Abstract
The Mre11/Rad50/Xrs2 (MRX) complex in Saccharomyces cerevisiae has well-characterized functions in DNA double-strand break processing, checkpoint activation, telomere length maintenance and meiosis. In this study, we demonstrate an involvement of the complex in the base excision repair (BER) pathway. We studied the repair of methyl-methanesulfonate-induced heat-labile sites in chromosomal DNA in vivo and the in vitro BER capacity for the repair of uracil- and 8-oxoG-containing oligonucleotides in MRX-deficient cells. Both approaches show a clear BER deficiency for the xrs2 mutant as compared to wildtype cells. The in vitro analyses revealed that both subpathways, long-patch and short-patch BER, are affected and that all components of the MRX complex are similarly important for the new function in BER. The investigation of the epistatic relationship of XRS2 to other BER genes suggests a role of the MRX complex downstream of the AP-lyases Ntg1 and Ntg2. Analysis of individual steps in BER showed that base recognition and strand incision are not affected by the MRX complex. Reduced gap-filling activity and the missing effect of aphidicoline treatment, an inhibitor for polymerases, on the BER efficiency indicate an involvement of the MRX complex in providing efficient polymerase activity.
INTRODUCTION
In all organisms, the genome is continuously damaged by endogenous and exogenous factors, such as reactive oxygen species (ROS) or alkylating and oxidizing agents. Frequently, purine and pyrimidine moieties are damaged yielding base lesions that can lead to mutations (1,2). 8-oxoguanine (8-oxo-G) is the most frequent base damage induced by hydrogen peroxide (H2O2) (3,4), whereas alkylating agents such as methyl methanesulfonate (MMS) modify bases by adding methyl groups to nucleophilic sites. The predominant forms of MMS-induced DNA damage are the N-methylation adducts 7-methylguanine and 3-methyladenine (5). Spontaneous depurination of methylated purines leads to the formation of abasic (AP) sites, which are heat-labile, due to breakage of the phosphodiester bond at clustered damage. Presence of unrepaired AP sites result in cytotoxicity and mutagenicity, as well as blocks in DNA replication and transcription (6).
Repair of damaged bases and AP sites is normally carried out by the base excision repair (BER) system. In patients with defects in BER the failure to repair base damage can lead to malignancies and is associated with age-related degenerative diseases (7). BER is initiated by specific DNA N-glycosylases that recognize and excise the damaged bases to produce AP sites (8). In the major BER subpathway in Saccharomyces cerevisiae AP sites are then incised by the AP-endonucleases Apn1 and Apn2 creating a 5′-deoxyribophosphate (5′-dRP) end at the site of damage (9,10). The removal of the blocked 5′-end is catalyzed by Rad27. This protein is a specific 5′-flap endonuclease extending the AP sites to gaps of up to 5 nt (11,12). New DNA synthesis and subsequent ligation by Cdc9 complete BER (13). In an additional BER subpathway the damaged bases are processed by a DNA N-glycosylase/AP-lyase. In S. cerevisiae three enzymes with combined N-glycosylase-/AP-lyase activities are known that catalyze the excision of oxidized bases, namely Ogg1, Ntg1 and Ntg2 (14). 7-methylformamide pyrimidine, produced by MMS treatment, is excised efficiently by Ntg1 and Ntg2 (15). These enzymes produce 3′-α,β-unsaturated aldehydic (3′-dRP) ends, which pose a problem for DNA integrity because they cannot be extended by polymerases. Genetic studies suggest that the AP-endonucleases Apn1 and Apn2 contribute to the removal of 3′-dRP by their 3′-phosphodiesterase activity (13). Afterwards, the gap is filled by polymerase activity and the original state is reconstituted by the ligase Cdc9 (13). In the case of the repair by AP-lyases, the repair patch length is not entirely clear (13). However, since elimination of the blocked 3′-end creates only a 1-nt gap, it is designated as the short-patch repair pathway (13,16). The nucleotide excision repair pathway components Rad1 and Rad10 provide a backup-pathway for 3′-dRP removal (13). Following the action of an AP-endonuclease, more than 1 nt is incorporated; therefore, this subpathway is called the long-patch BER (13). The polymerases that are involved in the BER process in yeast are not known. Wang et al. (17) could show that DNA synthesis during repair is carried out mainly by polymerase ε, but both, polymerase α and δ show modulating influences. In another report, polymerase δ was found to be the main enzyme for DNA synthesis after base damage by methylating agents (18).
In this article, we address the question of whether Xrs2 also has a direct role in the complex system of BER. Xrs2 is the yeast homolog to human Nbs1. Together with Mre11 and Rad50, it forms a trimeric complex (MRX) that is important for damage recognition and processing after DNA double-strand break (DSB) induction (19–22). Moreover, the complex plays a role in non-homologous end-joining (NHEJ) and homologous recombination (23,24). Besides its function in DSB repair, the MRX complex also affects many other cellular processes, including cell cycle checkpoint activation, telomere length maintenance and meiosis (25). Strains with mutations in RAD52 group genes, including XRS2, are shown to be sensitive to MMS and H2O2 treatment (26–29). Furthermore, it was shown that rad52 and xrs2 deletion strains exhibit an increased mutation frequency compared to wildtype cells (15,30). Nevertheless, these genes were never mentioned to be directly linked to the BER pathway. It was assumed that unrepaired base damage or BER intermediates can lead to stalled replication forks which can be converted into DSBs. In addition, clustered DNA base damage and AP sites can induce DSB (28,31,32). Therefore, it was supposed that recombination-deficient cells are sensitive to base-damaging agents due to their role in homologous recombination as tolerance pathway for unrepaired base damage (13,33,34).
In this study, we demonstrated a direct role of the yeast MRX complex in the BER process, which contributes to resistance against base-damaging agents and to the avoidance of mutations. We showed that the repair capacity of MMS-induced heat-labile sites in stationary haploid cells is reduced in the xrs2 mutant compared to wildtype, suggesting a BER defect. Consistently, decreased capacities in long-patch and short-patch BER were observed in cell extracts obtained from MRX deletion mutants using an in vitro assay. Subsequent analyses suggest the assignment of XRS2 into the NTG1/NTG2-mediated BER subpathway as well as a function in facilitating polymerase activity required in BER. Thus, our results show for the first time a direct role for the MRX complex in BER.
