Abstract
Systematic tandem-affinity-purification (TAP) of protein complexes was tremendously successful in yeast and has changed the general concept of how we understand protein function in eukaryotic cells. The transfer of this method to other model organisms has been difficult and may require specific adaptations. We were especially interested to establish a cell-type-specific TAP system for Caenorhabditis elegans, a model animal well suited to high-throughput analysis, proteomics and systems biology. By combining the high-affinity interaction between in vivo biotinylated target-proteins and streptavidin with the usage of a newly identified epitope of the publicly shared SB1 monoclonal antibody we created a novel in vivo fluorescent tag, the SnAvi-Tag. We show the versatile application of the SnAvi-Tag in Escherichia coli, vertebrate cells and in C. elegans for tandem affinity purification of protein complexes, western blotting and also for the in vivo sub-cellular localization of labelled proteins.
INTRODUCTION
Molecular identification of protein complexes with mass spectrometry (MS) has a high potential to answer many important biological questions. However, one must be able to purify specific hetero-multimeric protein complexes in sufficient amounts without contamination of unrelated proteins. In order to maintain the integrity of the native protein complex purification strategies employing non-denaturing buffer conditions are essentially required.
Tandem tags with variable combinations of small peptide-tags (1), protein-binding domains (2) or whole protein tags (3–5) are commonly used to obtain such conditions. Frequently, a recognition site for a sequence-specific protease is located between the two tags. The first described tandem tag, the tandem-affinity-purification (TAP)-tag, consists of a combination of an IgG binding domain and the calmodulin-binding peptide (CBP) separated by a Tobacco Etch Virus (TEV)-protease recognition site (2). This combination allows the purification of the tagged protein complexes in two steps: after the first purification step, the protein complexes can be eluted in mild buffer conditions using protease cleavage. In a subsequent step, the tagged protein can be enriched again with an affinity matrix that binds the second part of the tag.
This tag has earned many merits in yeast but there are some drawbacks in other types of cells, in which the respective combination of affinity tags may be sub-optimal or even fail. Possible reasons for this failure could be high concentrations of Ig- or calmodulin domains in the sample which might interact with the tag structure in an unfavourable way.
Thus, several groups have made modifications of the classical TAP-tag and substituted especially the CBP by other tags (6–8).
Another important feature is the in vivo visualization of the tagged protein within an organism or at the sub-cellular level. Mislocalization of a tagged protein could indicate that co-purified proteins are false-positive interactors, resulting from either ectopic expression or overexpression.
To include such a feature the ‘localization and affinity tag (LAP-tag) was developed (4) in which the green fluorescent protein (GFP, 9) is part of the tandem tag. Thus, the GFP can be used at first for in vivo analysis of a tagged protein. A co-immunoprecipitation with α-GFP antibody enables the first purification step with this LAP-tag. After its cleavage by the TEV-protease the S-tag (10) is bound to a S-protein matrix. In principle, this LAP-tag is functional but since the first purification step needs the usage of only commercially available α-GFP antibodies the whole procedure is expensive and cannot be used at large scales or in high-throughput applications. Alternatively, a modified GFP was developed in which several peptide tags are introduced into a loop of GFP (11).
Purification of low-abundant protein complexes requires a tag with a high binding affinity, because the tag’s dissociation constant determines the lowest applicable target protein concentration according to the law of mass action. One of the strongest interactions available in biochemistry is the interaction of biotin and chicken avidin or the homologous streptavidin protein from the bacteria Streptomyces avidinii (12). D-biotin is an essential vitamin for all organisms and is specifically transferred by biotin holoenyzme synthetases (BHS) to biotin carboxyl carrier proteins (BCCP, 13). The dissociation constant of the interaction between (strept-)avidin and biotin is considerably lower (10−14 to 10−16 M) than all other interactions commonly used for affinity purification. For example, the dissociation constant for glutathione and the Glutathione-S-Transferase (GST)-tag is only about 7 × 10 −9 M (14). An artificial target motif for biotinylation by the Escherichia coli biotin holoenzyme synthetase birA has already been described (15,16). This peptide consists of 13 amino acids and is known as AviTag. It has already been used in different expression systems including bacteria (17), yeast (18), Sf9 cells (19), mammalian cell culture (20) and even in mice (21).
