Abstract
DNA stretching in the nucleosome core can cause dramatic structural distortions, which may influence compaction and factor recognition in chromatin. We find that the base pair unstacking arising from stretching-induced extreme minor groove kinking near the nucleosome centre creates a hot spot for intercalation and alkylation by a novel anticancer compound. This may have far reaching implications for how chromatin structure can influence binding of intercalator species and indicates potential for the development of site selective DNA-binding agents that target unique conformational features of the nucleosome.
INTRODUCTION
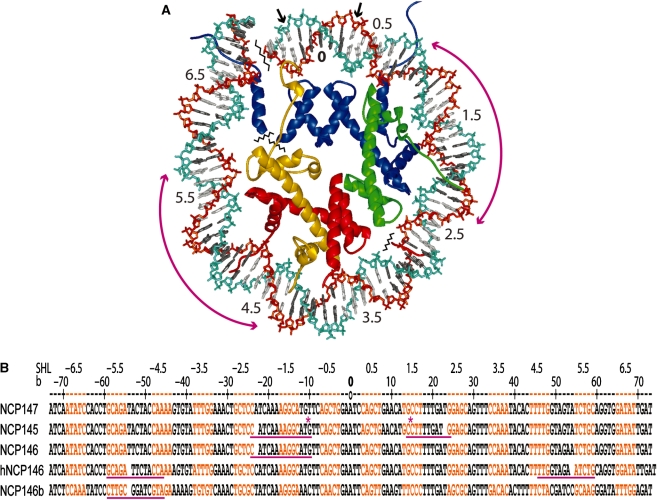
DNA structure in the nucleosome displays sequence- and context-dependent features, which could be potentially exploited for gene-specific drug targeting (1–3). DNA stretching in the nucleosome entails increased twist and unstacking of bases, resulting in a shift in histone–DNA register by displacement of a single base pair (Figure 1; 2). From crystal structure analysis and solution studies of different nucleosome core particle (NCP) constructs, there are four known sites of potential stretching at around 2 and 5 double helical turns from the nucleosome centre (dyad; 1,2,4–7). Comparison of average double helix twist values from crystal structures with those observed for NCP isolated from cellular chromatin (8) suggests that stretching has an average incidence of once or twice per nucleosome in vivo (1,2). Although its functional consequences in the genome are not fully understood, stretching can be associated with extreme DNA kinking at a site 1.5 turns from the nucleosome centre (2), which is a principal location for gene insertion by HIV-integrase that prefers highly distorted substrates (9,10). Moreover, by providing variability in DNA twist and length relationships, stretching may serve as a buffer against linker DNA constraints between nucleosomes to facilitate chromatin compaction (1,11).
Figure 1.
DNA wrapping and stretching in the nucleosome core (2). (A) View of the NCP147 crystal structure (1,5) down the DNA superhelix axis showing the major groove-inward (grey DNA bases) and minor groove-inward (white DNA bases) facing regions for approximately one-half of the particle. Numbers correspond to double-helical turns from the nucleosome centre (0), which coincides with the central base pair at the particle pseudo dyad axis where the major groove directly faces the histone octamer. Long magenta arrows indicate potential regions of DNA stretching around the 2-turn and 5-turn locations. Small arrows correspond to base pairs ±3 that are absent in NCP145 (2). The phosphodiester backbone of the DNA strands appears as cyan and orange. Histone proteins are colored blue for H3, green for H4, gold for H2A, and red for H2B. (B) Comparison of DNA sequence and histone–DNA register in NCP crystal structures described in the main text. NCP145, NCP146, NCP147 and NCP146b are particles with Xenopus laevis histones (1,2,4,5), whereas hNCP146 corresponds to a particle with the same DNA sequence as NCP146, but with histones from Homo sapiens (7) (altered crystal packing interactions between particles in the crystal apparently underlie the differences in stretching between NCP146 and hNCP146; 2). Minor groove-inward facing regions are highlighted in orange, and the base numbering scheme (b), relative to NCP147, corresponds to the 5′ (−) to 3′ (+) direction of either DNA strand in the duplex (only one strand is shown for each construct; SHL = superhelix location, turns from center). Observed regions of stretching are underlined in magenta, and a gap in the DNA sequence represents the resulting shift in histone-base pair register. Asterisks denote the extreme kinks into the major groove at the 1-turn location and into the minor groove at the 1.5-turn position associated with stretching in NCP145.