MATERIALS AND METHODS
Yeast strains and media
The haploid S. cerevisiae strains used in this study are listed in Table 1. Strains are isogenic derivates of MKP-0, originally obtained from B.A. Kunz (Geelong, Australia). Deletion strains were constructed by gene replacement of the open reading frame and in vivo recombination (35) using polymerase chain reaction (PCR) products of the cassettes KANMX6 and HIS3MX6 (36) or the selectable markers LEU2, TRP1 and URA3. All mutations were confirmed by PCR analysis. Construction and validation of mutants with truncated XRS2 alleles are described previously (24). Strains used to measure mutation frequencies are derivates of BY4741 (37).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| MKP-0 | MATα, can1-100, ade2-1, lys2-1, ura3-52, leu2-3-112, his3-Δ200, trp1-Δ901, RAD | BA. Kunz |
| MKP-0 Δxrs2 | Derivative of MKP-0 with XRS2::KANMX6 | (24) |
| MKP-0 xrs2-228M | Derivative of MKP-0 Δxrs2 with xrs2-228M | (24) |
| MKP-0 xrs2-630 | Derivative of MKP-0 Δxrs2 with xrs2-630 | (24) |
| MKP-0 Δapn1 | Derivative of MKP-0 with APN1::Leu2 | This study |
| MKP-0 Δapn1Δxrs2 | Derivative of MKP-0 with APN1::Leu2, XRS2::KANMX6 | This study |
| MKP-0 Δntg1Δntg2 | Derivative of MKP-0 with NTG1::URA3, NTG2::TRP1 | This study |
| MKP-0 Δntg1Δntg2Δxrs2 | Derivative of MKP-0 with NTG1::URA3, NTG2::TRP1, XRS2::HIS3MX6 | This study |
| MKP-0 Δapn1Δntg1Δntg2 | Derivative of MKP-0 with APN1::Leu2, NTG1::URA3, NTG2::TRP1 | This study |
| MKP-0 Δapn1Δntg1Δntg2Δxrs2 | Derivative of MKP-0 with APN1::Leu2, NTG1::URA3, NTG2::TRP1, XRS2::HIS3MX | This study |
| MKP-0 Δmre11 | Derivative of MKP-0 with MRE11::HIS3MX6 | (24) |
| MKP-0 Δrad50 | Derivative of MKP-0 with RAD50::LEU2 | (24) |
| BY4741 | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Euroscarf |
| BY4741 Δxrs2 | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 with xrs2::KANMX6 | Euroscarf |
Standard medium yeast extract peptone dextrose (YEPD), containing 2% dextrose, 1% yeast extract and 2% peptone, was used as complete growth medium for yeast. Synthetic complete (SC) medium (2% dextrose; 0.6% yeast nitrogen base and the appropriate nutrients added) was used for yeast transformations performed by the lithium acetate method (38). SC medium lacking one nutrient allowed selection for the corresponding wildtype gene. For solid media, 2% agar was added. Canavanine plates were prepared by dissolving 60 mg canavanine (Sigma) in 1 l SC medium lacking arginine. For MMS plates, the agent was dissolved in concentrations ranging from 0.0025 to 0.02% in YEPD agar prior to pouring. All chemicals for media were purchased from Difco.
Analysis of mutation frequency
To measure mutation frequencies we used the strain BY4741 that carries a wildtype CAN1 gene. Forward mutations in this gene lead to resistance to the arginine analog canavanine. For chemical-induced mutagenesis, overnight cultures were used to inoculate fresh YEPD medium with 2.5 × 106 cells/ml and incubated until they reached a cell titer of about 1 × 107 cells/ml. For MMS treatment, cells were incubated for 0, 10, 20 and 30 min with 0.1% MMS (Sigma) at 30°C. Afterwards, the cells were washed twice with and resuspended in potassium phosphate buffer (25 mM KPO4, pH 7.0). For H2O2 treatment, the cells were resuspended in M9 buffer (39) containing 0, 1, 3 and 5 mM H2O2 (Merck). Following incubation at 30°C for 20 min, the reaction was stopped by adding 500 U/ml catalase (Sigma). To score for cell survival, the cells were plated onto YEPD and, to screen for CanR (canavanine resistant) mutants, spread onto selective canavanine plates. The mutation frequencies are displayed as CanR mutants per 107 surviving cells.
Exposure to DNA-damaging agents
The sensitivity of yeast strains to MMS and H2O2 was measured in the exponential growth phase. Overnight cultures were diluted in fresh YEPD medium to a density of 3 × 106 cells/ml and allowed to grow at 30°C with agitation to about 1 × 107 cells/ml. Cells were then treated with 0.2% MMS for 0, 15, 30 and 45 min at 30°C, pelleted and washed once with YEPD. For the survival after H2O2 treatment, aliquots of 1 ml were incubated for 20 min with H2O2 concentrations ranging from 0 to 60 mM as described by Melo et al. (39). The addition of 500 U/ml catalase stopped the reaction. Appropriate dilutions were spread on YPED agar. After plating, cells were incubated for 3–5 days at 30°C. Survival was calculated by the ratio of the number of colonies after treatment versus the number of colonies without treatment.
For a crude analysis of the MMS sensitivity, cells in exponential growth phase were diluted serially in potassium phosphate buffer (25 mM KPO4, pH 7.0), and aliquots of 5 µl were spotted on YEPD agar supplemented with 0.0025–0.02% MMS.
Repair of heat-labile sites after MMS treatment
The efficiency to repair heat-labile sites after MMS treatment in vivo was measured in haploid yeast cells under non-growth conditions. To obtain highly stationary yeast cells with a maximum of 3% budding cells, a modified cultivation protocol of Pohlit and Heyder (40) was used. Cells were collected, diluted in potassium phosphate buffer (25 mM KPO4, pH 7.0) to a density of 7.5 × 108 cells/ml and treated for indicated time points with 0.1% MMS, followed by washing twice with potassium phosphate buffer. Afterwards, the samples were diluted in liquid holding recovery (LHR) buffer (100 mM glucose, 67 mM KPO4, pH 5.0) (41–43) to a final density of 7.5 × 106 cells/ml and incubated at 30°C. The cells were embedded into agarose plugs before and after LHR incubation and genomic DNA was prepared according to Friedl et al. (43). During incubation at 55°C for proteinase K digestion MMS-induced clustered heat-labile sites are converted into DSB (31). Incubation at 32°C for the digestion step was used to check the basal level of chromosomal degradation. Chromosomal DNA was separated by pulsed-field gel electrophoresis (PFGE). The evaluation of residual damage was carried out with the software Geltool, that calculates profile values (pv’s) representing the extent of chromosomal degradation. Details were described previously (24).