Our initial aim was to improve conditions for protein purification from the nematode Caenorhabditis elegans. Even though several proteomic studies on the purification of protein complexes from C. elegans have been published (22–24), there is still a need to ameliorate these methods. Proteomic approaches are often only practicable if highly specific antibodies against the protein or a tagged version of this protein are available (25,26). Unfortunately, many commercially available anti-peptide antibodies, such as α-Flag, α-myc or α-HA, are optimized for the use in vertebrate cells and result in a high cross-reactivity with non-vertebrate proteomes. This prevents an efficient usage of these antibodies, e.g. for protein complex purifications from C. elegans.
We describe here a novel tag combining several very advantageous features: the AviTag to enable high-affinity purifications, enhanced green fluorescent protein (EGFP) to enable in vivo localization studies, the TEV-protease recognition motif to allow elution under non-denaturing conditions, and the epitope for the publicly available peptide antibody SB1 that was mapped in this study. All these features are combined in a single peptide that can be fused to any protein for TAP from multiple organisms or expression systems.
MATERIALS AND METHODS
Cloning of the basic SnAvi-Tag coding vectors
The pEGFP-N1 vector backbone (GenBank Accession no. U55762; Clontech-Takara) was cut with BamHI and NotI to introduce a dsDNA linker. To obtain this linker the two oligonucleotides RB4444 (5′-GATCCACCGGTTACTACCATGGCTCCACGACCATCCAACAAACGTCTCCAGCAGT-3′) and RB4445 (5′-CCGGACTGCTGGAGACGTTTGTTGGATGGTCGTGGAGCCATGGTAGTAACCGGTG-3′) were allowed to anneal. The linker encodes one SB1 epitope. In addition, standard PCR conditions were used to amplify the EGFP coding sequence from the vector pEGFP-N1. The forward primer RB4432 (5′-ACCGGTATGCCACGACCCTCCAACAAGCGTCTTCAACAGGAG AACCTTTACTTTCAAGGTCAATTGGAAAATCTCTATTTCCAGGGA CCACCAGCGCCACCACAGATGGTGAGCAAGGGCGAGGAGCTGTTCACC-3′) includes the AgeI restriction site, the coding sequence for one additional SB1 epitope and two TEV-recognition motifs. The reverse primer RB4433 (5′-GCGGCCGCTTACTCGTGCCACTCGATCTTCTGGGCC TCGAAGATGTCGTTCAGGCCACCACCCTCACCCTGTGCT GCCAAATTCTCGCTTGCAGTAGTTGGAATATCATAATCCTTGTACAGCTCGTCCATGCCGA-3′) contains the coding sequence for the AviTag and a NotI restriction site.
The linker, the cut polymerase chain reaction (PCR)-fragment, and the BamHI and NotI cut EGFP-N1 vector were ligated to obtain the plasmid pBY2727 (CMV::SnAvi).
The plasmid pBY2887 was obtained by the ligation of a 977 bp fragment encoding the SnAvi-Tag into the AgeI and NotI restriction site of the pTriEx-5 vector (Novagen). This fragment was cut from pBY2727. Standard PCR conditions were used to amplify the GatewayTM-cassette from the pDEST53 plasmid (Invitrogen) with the primer pair RB4517 (5′-ACCGGTATGACAACAAGTTTGTACAAAAAAGC-3′) and RB4518 (5′-ACCGGTGGGACGACCACTTTGTACAAGAAAGC-3′). pBY2887 as well as the PCR-fragment were cut with AgeI and ligated to obtain the plasmid pBY2807.