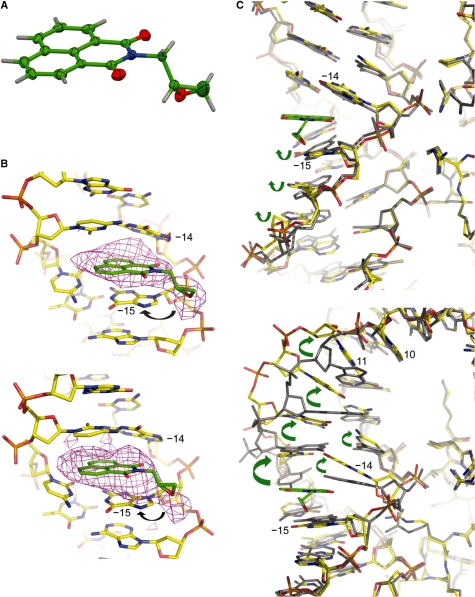
Figure 2.
X-ray crystallographic analysis of ENA and NCP crystals treated with ENA. (A) Crystal structure of ENA with thermal ellipsoids shown at 50% probability for non-hydrogen atoms (carbon, green; oxygen, red; nitrogen, blue; hydrogen, grey). (B and C) Crystal structure of an ENA-NCP145 complex (yellow carbon atoms, NCP; green carbon atoms, ENA) with an omit difference electron density map (B) or the native NCP145 crystal structure (grey atoms; C) superimposed. The image pairs correspond to pseudo-symmetry-related sites in the two particle halves, which display either a minor groove kink at the 1.5-turn GG dinucleotide (base −15 and −14; upper panels) or a major groove kink at the 1-turn CA dinucleotide (base 10 and 11; lower panels) in the native NCP145 structure (Figure 1). The Fo–Fc electron density map, contoured at 2.5σ, was calculated from an initial model prior to inclusion of ENA (B). Arrows designate the apparent primary site of attack by the ENA epoxy group (B) and conformational rearrangements accompanying binding of ENA (C).
Since the binding of even simple metal hydrates can be affected by single base pair changes of DNA orientation in the nucleosome core (12), factors which are able to stabilize or alter stretching could influence molecular recognition potential by modulating structure and dynamics. Such has been observed previously in the binding of polyamide minor groove ligands, which are capable of changing stretching location and thus histone–DNA register (13,14). Considering that stretching can also be associated with extreme DNA kinking in the nucleosome core (2), we investigated how it may influence reaction with a novel DNA-binding therapeutic candidate. N-(2,3-epoxypropyl)-1,8-naphthalimide (ENA) was recently discovered as a strong anti-proliferative agent, which displays potent anti-tumour and anti-fungal activity by inducing S phase arrest in the cell cycle (15).
MATERIALS AND METHODS
Material preparation
NCP was prepared from recombinant Xenopus laevis histones and 145, 146 or 147 base pair DNA fragments derived from human α-satellite DNA (2,4,5), using established methodologies (16). Crystals were grown as described previously (5). To screen for conditions allowing structural characterization of an ENA–NCP complex, NCP145 crystals were incubated for various durations in a stabilization buffer with a range of ENA concentrations. Results reported correspond to crystals soaked in buffer containing 37 mM MnCl2, 40 mM KCl, 20 mM K-Cacodylate (pH 6.0), 24% 2-methyl-2,4-pentanediol, 2% trehalose, 133 µM or 1 mM ENA and 2.7% or 10% dimethyl sulfoxide, respectively. ENA was allowed to react for 3–4 days prior to X-ray data collection.