Preparation of cell-free protein extracts
Fifty milliliters of YEPD was inoculated with a single colony and the culture was grown to stationary growth phase at 30°C. The pre-culture was diluted 10-fold with YEPD medium and incubated under vigorous shaking at 30°C to early logarithmic growth phase (5 × 106 −1 × 107 cells/ml). The cells were kept on ice for 20 min, centrifuged (4000 r.p.m., 4°C, 20 min) and washed twice with potassium phosphate buffer (25 mM KPO4, pH 8.0). Afterwards, the cell pellet was resuspended in 5 ml ice-cold extraction buffer (100 mM Tris–HCl, pH 8.0; 10 mM β-mercaptoethanol) and 1 ml protease inhibition mix (Sigma) was added per 20 g of cell pellet. Cells were crushed by using a pre-chilled French Press Cell (Aminco) applying an internal pressure of 20 000 psi. The crude extract was adjusted to pH 8.0 by adding 0.1 M NaOH and the cell debris was removed by centrifugation at 20 000 r.p.m. in a SS34 rotor for 40 min at 4°C. The supernatant was carefully removed without disturbing the pellet. Protein was precipitated by stepwise addition of ammonium sulfate to a concentration of 0.35 g/ml for at least 20 min on ice, collected by centrifugation (20 000 r.p.m., 30 min, 4°C), dissolved in 3 ml dialysis buffer (10 mM Tris–HCl, pH 7.5; 10 mM β-mercaptoethanol) and then dialyzed overnight at 4°C against the same buffer. The extract was further centrifuged for 10 min to remove insoluble particles. The crude extract was either used directly or stored for up to 6 months at –80°C before use. Protein concentrations were determined according to Bradford using BSA as standard.
Preparation of substrates for in vitro assays
For in vitro BER assays, single-strand (ss)DNA oligonucleotides (35-mer) containing a uracil residue (BER1 = 5′-GCC CTG CAG GTC GAU TCT AGA GGA TCC CCG GGT AC-3′) or an 8-oxoG (BER3 = 5′-GCC CTG CAG GTC GAG8-oxo TCT AGA GGA TCC CCG GGT AC-3′) at position 15 were annealed to their complementary synthetic strands (BER2 = 5′-GTA CCC GGG GAT CCT CTA GAG TCG ACC TGC AGG GC-3′ and BER4 = 5′-GTA CCC GGG GAT CCT CTA GAC TCG ACC TGC AGG GC-3′). Annealing was performed by mixing equimolar amounts of both oligonucleotides and incubating them for 10 min at 95°C followed by slow cooling down to room temperature. For the in vitro incision assay BER1 was labeled prior to annealing using T4 polynucleotide kinase (MBI-Fermentas) and [γ-32P]ATP according to the manufacturer’s instructions. Afterwards, the substrate was purified using G-25 Sephadex minicolumns (Roche) to remove unincorporated [γ-32P]ATP. To measure the gap-filling activity of cell extracts, a 14-nt ssDNA oligonucleotide (BER5 = 5′-GCC CTG CAG GTC GA-3′) was labeled, while the 5′-end of a 17–nt-long oligonucleotide (BER6 = 5′-AGA GGA TCC CCG GGT AC-3′) was non-radioactively phosphorylated. The annealing of BER5 and BER6 to BER2 generated a repair substrate lacking 4 nt in one strand.
In vitro repair assays
To determine the BER capacity of cell-free extracts, an in vitro BER assay was performed according to Harrigan et al. (44). Briefly, 20 µg protein were dissolved in BER reaction buffer (50 mM HEPES, pH 7.5; 0.5 mM EDTA; 2 mM dithiothreitol; 20 mM KCl; 4 mM ATP; 5 mM phosphocreatine; 0.5 mM NAD+; 0.1 mM ddTTP; 100 µg/ml freshly prepared phosphocreatine kinase) and preincubated with 250 nM oligonucleotide substrate (BER1 + BER2 or BER3 + BER4) for 5 min at room temperature. Addition of 2.2 µM [α-32P]dCTP and 10 mM MgCl2 initiated the repair reaction and the samples were incubated at 37°C for the indicated time points. The reactions were stopped by adding 5 mM EDTA and heating for 5 min to 72°C. Purification with G-25 Sephadex minicolumns (Roche) according to the manufacturer’s instructions was done to remove unincorporated [α-32P]dCTP. Afterwards, repair substrates were mixed with an equal volume of DNA loading dye (95% formamide; 20 mM EDTA; 0.02% bromphenol blue; 0.02% xylene cyanol) and incubated for 2 min at 72°C. For inhibition of polymerases protein extracts were preincubated for 30 min at 30°C with 2 µg/ml aphidicoline.
The in vitro incision assay was carried out on the basis of Wang et al. (45). Two-hundred and fifty nanomoles of oligonucleotide substrate ([γ-32P]BER1 + BER2) were incubated in 10 µl incision reaction buffer (50 mM HEPES, pH 7.7; 7.5 mM MgCl2; 0.5 mM EDTA; 1 mM dithiothreitol; 20 mM phosphocreatine; 2% glycerol; 100 µg/ml bovine serum albumin) with 1 µg cell free protein extract for the indicated time points at 23°C. The reactions were terminated by adding 20 µl stop solution (250 mM HEPES, pH 7.5; 1% sodium dodecyl sulfate (SDS); 500 µg/ml proteinase K) and heating for 20 min to 60°C. Repair products were ethanol-precipitated and solubilized in DNA loading dye as mentioned above.
For the gap-filling assay, 40 µg cell extract were mixed with 10 nM oligonucleotide substrate (BER2 + [γ-32P]BER5 + BER6) and reaction buffer (50 mM Tris, pH 7.4; 50 mM KCl; 1 mM dithiothreitol; 5 mM MgCl2; 5% glycerol). 2.5 mM dCTP and 2.5 mM dTTP were added. Reactions were stopped by adding 5 mM EDTA, mixed with 10 µl loading dye and heated for 5 min to 72°C.
All repair products were separated by polyacrylamide gel electrophoresis (24% acrylamide for the BER assay and the gap-filling assay and 18% acrylamide for the incision assay; 8 M urea; 89 mM Tris–HCl, pH 8.8; 89 mM boric acid; 2 mM EDTA), detected by autoradiography and quantified by digital imaging (TotalLab, Amersham).
RESULTS
Deletion of XRS2 leads to high mutation frequency and MMS sensitivity
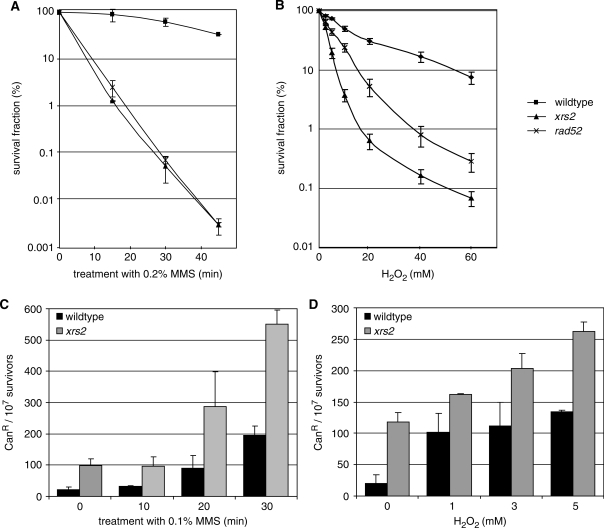
Mutations in RAD52 group genes lead to increased sensitivities to alkylating and oxidizing agents (26–28) and a genome-wide screen showed increased mutation frequencies after MMS treatment (15,30). We verified these findings in our experimental systems for the deletion of XRS2 and RAD52. Both, the xrs2 mutant and the recombination-deficient mutant rad52, conferred a high sensitivity to MMS and H2O2, whereas the xrs2 mutant shows an even higher sensitivity to H2O2 treatment as compared to the rad52 mutant (Figure 1A and B). Mutation frequencies were determined with a forward mutation system leading to canavanine resistance (Figure 1C and D). In wildtype cells, the spontaneous mutation frequency averaged 20 CanR mutants per 107 cells, while deletion of XRS2 increased the frequency 5–6-fold to 100–120 mutants per 107 cells. Furthermore, xrs2 mutants exhibited an elevated mutation frequency after MMS treatment. Following incubation for 30 min in 0.1% MMS the number of CanR mutants in the xrs2 strain was raised to 550 per 107 survivors, while in wildtype cells the number increased only to 200 CanR mutants. Mutation frequencies were also increased in the xrs2 mutant after H2O2 treatment. Incubation in 5 mM H2O2 yielded 130 mutants per 107 survivors in wildtype cells and 260 mutants per 107 survivors in the xrs2 deletion mutant. These results emphasized the importance of XRS2 and RAD52 to preserve genomic stability after treatment with base damaging agents.