To obtain pBY2946, pBY2727 was cut with NheI and HindIII. The vector pDP#MM016 (27) that contains the unc-119 rescue element was cut with HindIII and XbaI. A 5.6-kb fragment from pDP#MM016 and a 4.9-kb fragment from pBY2727 were ligated to obtain pBY2946.
All four described vectors contain the coding sequence for the SnAvi-Tag and can be used to generate arbitrary C-terminal protein fusions with the N-terminus of the SnAvi-Tag.
In pBY2727 and pBY2946 the SV40 polyadenylation site is located 3′ to the SnAvi-Tag coding sequence, whereas in pBY2887 and pBY2807 the rabbit globin 3′ untranslated region (UTR) is used.
Cloning of SnAvi-Tag fusions
The plasmids pBY3047 (rhgf-2::SnAvi in pTriEx) and pBY2824 (pink-1::SnAvi in pTriEx) were obtained by the GatewayTM-cloning procedure with the LR-Clonase II enzyme and the destination vector pBY2807 according to the manufacturer’s protocol (Invitrogen). The respective complementary DNAs (cDNAs) were obtained from the C. elegans cDNA library (Open Biosystems).
The plasmid pBY2730 (CMV::djr-1.1::SnAvi) was obtained by insertion of the djr-1.1 cDNA fragment in the NheI and SalI restriction sites of the vector pBY2727.
The lrk-1 cDNA sequence was cloned into the NheI and AgeI restriction sites of pBY2727 to yield the plasmid pBY2742.
To obtain the vector pBY2987 (myo-3::chn-1::SnAvi + unc-119 rescue) genomic DNA of the C. elegans chn-1 gene was amplified with the primer pair RB4885 (5′-CGGATCCGCAACCATGCCTCCGGGTTCAT-3′) and RB4886 (5′-GGGGTACCATGTCAAGCGGCGCCGAACA-3′). The PCR fragment was digested with BamHI and KpnI. The promoter of the C. elegans myo-3 gene was obtained from the plasmid pPD114.95 (gift from A. Fire, Stanford University, USA) after restriction with HindIII and KpnI. Both fragments were ligated with the HindIII and BamHI digested vector pBY2946 (SnAvi + unc-119 rescue element).
Cloning of the SB1 epitope for expression as EGFP fusion protein
The primer pair RB4881 (5′-AGCTTACCATGGACCCACGACCATCCAACAAACGTCTCCAGCAGAAG-3′) and RB4882 (5′-CTTCTGCTGGAGACGTTTGTTGGATGGTCGTGGGTCCATGGTAAGCT-3′) was allowed to anneal as a dsDNA linker. The EGFP-N1 vector was cut with HindIII and BamHI and ligated with the linker to obtain the plasmid pBY2947 that encodes SB1-EGFP under the control of the CMV-promoter.
Cloning of the birA::mCherry coding vectors
The plasmid pmCherry-N1 is a derivative of pEGFP-N1, in which the EGFP coding sequence was replaced with the coding sequence of the red-fluorescent protein mCherry (28). It served as a vector backbone. Genomic DNA of the TOP10 E. coli strain (Invitrogen) was used to amplify the birA coding sequence with the primer pair RB4883 (5′-CCCGGGATCCAATGAAGGATAACACCGTGCCAC-3′) and RB4884 (5′-CCACCGGTGTGGCGATGCTCCTTTTTTCTGCACTACGCAGG-3′). The forward primer contains a BamHI restriction site, the reverse primer an AgeI restriction site and the coding sequence for a small peptide spacer (RSIAT). The cut PCR fragment was inserted with BamHI and AgeI in pmCherry-N1. The resulting plasmid pBY2982 (CMV::birA::mCherry) encodes a red fluorescent birA::mCherry fusion protein at the 3′-end of a CMV promoter. pBY2982 was excised with NheI and HindIII, a 5.6-kb fragment from pDP#MM016 was digested with XbaI and HindIII, and a 2.4-kb-long fragment of the promoter for the C. elegans myo-3 gene was cut from the plasmid pPD114.95 (gift from A. Fire, Stanford University). All three fragments were ligated to obtain the plasmid pBY2983 (Pmyo-3::birA::mCherry + unc-119 (+) rescue element).