Structural analysis
X-ray diffraction data for ENA-treated NCP (Table 1) were recorded as described previously (2) at the X06SA beam line of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) using the PILATUS detector (NCP + 1 mM, λ=1.07 Å) and on a Rigaku MicroMax-007HF diffractometer with a Rigaku Raxis IV++ image plate (NCP + 0.1 mM, λ=1.54 Å; Nanyang Technological University, Singapore). Data were processed using MOSFLM (17) and SCALA from the CCP4 suite (18). The initial model for solution of the ENA-NCP145 complex by molecular replacement consisted of the DNA, histones, and one Mn2+ ion at the major interparticle interface from the structure of native NCP145 (pdb code 2NZD; 2). Structural refinement and model building were carried out with routines from the CCP4 suite (18). Graphic figures were prepared with PyMOL (DeLano Scientific LLC, San Carlos, CA, USA). Atomic coordinates and diffraction data have been deposited in the RCSB Protein Data Bank under accession code 3KUY.
Table 1.
Data collection and refinement statistics for ENA-treated NCP
| NCP + 0.1 mM ENA | NCP + 1 mM ENA | |
|---|---|---|
| Data collectiona | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 106.2, 109.6, 182.4 | 104.1,107.3,176.4 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 2.90–54.8 (2.90–3.06) | 2.85–51.4 (2.85–3.00) |
| Rmerge | 6.8% (48.3%) | 10.5% (47.4%) |
| I/σI | 22.6 (2.9) | 16.7 (2.3) |
| Completeness (%) | 99.9 (99.2) | 98.1 (97.3) |
| Redundancy | 7.4 (5.3) | 5.6 (5.4) |
| Refinement | ||
| Resolution (Å) | 2.90–54.8 | 2.85–51.4 |
| No. reflections | 46 854 | 44 886 |
| Rwork/Rfree | 23.8%/28.6% | 41.6%/49.5% |
| No. atoms | 12 056 | 12 018 |
| Protein | 6078 | 6078 |
| DNA | 5939 | 5939 |
| ENA | 38 | 0 |
| B-factors (Å2) | 76.1 | 66.4 |
| Protein | 49.4 | 44.8 |
| DNA | 103.4 | 88.6 |
| ENA | 135.6 | ‐ |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.013 |
| Bond angles (°) | 1.39 | 1.88 |
aData sets are based on single crystal diffraction, and values in parentheses are for the highest-resolution shell.
Crystals of ENA were obtained by slow evaporation from a saturated acetone solution. Single crystal (0.18 × 0.18 × 0.13 mm) diffraction data were recorded at −163°C with a Rigaku Saturn CCD area detector diffractometer using graphite monochromated Mo-Kα radiation (λ = 0.71 Å). Data were collected and processed using CrystalClear (Rigaku/MSC), and the structure was solved by direct methods (SHELX97, 19) and expanded using the Fourier technique (DIRDIF-99, 20). All calculations were carried out using the CrystalStructure crystallographic software package (Rigaku/MSC), and the crystal structure image was generated with Mercury (Cambridge Crystallographic Data Centre). Crystal data for ENA: space group P21/c (#14), a = 7.750(4) Å, b = 12.748(6) Å, c = 11.584(8) Å, β = 95.65(3)°, V = 1138.9(12) Å3, Z = 4, Dc = 1.477 g/cm3, 6353 reflections collected, 2980 independent reflections, R1 = 0.0714, wR2(I > 2σ(I)) = 0.0655. Atomic coordinates have been deposited in the Cambridge Crystallographic Data Centre under deposition code CCDC 756223.
DNA footprinting
ENA, from a stock solution of 4 mM in 100% dimethyl sulfoxide, was added to 2.5 µM DNA or NCP in TE buffer [10 mM Tris (pH 7.4) and 0.1 mM EDTA] at molar stoichiometry given in Figure 3. Samples were allowed to incubate overnight at room temperature prior to the addition of 4 M NaCl and phenol–chloroform extraction to remove unreacted ENA and histone proteins. DNA was subsequently ethanol precipitated, resuspended in TE buffer and 5′ end-labeled (32P) with polynucleotide kinase. Thermal depurination and strand cleavage reactions were effected by 30-min incubation at 99°C, followed by an additional 30-min incubation with 10% piperidine (21). Maxam–Gilbert purine sequencing standards (22) were prepared as markers. DNA fragments were resolved by denaturing PAGE (10% polyacrylamide, 8 M urea, 88 mM Tris–borate, 2 mM EDTA, pH 8.3), followed by phosphorimage (BioRad) analysis of the dried gel.