Figure 1.
Sensitivity to base-damaging agents and mutation frequencies. (A, B) Sensitivity of the xrs2 deletion mutant to MMS and H2O2 compared to MKP-0 wildtype and the rad52 deletion mutant. Survival was plotted as the ratio of colonies obtained after treatment versus colonies obtained without treatment. (C, D) Induced mutation frequencies of BY4741 wildtype and its isogenic xrs2 deletion mutant after MMS and H2O2 treatment shown as canavanine-resistant (CanR) mutants per 107 survivors. Results are an average of three independent experiments ± SD.
Xrs2 plays a role in the repair of heat-labile sites after MMS treatment
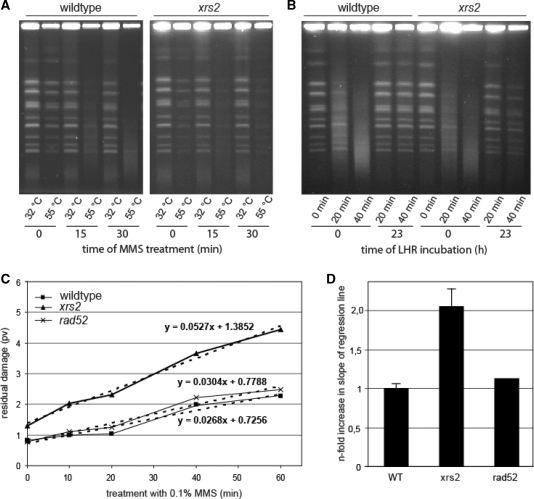
For many years, MMS was used as radiomimetic agent, to introduce DSB into DNA (46). New results, however, showed that MMS-induced DSB, which can be detected by PFGE, are due to elevated temperatures in DNA preparation where MMS-induced heat-labile sites are converted to DSB (31,47). We used this method to investigate the repair of MMS-induced heat-labile sites in stationary cells from wildtype, xrs2 and rad52 deletion strains. We determined the residual DNA damage 23 h after MMS treatment with 0.1% MMS for 0, 10, 20, 40 and 60 min. DNA damage repair was facilitated by incubating the cells under non-growth conditions (LHR), which allows DNA repair while proliferation is suppressed (41–43). Residual heat-labile sites were converted into DSB during proteinase K incubation at 55°C for DNA agarose plug preparation. As a control, DNA plugs were also prepared at 32°C. The resulting gel showed distinct bands of chromosomal DNA and little background in untreated and MMS-treated samples prepared at 32°C from both, wildtype and xrs2 strains (Figure 2A). This result indicated that there were equally low levels of DSB in wildtype and in the xrs2 mutant, and no direct induction of DSB by MMS was detectable in both strains. The results for DNA preparation at 55°C 0 h and 23 h after irradiation are shown in Figure 2B. Immediately after MMS treatment, the DNA fragmentation increased dependent on the length of MMS treatment. Twenty-three hours after incubation under LHR conditions, DNA repair was visible as the reappearance of chromosomal bands. To quantify the extent of chromosomal degradation we used the software program Geltool (24) and the residual damage was plotted against the time of MMS treatment (Figure 2C). Dependent on treatment time, the amount of residual damage increased moderately in wildtype (slope of regression line 0.0268), while there was a more pronounced accumulation of residual damage in the xrs2 mutant (slope of regression line 0.0527). In line with a recent publication (31), we found that the HR-deficient rad52 mutant shows a repair activity comparable to wildtype cells (slope of regression line 0.0304). Relative quantification of slopes of regression lines showed a 2-fold enhanced increase in the xrs2 mutant (Figure 2D). Furthermore, the xrs2 mutant exhibited a marginally increased amount of heat-labile sites in untreated cells. Therefore, the reduced ability of the xrs2 mutant to repair MMS-induced heat-labile sites was not due to reduced recombinational repair, a backup pathway for the repair of DNA base damage.
Figure 2.
Analysis of the repair of heat-labile sites. (A) Induction of chromosomal degradation in wildtype and the xrs2 mutant visualized by pulsed-field gel electrophoresis. Cells were treated for 0, 15 and 30 min with 0.1% MMS. During preparation of chromosomal DNA, the incubation with proteinase K was carried out at 32°C (heat-labile sites remain stable) as well as 55°C (heat-labile sites are converted into DSB). (B) Analysis of chromosomal degradation (proteinase K incubation at 55°C) immediately after treatment for 0, 20 and 40 min with 0.1% MMS as well as after 23 h incubation of the cells under LHR conditions. (C) Residual DNA fragmentation plotted against the time of MMS treatment. The degree of residual chromosomal degradation was quantified using the software program Geltool and is shown as profile value (pv). Linear equations are calculated from regression lines (dashed lines). One representative experiment is shown. (D) Slope of regression lines obtained from residual chromosomal degradations from the xrs2 and rad52 mutant relative to wildtype. Mean values from three independent experiments for the xrs2 mutant and for wildtype are shown. The rad52 mutant was analyzed once.
