Mellor J. The dynamics of chromatin remodeling at promoters. Mol. Cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- 4.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 5.Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 6.Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuras L, Kosa P, Mencia M, Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 8.Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 9.Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang TH, Young RA, Green MR. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mencia M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- 13.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 14.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 15.Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Kruk JA, Reese JC. Dissection of coactivator requirement at Rnr3 reveals unexpected contributions from TFIID and SAGA. J. Biol. Chem. 2008;283:27360–27368. doi: 10.1074/jbc.M803831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanton SJ, Pugh BF. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl Acad. Sci. USA. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huisinga KL, Pugh BF. A TATA binding protein regulatory network that governs transcription complex assembly. Genome Biol. 2007;8:R46. doi: 10.1186/gb-2007-8-4-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner DB, vom Baur E, Thibault C, Sanders SL, Gangloff YG, Davidson I, Weil PA, Tora L. Distinct mutations in yeast TAF(II)25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol. Cell Biol. 2002;22:3178–3193. doi: 10.1128/MCB.22.9.3178-3193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durant M, Pugh BF. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:2791–2802. doi: 10.1128/MCB.26.7.2791-2802.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irvin JD, Pugh BF. Genome-wide transcriptional dependence on TAF1 functional domains. J. Biol. Chem. 2006;281:6404–6412. doi: 10.1074/jbc.M513776200. [DOI] [PubMed] [Google Scholar]
- 24.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 25.Demeny MA, Soutoglou E, Nagy Z, Scheer E, Janoshazi A, Richardot M, Argentini M, Kessler P, Tora L. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS ONE. 2007;2:e316. doi: 10.1371/journal.pone.0000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 27.Cler E, Papai G, Schultz P, Davidson I. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol. Life Sci. 2009;66:2123–2134. doi: 10.1007/s00018-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gegonne A, Weissman JD, Zhou M, Brady JN, Singer DS. TAF7: a possible transcription initiation check-point regulator. Proc. Natl Acad. Sci. USA. 2006;103:602–607. doi: 10.1073/pnas.0510031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann A, Oelgeschlager T, Roeder RG. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc. Natl Acad. Sci. USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oelgeschlager T, Chiang CM, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 32.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman PM, Berk AJ. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 34.Liu WL, Coleman RA, Ma E, Grob P, Yang JL, Zhang Y, Dailey G, Nogales E, Tjian R. Structures of three distinct activator-TFIID complexes. Genes Dev. 2009;23:1510–1521. doi: 10.1101/gad.1790709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, Nogales E. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 2006;14:511–520. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, Timmers HT. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 2008;22:2359–2369. doi: 10.1101/gad.1682308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J. 2007;26:944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papai G, Tripathi MK, Ruhlmann C, Werten S, Crucifix C, Weil PA, Schultz P. Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure. 2009;17:363–373. doi: 10.1016/j.str.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 42.Kokubo T, Swanson MJ, Nishikawa JI, Hinnebusch AG, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA- binding protein. Mol. Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahata S, Kasahara K, Kawaichi M, Kokubo T. Autonomous function of the amino-terminal inhibitory domain of TAF1 in transcriptional regulation. Mol. Cell Biol. 2004;24:3089–3099. doi: 10.1128/MCB.24.8.3089-3099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahata S, Ryu H, Ohtsuki K, Kasahara K, Kawaichi M, Kokubo T. Identification of a novel TATA element-binding protein binding region at the N terminus of the Saccharomyces cerevisiae TAF1 protein. J. Biol. Chem. 2003;278:45888–45902. doi: 10.1074/jbc.M306886200. [DOI] [PubMed] [Google Scholar]
- 45.Tsukihashi Y, Miyake T, Kawaichi M, Kokubo T. Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell Biol. 2000;20:2385–2399. doi: 10.1128/mcb.20.7.2385-2399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longtine MS, McKenzie A., III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Southern JA, Young DF, Heaney F, Baumgartner WK, Randall RE. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 1991;72(Pt 7):1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- 48.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 49.Katou Y, Kaneshiro K, Aburatani H, Shirahige K. Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol. 2006;409:389–410. doi: 10.1016/S0076-6879(05)09023-3. [DOI] [PubMed] [Google Scholar]