Cloning of the plasmids for the fluorescence energy transfer experiment
The myo-3 promoter was excised from the plasmid pBY2983 with HindIII and KpnI. The chn-1 coding sequence was cut from the plasmid pBY2987 with KpnI and BamHI. Both fragments were ligated in the HindIII/BamHI sites of an EGFP-N1 (Clontech) analogous vector, in which the EGFP coding sequence was exchanged with the coding sequence of the cyan fluorescence protein Cerulean (29) to yield the plasmid pBY3144. ubc-25::Venus (30) was generated by replacing the EGFP encoding sequence of ubc-25::GFP (described in 31) with the coding sequence of the flurophore Venus using NotI and BamHI. In addition, the unc-119 rescue sequence was introduced in the NheI and HindIII sites of the vector to obtain the plasmid pBY3145.
The anti C. elegans synaptobrevin antibody SB1
The mouse monoclonal SB1 antibody was obtained as a cell culture supernatant from the Developmental Studies Hybridoma Bank, University of Iowa. From the same institution we also obtained the SB1 hybridoma cell line that was developed by Michael Nonet and Gayla Hadwiger (Washington University in St Louis). The cell line was re-cloned by Nanotools, Teningen, Germany, and a production-run of 1 l of cell culture supernatant was performed.
Antibody interaction measurements
The interaction kinetics of the SB1 antibody with the epitope sequence PRPSNKRLQQTQAQ fused to the C-terminus of the C. elegans HMG-11 protein were determined using surface plasmon resonance (SPR) technology with a Biacore 3000 (Biacore/GE Healthcare) instrument. The antibody (6.9 ng/ml) was adsorbed in 10 mM acetate pH 3.5 on a CM5 chromatin immunoprecipitation (CHIP) with a flow rate of 10 µl/min and a contact time of 25 s. After immobilization with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS) for 7 min the surface was blocked with ethanolamine and finally 1270 resonance units (RUs) were deposited on the CHIP. To enable background detection within a measurement, a reference surface was activated and blocked with ethanolamine and the sample was first directed over the reference and then over the antibody surface. The tagged protein (stock 20 µg/ml) was diluted in 50 mM Na-phosphate 150 mM NaCl, 0.005% Tween, pH 7.2 and applied in a dilution series of 79 nM, 159 nM, 317 nM, 635 nM and 1270 nM. Between each sample the surface was regenerated with a low pH step. Measurements were background corrected and evaluated with the Biacore evaluation software version 3.1 using the standard Langmuir two molecule (A + B ↔ AB) interaction model.
Lysis of C. elegans for western blot analysis
For western blot analysis, worms were washed from the plates with M9 and pelleted by centrifugation. The pellets were washed with M9 until the supernatant was cleared from E. coli. Standard RIPA buffer supplemented with 3× concentrated Roche Protease Inhibitor cocktail was added (about 1:2 worm volume to volume of RIPA). The animals were lysed in the presence of glass beads (1-mm diameter) by alternating vortexing and freezing in liquid nitrogen.
Western blot analyses
The membranes were stained with the SB1 antibody (mouse, pure hybridoma supernatant or 4.6 µg/ml of the purified antibody), the Roche α-GFP antibody (#11814460001, mouse, 0.4 µg/ml), the Abcam α-GFP antibody (abcam290, rabbit, 1: 2000) or the α–β-actin antibody (#8691002, mouse, MPBio) overnight at 4°C.