Xrs2 is directly involved in the BER pathway
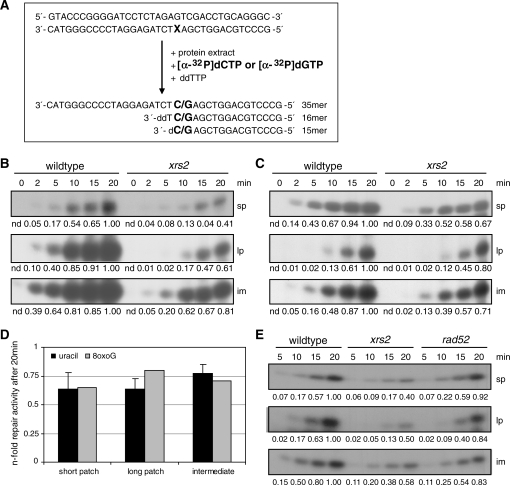
The results from the analysis of MMS-induced heat-labile sites suggested an involvement of XRS2 in the repair of DNA base damage. To further elucidate this assumption we employed an in vitro BER assay according to Harrigan et al. (44) (Figure 3A). We analyzed the repair of 35-bp double-stranded oligonucleotides containing a uracil or an 8-oxoG at position 15. These substrates were incubated with the same amounts of protein extracted from wildtype cells and xrs2 mutant cells in logarithmic growth phase. During the repair process damaged bases (uracil and 8-oxoG) are removed from the substrate and a [α-32P]dCTP or a [α-32P]dGTP was incorporated opposite G or C, respectively. Before ligation, this intermediate has a size of 15 nt; after ligation the original size of 35 nt was restored. To distinguish between short-patch repair, where only 1 nt is substituted, and long-patch repair, where a number of nucleotides were replaced, ddTTP is added to the reaction mixture to abort elongation. This allowed detecting long-patch repair products with a size of 16 nt. Using an error-free oligonucleotide, we observed no repair products (data not shown). Figure 3B shows a representative experiment for the repair of a uracil-containing oligonucleotide by cell extracts from wildtype cells and from the xrs2 mutant. It has to be noted that exposure times were different for monitoring long-patch and short-patch repair. In line with current publications usage of equal exposure times revealed clearly higher amounts of long-patch products (13,48), but quantification was hardly possible (data not shown). The amount of all three detectable repair products (intermediate, long patch and short patch) were reduced in extracts obtained from the xrs2 mutant. The formation of repair products was similarly reduced for an 8-oxoG containing oligonucleotide in the BER assay (Figure 3C). The quantitative examination of at least three independent extracts showed that the amount of short-patch and long-patch products synthesized by xrs2 extracts within 20 min was reduced to 0.64-fold the amount produced by wildtype extracts. The intermediate constituted 0.77-fold of the amount synthesized by wildtype extracts (Figure 3D). In contrast, extracts from the rad52 mutant remained BER proficient (Figure 3E). For the first time, we demonstrated reduced BER capacity in cell free extracts from the xrs2 mutant for the repair of both, uracil- and 8-oxoG-containing oligonucleotides.
Figure 3.
In vitro analysis of short-patch and long-patch BER with whole-cell extracts from wildtype cells and from the xrs2 mutant. (A) Principle of the in vitro assay (X = uracil or 8-oxoG). Labeled repair products are generated by BER activity of whole cell extracts. (B, C) Repair capacities for the repair of the uracil-containing oligonucleotide (B) and the 8-oxoG-containing oligonucleotide (C) of cell extracts obtained from wildtype cells and from the xrs2 mutant. The number below the images indicates the relative intensity of the corresponding band relative to the strongest band in that line. (D) Relative BER activity of the xrs2 mutant after a 20-min repair time. The diagram shows the average of three independent experiments using the uracil-containing oligonucleotide; the results for the 8-oxoG-containing substrate were obtained from one experiment. (E) Analysis of the BER capacity in cell extracts from the rad52 mutant in comparison to cell extracts from wildtype cells and the xrs2 mutant using the uracil-containing substrate. One representative experiment out of three is shown. nd., not determined.
The new role of Xrs2 in BER is a function of the whole MRX complex
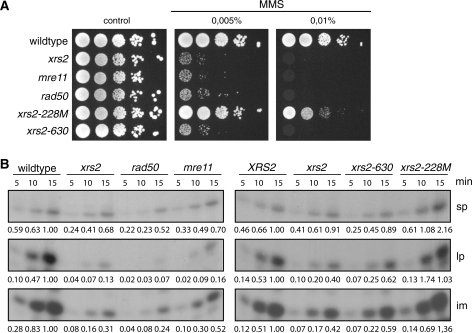
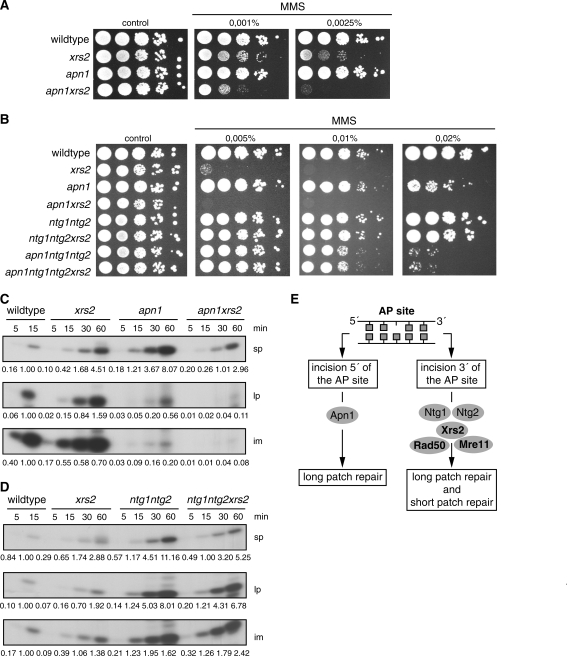
Xrs2 forms a trimeric complex with Mre11 and Rad50 (49). To analyze whether the newly detected role of Xrs2 in BER is a function of Xrs2 alone, or of the whole MRX complex, we analyzed MMS sensitivity and in vitro BER capacity in mre11 and rad50 deletion mutants. Since it was previously shown that several N- and C-terminal truncated versions of XRS2 exhibit MMS sensitivity (50), we also analyzed two recently constructed xrs2 mutants (24) either lacking the FHA- and BRCT-domains at the N-terminus (xrs2-228M) or the Tel1- and Mre11-binding domains at the C-terminus (xrs2-630) for their BER capacity. Figure 4 shows that all mutants except the xrs2-228M mutant behaved like the xrs2 mutant. Reduced BER capacities and increased sensitivity against MMS were equally evident for the mre11, rad50 and xrs2-630 mutant, and comparable to that seen in the xrs2 deletion mutant. Thus, we concluded that the complete MRX complex is involved in the BER pathway and especially the Mre11-binding domain of Xrs2 is important, while the FHA- and the BRCT-domains have no function in BER.
Figure 4.
MMS sensitivity and BER capacity of MRX-deficient cells and mutants carrying truncated versions of the Xrs2 protein. (A) Analysis of the MMS sensitivity. Shown are serial dilutions of cells on plates containing MMS as indicated. (B) Repair capacity of cell extracts obtained from the mre11, the rad50 mutant and from strains expressing truncated versions of XRS2 compared to wildtype and the xrs2 deletion mutant. The numbers below the images indicate the relative intensity of the corresponding band relative to the strongest band in that line. One of three independent experiments is shown.