- 50.Kasahara K, Ohtsuki K, Ki S, Aoyama K, Takahashi H, Kobayashi T, Shirahige K, Kokubo T. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:6686–6705. doi: 10.1128/MCB.00876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kasahara K, Ki S, Aoyama K, Takahashi H, Kokubo T. Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II. Nucleic Acids Res. 2008;36:1343–1357. doi: 10.1093/nar/gkm1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meisterernst M, Roy AL, Lieu HM, Roeder RG. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 53.Kamada K, Shu F, Chen H, Malik S, Stelzer G, Roeder RG, Meisterernst M, Burley SK. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell. 2001;106:71–81. doi: 10.1016/s0092-8674(01)00417-2. [DOI] [PubMed] [Google Scholar]
- 54.Masson P, Leimgruber E, Creton S, Collart MA. The dual control of TFIIB recruitment by NC2 is gene specific. Nucleic Acids Res. 2008;36:539–549. doi: 10.1093/nar/gkm1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albert TK, Grote K, Boeing S, Stelzer G, Schepers A, Meisterernst M. Global distribution of negative cofactor 2 subunit-alpha on human promoters. Proc. Natl Acad. Sci. USA. 2007;104:10000–10005. doi: 10.1073/pnas.0703490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 57.Durant M, Pugh BF. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell Biol. 2007;27:5327–5335. doi: 10.1128/MCB.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daniel JA, Grant PA. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat. Res. 2007;618:135–148. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reina JH, Hernandez N. On a roll for new TRF targets. Genes Dev. 2007;21:2855–2860. doi: 10.1101/gad.1623207. [DOI] [PubMed] [Google Scholar]
- 60.Torres-Padilla ME, Tora L. TBP homologues in embryo transcription: who does what? EMBO Rep. 2007;8:1016–1018. doi: 10.1038/sj.embor.7401093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hart DO, Green MR. Targeting a TAF to make muscle. Mol. Cell. 2008;32:164–166. doi: 10.1016/j.molcel.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 62.Gegonne A, Weissman JD, Lu H, Zhou M, Dasgupta A, Ribble R, Brady JN, Singer DS. TFIID component TAF7 functionally interacts with both TFIIH and P-TEFb. Proc. Natl Acad. Sci. USA. 2008;105:5367–5372. doi: 10.1073/pnas.0801637105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MM, Yatherajam G, Ranallo RT, Bric A, Paule MR, Stargell LA. Mapping and functional characterization of the TAF11 interaction with TFIIA. Mol. Cell Biol. 2005;25:945–957. doi: 10.1128/MCB.25.3.945-957.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erlich RL, Fry RC, Begley TJ, Daee DL, Lahue RS, Samson LD. Anc1, a protein associated with multiple transcription complexes, is involved in postreplication repair pathway in S. cerevisiae. PLoS ONE. 2008;3:e3717. doi: 10.1371/journal.pone.0003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250- TAF(II)150 complex recognizes the initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaufmann J, Verrijzer CP, Shao J, Smale ST. CIF, an essential cofactor for TFIID-dependent initiator function. Genes Dev. 1996;10:873–886. doi: 10.1101/gad.10.7.873. [DOI] [PubMed] [Google Scholar]
- 67.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neil H, Malabat C, d'A;ubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 69.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Creton S, Svejstrup JQ, Collart MA. The NC2 alpha and beta subunits play different roles in vivo. Genes Dev. 2002;16:3265–3276. doi: 10.1101/gad.234002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 72.Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y, Pursell ZF, Linn S. Identification and cloning of two histone fold motif-containing subunits of HeLa DNA polymerase epsilon. J. Biol. Chem. 2000;275:23247–23252. doi: 10.1074/jbc.M002548200. [DOI] [PubMed] [Google Scholar]
- 74.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim J, Iyer VR. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol. Cell Biol. 2004;24:8104–8112. doi: 10.1128/MCB.24.18.8104-8112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harismendy O, Gendrel CG, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl Acad. Sci. USA. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gilfillan S, Stelzer G, Piaia E, Hofmann MG, Meisterernst M. Efficient binding of NC2. TATA-binding protein to DNA in the absence of TATA. J. Biol. Chem. 2005;280:6222–6230. doi: 10.1074/jbc.M406343200. [DOI] [PubMed] [Google Scholar]
- 79.Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M. NC2 mobilizes TBP on core promoter TATA boxes. Nat. Struct. Mol. Biol. 2007;14:1196–1201. doi: 10.1038/nsmb1328. [DOI] [PubMed] [Google Scholar]
- 80.Hsu JY, Juven-Gershon T, Marr M.T., 2nd, Wright KJ, Tjian R, Kadonaga JT. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–2358. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deato MD, Tjian R. An unexpected role of TAFs and TRFs in skeletal muscle differentiation: switching core promoter complexes. Cold Spring Harb. Symp. Quant. Biol. 2008;73:217–225. doi: 10.1101/sqb.2008.73.028. [DOI] [PubMed] [Google Scholar]
- 82.Li XY, Bhaumik SR, Zhu X, Li L, Shen WC, Dixit BL, Green MR. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 2002;12:1240–1244. doi: 10.1016/s0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- 83.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma VM, Li B, Reese JC. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 2003;17:502–515. doi: 10.1101/gad.1039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 87.Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol. Cell Biol. 2007;27:297–311. doi: 10.1128/MCB.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.