Dependent on the detection method either α-mouse Alexa 680 (goat, 1:10 000 in Odyssey blocking solution, Molecular Probes), α-mouse Alexa 800 (goat, 1:10 000 in Odyssey blocking solution, LI-COR), α-rabbit Alexa 800 (goat, 1: 10 000, LI-COR, in Odyssey blocking solution) or α-mouse-HRP (goat, 1: 6000 in 3% milk powder in PBST, Dianova, for detection with ECL) were used as secondary antibodies. Alternatively, biotinylated proteins were directly detected with streptavidin coupled to Alexa 680 (Molecular Probes).
The detection of the fluorophore coupled secondary antibodies and Alexa 680 coupled streptavidin was performed with the Odyssey Scanner (LI-COR). The detection of the HRP-coupled secondary antibody was performed with the chemiluminescence detecting LAS-4000 image reader (Fujifilm) and SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Expression of proteins in E. coli
Escherichia coli of the strain Rosetta were transformed with pBY3047 (rhgf-2::SnAvi). Transformed bacteria were grown in 5 ml LB-medium at 37°C over-night. The next day the overnight culture was diluted 1: 100 in LB-medium. The culture was grown until it reached an OD600 of 0.6. At this stage isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to an end concentration of 100 µM in order to induce protein expression.
The E. coli were incubated for another 6 h at 30°C before being pelleted by centrifugation (20 min, 4°C, 6000g). The pellet was lysed in BugBusterTM (Novagen) supplemented with protease inhibitor cocktail tablets (Roche) according to the manufacturer’s protocol.
Expression of tagged proteins in vertebrate cells
HEK293T cells were grown in MEM with Glutamax supplemented with Pen/Strep and 10% fetal bovine serum (Gibco). Ten micrograms pBY3047 (RHGF-2::SnAvi), 5 µg pBY2955 (DJR-1.1::SnAvi), 20 µg pBY2824 (PINK-1::SnAvi) or 20 µg pBY2742 (LRK-1::SnAvi) were co-transfected with 2.5 µg pBY2982 (birA::mCherry) each in HEK293T cells that were plated on 10-cm dishes. For transfection, the lipofection reagent GeneJuiceTM was used according to the manufacturer’s protocol (Novagen). Additionally, free D-biotin (Sigma-Aldrich) dissolved in MEM was added prior to transfection to an end concentration of 100 µM plus the endogenous free D-biotin within the medium as described (20). Twenty-four to forty-eight hours post-transfection the cells were washed once with phosphate-buffered saline (PBS) and lysed in CytoBusterTM supplemented with protease inhibitor cocktail tablets (Roche) according to the manufacturer’s protocol (Novagen). For the analysis of the cross-reactivity of the SB1 antibody with different cell lines HeLa cells were grown in the medium described above. Chicken DT40 cells were cultured in RPMI 1640 Medium supplemented with 1 µM l-glutamine, 10% fetal bovine serum, 1% chicken serum, 50 µM 2-mercaptoethanol and Pen/Strep (Gibco). The lysates for both cell lines were prepared as described for HEK293T cells.
Purification of SnAvi-tagged proteins from HEK293T cells
SnAvi-tagged proteins expressed in HEK293T cells were allowed to bind to TetraLink-beads (Promega) on a rotating shaking incubator at 4°C for at least 4 h or overnight. Fifty-percent slurry of the beads was used in a ratio of 1: 2 to the volume of the respective lysates. The beads were washed with the TEV elution buffer (0.5 mM EDTA, 1 mM DTT, 150 mM NaCl, 50 mM Tris, pH 8). For the TEV digestion a ratio of 3:1 bead volume: volume of TEV elution buffer was used. AcTEV protease (Invitrogen) was added to a final concentration of 0.5 U/µl. The digestion was performed at 16–23°C for at least 2 h or overnight. The TEV eluates were either concentrated in a vacuum centrifuge or directly used for western blot analysis.