Epistatic relationship of XRS2 to other genes involved in BER
To assign the MRX complex to the AP-endonuclease or the AP-lyase subpathway we analyzed the epistatic relationship of XRS2 and other BER genes concerning survival and in vitro BER capacity. We deleted XRS2 in an apn1 mutant, where the major BER pathway in yeast via the AP-endonuclease Apn1 is blocked. The AP-lyase pathway remains unaffected because in this mutant Apn2 can act as an efficient 3′-phosphodiesterase and compensate for the APN1 deletion in this function (13). Furthermore, we deleted XRS2 in the AP-lyase deficient mutant ntg1ntg2. Finally, we compared the quadruple mutant xrs2apn1ntg1ntg2 to the triple mutant apn1ntg1ntg2. The apn1 mutant exhibited a moderate MMS sensitivity (Figure 5A and B), while survival was hardly influenced by the deletion of NTG1 and NTG2 (Figure 5B). An effect of the ntg1ntg2 deletion appeared after additional deletion of apn1, when both the AP-endonuclease and the AP-lyase subpathways were impaired. Additional deletion of XRS2 in the apn1 background enhanced the sensitivity. In contrast, deletion of NTG1 and NTG2 suppressed the high MMS sensitivity of the xrs2 single mutant, resulting in the phenotype of the ntg1ntg2 double mutant. Similarly, the xrs2 phenotype was suppressed in the quadruple mutant xrs2apn1ntg1ntg2 (Figure 5B).
Figure 5.
Epistatic relationship of XRS2 to other BER genes. (A, B) Analysis of MMS sensitivity. Shown are serial dilutions of cells on plates containing MMS as indicated. (C) Repair capacity for the repair of the uracil-containing oligonucleotide of cell extracts obtained from the apn1 and the apn1xrs2 mutant compared to cell extracts from wildtype cells and from the xrs2 single mutant. (D) Repair capacity for the repair of the uracil-containing oligonucleotide of cell extracts derived from the ntg1ntg2 double mutants and ntg1ntg2xrs2 triple mutant compared to cell extracts derived from wildtype cells and from the xrs2 single mutant. In each case, one representative experiment out of three is shown. (E) Proposed model for the allocation of XRS2 to the AP-lyase-mediated repair pathway. The phenotypes of the apn1 and xrs2 single mutants are combined in the apn1xrs2 double mutant; otherwise, the effect of xrs2 is suppressed in the ntg1ntg2 mutant. These findings assign the MRX complex downstream of the AP-lyases where short-patch and long-patch repair products can be synthesized.
Similar effects were observed in the in vitro BER assays (Figure 5C and D). The apn1 mutant produced less intermediate and long-patch products when compared to wildtype, while the short-patch capacity was not influenced. The apn1xrs2 double mutant showed an additive effect on long-patch BER activity, while the short-patch BER activity of the xrs2 phenotype was observed. Compared to wildtype cells the ntg1ntg2 mutant showed no BER defect. Moreover, the additional deletion of XRS2 hardly influenced the repair capacity. For all three repair products the xrs2 phenotype was almost completely suppressed. We also tried to analyze the apn1ntg1ntg2 triple mutant and the apn1ntg1ntg2xrs2 quadruple mutant for their BER capacity, but insufficient repair products were generated (data not shown). Therefore, it was not possible to draw a conclusion for the influence of an additional XRS2 deletion. However, due to the additive effect of APN1 and XRS2, as well as the suppressive effect, of the NTG1 and NTG2 deletion on the xrs2 phenotype, we propose an involvement of the MRX complex downstream of the AP-lyases.
Functional analysis of the BER mechanism after deletion of XRS2
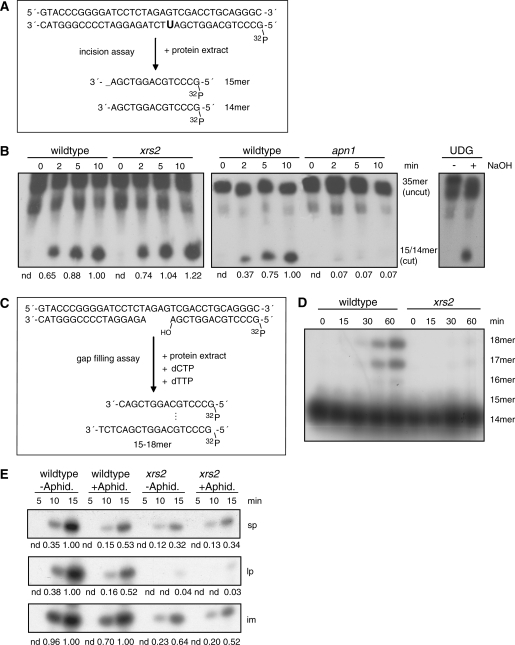
To analyze the efficiencies of base recognition and DNA incision at the AP site we carried out an in vitro base incision assay using the same uracil-containing substrate as in the BER assay labeled at the 5′-end. If the uracil is recognized and removed and the DNA is incised, a 14-nt and a 15-nt-long product is generated (Figure 6A). The specificity of the reaction was tested with different control approaches. Without cell extract no incision could be detected. The addition of exogenous recombinant uracil glycosylase (UDG) from Escherichia coli caused the removal of uracil, while the phosphate backbone was hydrolyzed by NaOH (Figure 6B, right part of image). Figure 6B (left part of image) shows that an extract from the xrs2 mutant produced the same amount of incision product compared to an extract from wildtype cells. However, deletion of APN1 led, as expected, to a clear defect in the incision assay (Figure 6B, middle part of image). Therefore, the first steps in BER, base recognition and DNA incision, were not influenced by Xrs2.
Figure 6.
Analysis of individual steps in the BER process after deletion of XRS2. (A) Incision assay using the uracil-containing oligonucleotide. Generation of an AP site and incision activity can be monitored through the appearance of cut products of the labeled substrate. (B) Incision assay with whole-cell extracts derived from wildtype cells and form the xrs2 (left part of image) and the apn1 (middle part of image) mutant. Stability of the phosphodiester bond was demonstrated by using an UDG-treated (Uracil-DNA-Glycosylase) substrate. The successful generation of AP-sites after UDG-treatment was shown by hydrolyzing the phosphodiester bond under alkaline conditions (right part of image). (C) Four-nucleotide gap-filling assay; gap-filling activity is visible through the extension of the 5′-labeled substrate. (D) Gap-filling activity of cell extracts derived from the xrs2 mutant compared to cell extracts from wildtype cells. (E) In vitro BER assay of whole-cell extracts from wildtype cells and from the xrs2 mutant preincubated with 2 µg/ml aphidicoline for 30 min. In each case, one representative experiment out of three is shown.