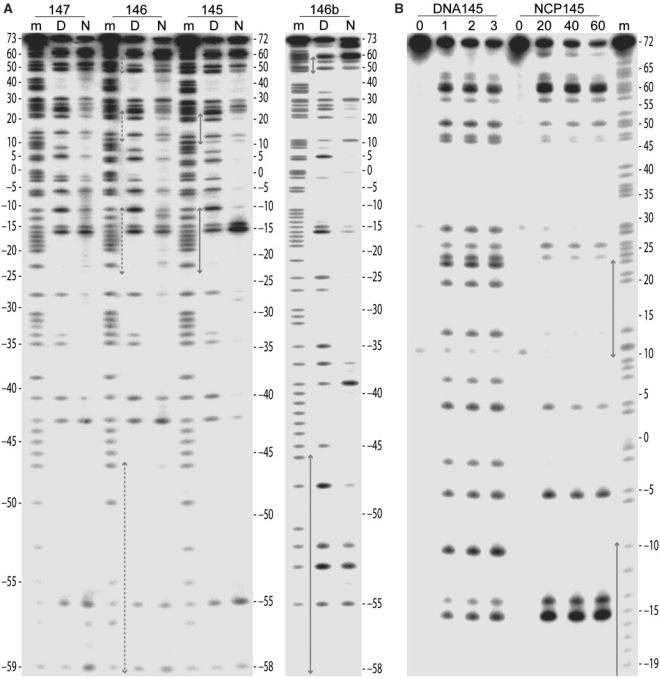
Figure 3.
Footprinting analysis of ENA-DNA alkylation sites by thermally induced strand breakage. (A and B) Thermal cleavage reactions were conducted with DNA samples isolated from naked DNA and NCP, which had been incubated with ENA. The ENA:DNA molar stoichiometry was 2:1 and 20:1 for the naked (D) and nucleosomal (N) samples, respectively (A), and it corresponds to the number shown above the lanes for the naked (DNA145) and nucleosomal (NCP145) samples (B). Fragments from four different DNA constructs (145, 146, 147 and 146b) together with the respective Maxam–Gilbert purine sequencing standards (m) were separated by denaturing PAGE, revealing alkylation site preference. Numbers flanking the gels represent base position with respect to that at the nucleosome centre (0). Grey arrows designate regions of stretching observed in the nucleosome core (Figure 1).
RESULTS
ENA site selectivity
Crystal structure determination of ENA reveals that the chiral epoxy carbon is in the R configuration (Figure 2A). The planar naphthalimide group intercalates between DNA base pairs, placing the epoxy group in a position to potentially undergo electrophilic attack at base substituents on the major groove side (Figure 2B and C, Table 1 and Supplementary Figure S1). For investigating the reactivity and site selectivity of ENA, we utilized a footprinting method, which functions for DNA-alkylating agents that yield base modifications rendering the phosphodiester backbone susceptible to thermally induced cleavage. We find that ENA is indeed an alkylating agent, reacting exclusively with guanine bases (Figure 3). For naked DNA, reaction is apparent at all guanine sites, with a modest degree of preference for certain sequence motifs.
In order to elucidate potential influences on ENA reactivity from histone association, we conducted footprinting analysis with four different NCP constructs, which have displayed differing incidences of DNA stretching (Figures 1, 3 and 4; 2,4–7). Nucleosome packaging reduces the overall DNA reactivity towards ENA by roughly 10-fold relative to the naked state, consistent with histone-imposed restraints on DNA distortability, which would tend to oppose intercalation (3,23,24). In addition, the general reactivity of guanine sites is maximal at the nucleosome termini, decreasing inward towards the centre. This is in accordance with the profiles of differential inaccessibility and histone–DNA interaction strength, which become greatest at the nucleosome centre (5,25–28). In stark contrast to these general trends, the most preferred site of alkylation in a 145 base pair core particle (NCP145) is located 1.5 turns from the centre. Reaction at this minor groove inward-facing AAGGC element is strongly selective for the 5′ guanine (base −15; Figure 3).
Figure 4.
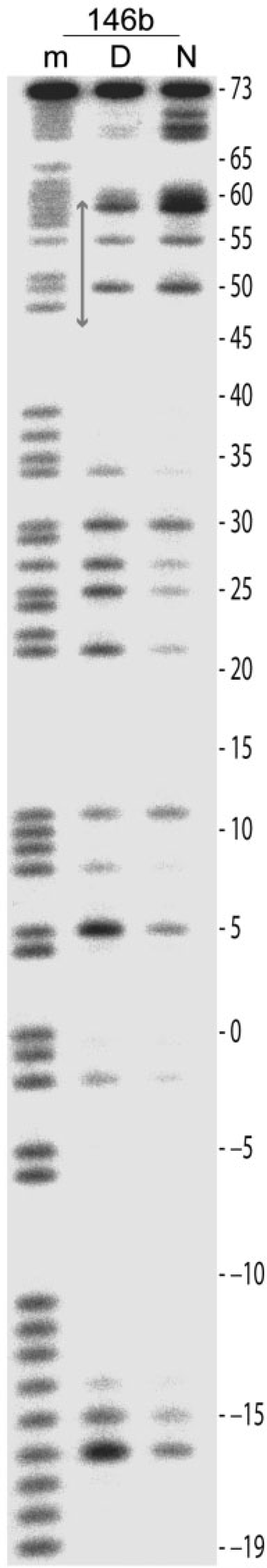
Footprinting analysis of ENA-DNA alkylation sites in the 146b construct by thermally induced strand cleavage. The gel has been run out to resolve detail at the DNA 3′-end. The labeling scheme and experimental conditions are identical to those for Figure 3A.
Modulation of ENA association by DNA stretching
The crystal structure of NCP145 displays two incidences of stretching around the 2-turn locations in both pseudo symmetry-related halves of the particle (Figures 1 and 2C; 2). In one half, stretching is accompanied by a massive, −55° kink into the minor groove at the 1.5-turn GG = CC dinucleotide. This pronounced base pair unstacking underlies the translational component of the stretch, which in the opposing half of NCP145 instead coincides with a 35° kink into the major groove at the 1-turn CA = TG dinucleotide. Although heavy derivatization of NCP145 crystals with ENA gives rise to uninterpretable X-ray diffraction electron density apparently from DNA disordering (Table 1), data collected from milder treatment reveals intercalation of the naphthalimide group exclusively within the 1.5-turn GG dinucleotide sites in either half of the particle (Figure 2B and C). The crystal seems to contain a mixture of pre- and post-reaction states, with the epoxy group situated on the major groove side of the 5′ guanine within 3–4 Å of the N7 ring nitrogen atom—the apparent site of alkylation. This is consistent with the footprinting results showing absolute selectivity for guanine and preferential alkylation at the 5′ guanine of the 1.5-turn GG element (Figure 3). Moreover, in the one half with the preexisting kink at GG, intercalation results in relatively minor structural rearrangements stemming largely from realignment of the 5′ guanine to stack in a coplanar fashion with the naphthalimide group (Figure 2C, upper panel). In the other particle half, there is a substantial rearrangement from intercalation, wherein the 1-turn CA kink is replaced by kinking/unstacking at the 1.5-turn GG to yield a very similar configuration in both halves of the NCP (Figure 2B and C, lower panel). This emphasizes the conformational selectivity of ENA intercalation, which in this instance likely relates to the greater steric access provided by base pair unstacking at the major groove side (1.5-turn GG kink) compared to that at the minor groove edge (1-turn CA kink).
Although the two 1.5-turn sites settle into a very similar conformation subsequent to ENA binding, differences in the electron density profiles suggest that the site with the initial 1-turn CA kink, which displays more density near the −15 guanine N7 atom, may contain a greater fraction of alkylation product (Figure 2B). This could come about if there were a strong requirement for conformational flexibility in the alkylation event, in which case the site with the preexisting GG kink may readily favor intercalation, but otherwise lack the structural freedom seemingly inherent in the opposing half. Alternatively, the two halves may share a similar fraction of alkylation product, but differ in the portion of bound unreacted ENA. Thus, whereas the pre-kinked GG site would be predisposed for the intercalator-bound state, the other location may require covalent attachment of ENA in order to provide sufficient stability for sustained intercalation.