Following removal of the damaged base, the gap has to be filled by polymerase activity. To measure strand elongation reactions we carried out a gap-filling assay where the substrate mimics an intermediate of the long-patch pathway with a gap of 4 nt in one strand. Since no ATP for ligase activity was added the products can only reach a length of 15–18 nt (Figure 6C). Compared to wildtype the xrs2 mutant showed decreased gap-filling activity (Figure 6D). While extracts from wildtype cells were able to fill in the missing 4 nt within 15 min, the xrs2 mutant showed no strand elongation within 60 min incubation time. A further indication for reduced polymerase activity in the xrs2 mutant can be seen in the in vitro BER assay with cell extracts supplemented with 2 µg/ml aphidicoline for the inhibition of polymerases (Figure 6E). In the wildtype extract, aphidicoline treatment reduced the amount of both, short-patch and long-patch product, 0.5-fold compared to mock-treated samples. In samples with xrs2 extract, however, aphidicoline treatment had no impact on strand elongation and polymerization activity was equally low in treated and untreated xrs2 extracts. This indicates that the main part of new DNA synthesis in wildtype cells is executed by an aphidicoline sensitive polymerase, while the residual amount of repair synthesis in the xrs2 mutant is done by an aphidicoline insensitive polymerase.
DISCUSSION
In this study, we demonstrated that fully active BER is dependent on an intact MRX complex activity in S. cerevisiae. This was evidenced by an impaired repair of chromosomal heat-labile sites, which are clusters of base damage, in the xrs2 deletion strain. Furthermore, whole-cell extracts from strains containing deletions in MRX component genes displayed reduced repair efficiency in an in vitro BER assay as well as decreased gap-filling activity.
Deletion of XRS2 led to an elevated spontaneous mutation frequency as well as to increased induced mutation frequency and increased sensitivities after MMS and H2O2 treatment (15,30). So far, this was explained by the role of the RAD52 epistasis group genes facilitating recombinational repair as part of a tolerance pathway for stalled replication forks after base damage (28,31,32). However, we detected differences in the repair of MMS-induced chromosomal DNA damage between the rad52 deletion mutant and the xrs2 mutant. The rad52 mutant repaired the induced damage within 23 h as efficiently as wildtype cells, consistent with the assumption that replication bypass and recombinational repair were less important in G1 haploid cells. In contrast, in the xrs2 deletion mutant the residual damage was increased about 2-fold as compared to wildtype and the rad52 mutant. As the assay detected damaged bases, abasic sites and intermediate breaks arising during BER (31,47), this observation hinted at a role for XRS2 in BER that is independent of RAD52.
This assumption was further supported by the results of an in vitro BER assay modified from Harrigan et al. (44). Whole-cell extracts from the xrs2 mutant showed only 60% of the repair capacity of wildtype or the rad52 mutant within 20 min. Since it was possible to distinguish between short-patch and long-patch BER with this assay, we could show that both subpathways were equally affected by deletion of XRS2. Therefore, the function of XRS2 in BER should be upstream of the branching of the two pathways. Alternatively, XRS2 could be independently required for both subpathways. This BER defect can be seen in the repair of both, 8-oxoG and uracil, indicating a function for XRS2, independent of the native of the damaged base. Further analysis of mre11 and rad50 mutants demonstrated that the new function of Xrs2 is a function of the complete MRX complex. The in vitro BER capacity in all three mutants was decreased to a similar extend. In addition, we analyzed BER activity in mutants lacking the Mre11-binding domain or the N-terminus, including the FHA and BRCT domain of Xrs2, which were shown to be important for NHEJ (51). These experiments revealed that the truncation of the Mre11-binding domain was sufficient for the inactivation of the Xrs2 protein in BER.
To further elucidate the involvement of Xrs2 in BER, we investigated the epistatic relationship of XRS2 to both BER subpathways. We combined the deletion of XRS2 with deletions blocking either the AP-endonuclease-dependent long-patch repair pathway, which is most important in S. cerevisiae or the mechanistically less defined AP-lyase-mediated subpathway (13). In agreement with the current model of BER, the APN1 deletion displayed a clear defect in long-patch BER. The apn1xrs2 double mutant showed an additive effect in the long-patch repair and the xrs2 defect in the short-patch repair. An additive effect was also observed for MMS sensitivity. Therefore, we concluded that XRS2 and APN1 act on different pathways concerning DNA base damage removal. Deletion of NTG1 and NTG2 had no effect on BER and survival after MMS. This was consistent with the minor importance of the AP-lyase-mediated BER pathway (7,13). Otherwise, the function of Ntg1 and Ntg2 could be replaced by other lyases, like Ogg1. Interestingly, the effects of the XRS2 deletion were almost completely suppressed by the additional deletion of NTG1 and NTG2. In conclusion, our data suggested that the MRX complex acts downstream of Ntg1 and Ntg2 in the AP-lyase-mediated repair pathway and that intermediates resulting from incomplete processing in this pathway due to the XRS2 deletion were more severe than blocking the whole subpathway. In addition to the current model, our data suggest that in this pathway short-patch and long-patch repair products were synthesized instead of short-patch products only.
The lack of influence of XRS2 on the incision assay demonstrated that base recognition and the generation of single-strand breaks were independent of XRS2 and put the role of the MRX complex downstream of damage recognition and strand incision, which fitted with the fact that both, the repair of a uracil-containing and an 8-oxoG-containing oligonucleotide, were affected in the xrs2 mutant. Most likely, the role of the MRX complex is the enabling of an efficient polymerase activity. This assumption was substantiated by two findings: (i) the gap-filling assay directly showed that protein extracts from the xrs2 mutant display a decreased polymerization activity and (ii) the inhibition of polymerases by aphidicoline demonstrated that a major part of DNA synthesis in BER was executed by an aphidicoline-sensitive polymerase(s) in wildtype, whereas aphidicoline treatment had no impact in xrs2 extracts, indicating that this (these) polymerase(s) was already inactive as a consequence of the XRS2 deletion.
In mammalian cells, the WRN protein, a member of the RecQ helicase family, was demonstrated to activate BER by interacting with key players of this pathway. WRN cooperated especially with Polβ, the most important polymerase for BER in mammalian cells that is needed for the insertion of the first nucleotide in the gap-filling step (44,52). WRN also interacts with Nbs1 (53), the mammalian homolog of Xrs2, which was recently shown to be involved in BER (54). The authors suggested a role of Nbs1 in the recruitment step of Polβ. In yeast cells it was already shown that the RecQ helicase Sgs1 forms a complex with Mre11 following MMS treatment (55). We speculate that, like in mammalian cells, a large complex, composed of MRX and further BER factors, is needed to activate BER by managing the recruitment of polymerases for the repair process. Our results demonstrate that the MRX complex is a requisite for the efficient repair of DNA base damage, in addition to its better-known functions in DNA DSB repair. Further studies will be needed to identify the exact role and the interaction partners of the MRX complex in the BER pathway.