Considering both the footprinting and crystallographic analysis, it is apparent that the base unstacking at the major groove side promoted by DNA stretching facilitates intercalation and reaction of ENA. Although the effect is most prominent in NCP145, some degree of relative enhancement at the 1.5-turn GG site can also be seen in NCP146 and NCP147, which are composed of 146 and 147 base pair sequences nearly identical to that of NCP145 yet display either a mixture of stretched-versus-unstretched states or no incidence of stretching, respectively (Figures 1 and 3A; 2,4–7). Thus, even in the absence of sustained stretching, DNA at the 1.5-turn location appears to have special reactivity attributes as we have previously also observed distinctions towards platinum drug binding at this site in NCP147 (29). Conversely, the 5-turn region is proximal to the nucleosome termini and appears to impose fewer conformational constraints on the DNA (5). It is thus likely that stretching at this location has distinct structural–dynamic character relative to the 2-turn position. Nonetheless, in a 146 base pair particle (NCP146b) composed of a distinct sequence and displaying stretching only at the 5-turn location in one half (5), there is some degree of relative enhancement of ENA reactivity apparent at a guanine site nearby the region of stretching observed in the crystal (base −39; Figures 1 and 3A). In any case, since ENA alkylates only guanine, and we find conformation-dependent reactivity in NCP145, the degree of stretching-induced enhancement likely depends on the precise location of potential guanine sites.
DISCUSSION
DNA packaging into nucleosomes has previously been shown to allow unrestricted monoalkylation by certain minor groove-associating compounds (21,30). In contrast, multifunctional adduct formation or non-covalent binding, such as intercalation, which necessitate pronounced conformational change in the DNA are typically suppressed by the nucleosomal state (3,23,24,30,31). As an apparent prerequisite for guanine alkylation, intercalation of ENA is also more favoured overall for naked DNA, but the hot spot for intercalation and reaction created by DNA stretching would promote adduct accumulation near the nucleosome centre. Moreover, ENA’s cytotoxic activity appears to arise through inhibition of DNA synthesis by halting fork progression at or near replication origins (15). Since the increased twist downstream accompanying unwinding of the double helix would be expected to favour stretching, it raises the possibility that this overtwisting ahead of replication forks further promotes ENA attack at nucleosomal sites. This is significant since the cellular response to adducts located on histone-free versus nucleosomal regions in the genome can be distinct. For instance, the repair of different types of DNA lesions has been found to be suppressed by histone octamer association (32,33), and the inhibitory effect may be maximal at the most inaccessible sites nearest the nucleosome centre (3,33).
The acquisition of superior medicinal agents will ultimately depend on improving site discrimination attributes. Considering their context-dependent DNA conformation and histone proximity characteristics, nucleosomes appear to have a targeting potential greater than that of naked DNA (3). One example is small molecule recognition of adjacent DNA duplex sections within a superhelix, the nucleosomal ‘supergroove’ (34). The reactivity behaviour observed for ENA suggests possibilities for exploiting DNA stretching and other unique—perhaps yet undiscovered—features of the nucleosome for drug development.
ACCESSION NUMBERS
RCSB Protein Data Bank accession code 3KUY, Cambridge Crystallographic Data Centre deposition code CCDC 756223.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by Academic Research Council grant 19/08 from the Ministry of Education, Singapore. Funding for open access charge: Ministry of Education (Academic Research Council grant 19/08).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Synchrotron crystallographic experiments were performed on the X06SA and X06DA beamlines at the Swiss Light Source, Paul Scherrer Institute (Villigen, Switzerland). We thank C. Schulze-Briese, M. Wang, V. Olieric, T. Tomizaki, R. Bingel-Erlenmeyer, M. Fuchs and A. Pauluhn at the Swiss Light Source who ensured success with data collection. We are grateful to P. Dröge for input on the project and to W.K. Ng and C.H. Wang for assistance with crystal structure determination of ENA.