FUNDING
Funding for open access charge: Helmholtz Centre Munich—German Research Centre for Environmental Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Cadet J, Berger M, Douki T, Ravanat JL. Oxidative damage to DNA: formation, measurement, and biological significance. Rev. Physiol. Biochem. Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 4.Boiteux S, Gellon L, Guibourt N. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radic. Biol. Med. 2002;32:1244–1253. doi: 10.1016/s0891-5849(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 5.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 6.Guillet M, Boiteux S. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson DM, III, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst.) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325 (Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramotar D, Popoff SC, Gralla EB, Demple B. Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol. Cell. Biol. 1991;11:4537–4544. doi: 10.1128/mcb.11.9.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander M, Ramotar D. Partial purification of Pde1 from Saccharomyces cerevisiae: enzymatic redundancy for the repair of 3'-terminal DNA lesions and abasic sites in yeast. Biochemistry. 1997;36:6100–6106. doi: 10.1021/bi970048y. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Wang Z. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 1999;27:956–962. doi: 10.1093/nar/27.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao HI, Henricksen LA, Liu Y, Bambara RA. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 13.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst.) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Girard PM, Boiteux S. Repair of oxidized DNA bases in the yeast Saccharomyces cerevisiae. Biochimie. 1997;79:559–566. doi: 10.1016/s0300-9084(97)82004-4. [DOI] [PubMed] [Google Scholar]
- 15.Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjoras M. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alseth I, Osman F, Korvald H, Tsaneva I, Whitby MC, Seeberg E, Bjoras M. Biochemical characterization and DNA repair pathway interactions of Mag1-mediated base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2005;33:1123–1131. doi: 10.1093/nar/gki259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Wu X, Friedberg EC. DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase epsilon and is influenced by DNA polymerases alpha and delta in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:1051–1058. doi: 10.1128/mcb.13.2.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blank A, Kim B, Loeb LA. DNA polymerase delta is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1994;91:9047–9051. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grenon M, Gilbert C, Lowndes NF. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 2001;3:844–847. doi: 10.1038/ncb0901-844. [DOI] [PubMed] [Google Scholar]
- 23.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu. Rev. Genet. 2005;39:431451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 24.Steininger S, Gomez-Paramio I, Braselmann H, Fellerhoff B, Dittberner D, Eckardt-Schupp F, Moertl S. Xrs2 facilitates crossovers during DNA double-strand gap repair in yeast. DNA Repair (Amst) 2008;7:1563–1577. doi: 10.1016/j.dnarep.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 25.D'A;mours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 26.Mallory JC, Bashkirov VI, Trujillo KM, Solinger JA, Dominska M, Sung P, Heyer WD, Petes TD. Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair (Amst.) 2003;2:1041–1064. doi: 10.1016/s1568-7864(03)00115-0. [DOI] [PubMed] [Google Scholar]
- 27.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl Acad. Sci. USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letavayova L, Markova E, Hermanska K, Vlckova V, Vlasakova D, Chovanec M, Brozmanova J. Relative contribution of homologous recombination and non-homologous end-joining to DNA double-strand break repair after oxidative stress in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:602–610. doi: 10.1016/j.dnarep.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Tucker CL, Fields S. Quantitative genome-wide analysis of yeast deletion strain sensitivities to oxidative and chemical stress. Comp Funct Genom. 2004;5:216–224. doi: 10.1002/cfg.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl Acad. Sci. USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendricks CA, Razlog M, Matsuguchi T, Goyal A, Brock AL, Engelward BP. The S. cerevisiae Mag1 3-methyladenine DNA glycosylase modulates susceptibility to homologous recombination. DNA Repair (Amst.) 2002;1:645–659. doi: 10.1016/s1568-7864(02)00072-1. [DOI] [PubMed] [Google Scholar]
- 33.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao W, Chow BL, Rathgeber L. The repair of DNA methylation damage in Saccharomyces cerevisiae. Curr. Genet. 1996;30:461–468. doi: 10.1007/s002940050157. [DOI] [PubMed] [Google Scholar]
- 35.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 37.Myslinski E, Segault V, Branlant C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science. 1990;247:1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- 38.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 39.Melo RG, Leitao AC, Padula M. Role of OGG1 and NTG2 in the repair of oxidative DNA damage and mutagenesis induced by hydrogen peroxide in Saccharomyces cerevisiae: relationships with transition metals iron and copper. Yeast. 2004;21:991–1003. doi: 10.1002/yea.1144. [DOI] [PubMed] [Google Scholar]
- 40.Pohlit W, Heyder IR. Growth of cells on solid culture medium. II. Cell physiological data of stationary yeast cells and the initiation of cell cycle in nutrient free buffer solution. Radiat. Environ. Biophys. 1977;14:213–230. doi: 10.1007/BF01323940. [DOI] [PubMed] [Google Scholar]
- 41.Dardalhon M, Nohturfft A, Meniel V, Averbeck D. Repair of DNA double-strand breaks induced in Saccharomyces cerevisiae using different gamma-ray dose-rates: a pulsed-field gel electrophoresis analysis. Int. J. Radiat. Biol. 1994;65:307–314. doi: 10.1080/09553009414550361. [DOI] [PubMed] [Google Scholar]
- 42.Frankenberg-Schwager M, Frankenberg D, Blocher D, Adamczyk C. Repair of DNA double-strand breaks in irradiated yeast cells under nongrowth conditions. Radiat. Res. 1980;82:498–510. [PubMed] [Google Scholar]
- 43.Friedl AA, Kraxenberger A, Eckardt-Schupp F. Use of pulsed-field gel electrophoresis for studies of DNA double-strand break repair in the yeast Saccharomyces cerevisiae. Methods: A Companion to Methods in Enzymology. 1995;7:205–218. [Google Scholar]
- 44.Harrigan JA, Wilson D.M., III, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Wu X, Friedberg EC. Molecular mechanism of base excision repair of uracil-containing DNA in yeast cell-free extracts. J. Biol. Chem. 1997;272:24064–24071. doi: 10.1074/jbc.272.38.24064. [DOI] [PubMed] [Google Scholar]
- 46.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma W, Resnick MA, Gordenin DA. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008;36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelley MR, Kow YW, Wilson D.M., III Disparity between DNA base excision repair in yeast and mammals: translational implications. Cancer Res. 2003;63:549–554. [PubMed] [Google Scholar]
- 49.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 50.Tsukamoto Y, Mitsuoka C, Terasawa M, Ogawa H, Ogawa T. Xrs2p regulates Mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol. Biol. Cell. 2005;16:597–608. doi: 10.1091/mbc.E04-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmbos PL, Daley JM, Wilson TE. Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol. Cell. Biol. 2005;25:10782–10790. doi: 10.1128/MCB.25.24.10782-10790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J. Biol. Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 53.Cheng WH, von Kobbe C, Opresko PL, Arthur LM, Komatsu K, Seidman MM, Carney JP, Bohr VA. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J. Biol. Chem. 2004;279:21169–21176. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- 54.Sagan D, Muller R, Kroger C, Hematulin A, Mortl S, Eckardt-Schupp F. The DNA repair protein NBS1 influences the base excision repair pathway. Carcinogenesis. 2009;30:408–415. doi: 10.1093/carcin/bgp004. [DOI] [PubMed] [Google Scholar]
- 55.Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]