REFERENCES
- 1.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 2.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J. Mol. Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 3.Davey GE, Davey CA. Chromatin – a new, old drug target? Chem. Biol. Drug Des. 2008;72:165–170. doi: 10.1111/j.1747-0285.2008.00697.x. [DOI] [PubMed] [Google Scholar]
- 4.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Davey CA, Sargent DF, Luger K, Mäder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 6.Edayathumangalam RS, Weyermann P, Dervan PB, Gottesfeld JM, Luger K. Nucleosomes in solution exist as a mixture of twist-defect states. J. Mol. Biol. 2005;345:103–114. doi: 10.1016/j.jmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Travers AA, Klug A. The bending of DNA in nucleosomes and its wider implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1987;317:537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- 9.Pryciak PM, Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 10.Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl Acad. Sci. USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 12.Davey CA, Richmond TJ. DNA-dependent divalent cation binding in the nucleosome core particle. Proc. Natl Acad. Sci. USA. 2002;99:11169–11174. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottesfeld JM, Melander C, Suto RK, Raviol H, Luger K, Dervan PB. Sequence-specific recognition of DNA in the nucleosome by pyrrole-imidazole polyamides. J. Mol. Biol. 2001;309:615–629. doi: 10.1006/jmbi.2001.4694. [DOI] [PubMed] [Google Scholar]
- 14.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 15.Krishnan V, Dirick L, Lim HH, Lim TS, Si-Hoe SL, Cheng CS, Yap KL, Ting A, Schwob E, Surana U. A novel cell cycle inhibitor stalls replication forks and activates S phase checkpoint. Cell Cycle. 2007;6:1621–1630. doi: 10.4161/cc.6.13.4373. [DOI] [PubMed] [Google Scholar]
- 16.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 17.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 18.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 20.Beurskens PT, Beurskens G, de Gelder R, Garcia-Granda S, Gould RO, Israel R, Smits JMM. The DIRDIF-99 program system. The Netherlands: Crystallography Laboratory, University of Nijmegen; 1999. [Google Scholar]
- 21.Trzupek JD, Gottesfeld JM, Boger DL. Alkylation of duplex DNA in nucleosome core particles by duocarmycin SA and yatakemycin. Nat. Chem. Biol. 2006;2:79–82. doi: 10.1038/nchembio761. [DOI] [PubMed] [Google Scholar]
- 22.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 23.McMurray CT, van Holde KE. Binding of ethidium to the nucleosome core particle. 1. Binding and dissociation reactions. Biochemistry. 1991;30:5631–5643. doi: 10.1021/bi00237a001. [DOI] [PubMed] [Google Scholar]
- 24.Moyer R, Marien K, van Holde K, Bailey G. Site-specific aflatoxin B1 adduction of sequence-positioned nucleosome core particles. J. Biol. Chem. 1989;264:12226–12231. [PubMed] [Google Scholar]
- 25.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 26.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 27.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 28.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu B, Dröge P, Davey CA. Site selectivity of platinum anticancer therapeutics. Nat. Chem. Biol. 2008;4: 110–112. doi: 10.1038/nchembio.2007.58. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian V, Ducept P, Williams RM, Luger K. Effects of photochemically activated alkylating agents of the FR900482 family on chromatin. Chem. Biol. 2007;14:553–563. doi: 10.1016/j.chembiol.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millard JT, Spencer RJ, Hopkins PB. Effect of nucleosome structure on DNA interstrand cross-linking reactions. Biochemistry. 1998;37:5211–5219. doi: 10.1021/bi972862r. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Hara R, Singh G, Sancar A, Lippard SJ. Nucleotide excision repair from site-specifically platinum-modified nucleosomes. Biochemistry. 2003;42:6747–6753. doi: 10.1021/bi034264k. [DOI] [PubMed] [Google Scholar]
- 33.Thoma F. Repair of UV lesions in nucleosomes–intrinsic properties and remodeling. DNA Repair. 2005;4:855–869. doi: 10.1016/j.dnarep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Edayathumangalam RS, Weyermann P, Gottesfeld JM, Dervan PB, Luger K. Molecular recognition of the nucleosomal “supergroove”. Proc. Natl Acad. Sci. USA. 2004;101:6864–6869. doi: 10.1073/pnas.0401743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.