Worm strains
Caenorhabditis elegans strains were cultured as described by Brenner (32). The Bristol strain N2 was used as standard wild-type strain. The strain BR3771 carrying the integrated transgene byIs143 was obtained by microinjection (33) of the N2 wild-type strain with myo-2::dsRed2 (pBY2000) and snb-1::snb-1-GFP (pSB120.65, kindly provided by M. Nonet), followed by integration with ultraviolet (UV)-irradiation and eight outcrossing steps. The strain BR5072 carries the extrachromosomal array byEx715. This array was obtained by microinjection of the strain DP38 (unc-119(ed3)) with the plasmids myo-3::birA::mCherry (pBY2983) and myo-3::chn-1::SnAvi (pBY2987).
For the fluorescence energy transfer (FRET) experiment, the strain BR5518 (unc-119(ed3);byEx780) was generated by microinjection of the plasmids pBY3144 and pBY3145 in the strain DP38 (unc-119(ed3)). The transgene rescues the unc-119 mutation.
Tandem-affinity protein-complex purification from C. elegans
The lysates of the worm strain BR5072 were prepared in IPP150 buffer (10 mM Tris–HCl, pH 8, 150 mM NaCl, 0.1% NP-40, supplemented with 3× concentrated Roche protease inhibitor cocktail). Five millilitres of IPP150 buffer was used for the lysis of 0.5g worms of the strain BR5072. The lysates were prepared in the French press (8000 psi). The same lysate was used sequentially 3× to lyse all animals completely. The lysate was incubated with 400 µl TetraLink beads (Promega) for 30 min before the beads were washed five times with IPP150 avidin buffer with 1× Roche protease inhibitor cocktail. Two additional washing steps were performed to exchange the buffer with TEV-cleavage buffer [10 mM Tris–HCl, (pH 8), 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM DTT]. Two-hundred units of AcTEV protease (Invitrogen) were used in a volume of 1.5 ml TEV-cleavage buffer to specifically cut SnAvi-tagged proteins from the TetraLink beads (incubation at 16°C, 2 h).
The supernatant was incubated with 15 µl SB1-coupled Protein G agarose beads at 4°C overnight. The coupling of the SB1 antibody was performed using the Protein Size X protein G immunoprecipitation kit (Pierce) according to the manufacturer’s protocol.
After being washed three times with TEV-cleavage buffer, the beads were boiled in 15 µl modified Laemmli buffer [300 mM Tris–HCl (pH 6.8), 12 mM EDTA, 6% SDS (w/v), 30% glycerol].
The protein eluate was then allowed to run a few millimetres into a 12% sodium dodecyl sulphate (SDS) separation gel. The gel was stained with Coomassie blue and all co-purified proteins were excised as a single band from the gel. This band was used completely for analysis by MS.
MS
In-gel digests were performed as described in standard protocols. Briefly, the excised gel bands were destained with 30 % ACN, shrunk with 100% ACN, and dried in a Vacuum Concentrator (Concentrator 5301, Eppendorf, Hamburg, Germany). Digests with trypsin were performed overnight at 37°C in 0.05 M NH4HCO3 (pH 8). About 0.1 µg of protease was used for one gel band. Peptides were extracted from the gel slices with 5% formic acid.
All liquid chromatography/mass spectroscopy (LC-MS)/MS analyses were performed on an ion trap mass spectrometer (Agilent 6340, Agilent Technologies) coupled to an 1200 Agilent nanoflow system with a high-performance liquid chromatography (HPLC)-CHIP cube ESI interface. Peptides were separated on a HPLC-CHIP with an analytical column of 75 µm i.d. and 150 mm length and a 40-nl trap column, both packed with Zorbax 300SB C-18 (5-µm particle size). Peptides were eluted with a linear acetonitrile gradient with 1%/min at a flow rate of 300 nl/min (starting with 3% acetonitrile).
MS/MS analyses were performed using data-dependent acquisition mode. After an MS scan (standard enhanced mode), a maximum of three peptides were selected for MS/MS (CID, standard enhanced mode). The automated gain control was set to 350 000. The maximum accumulation time was set to 300 ms.
Mascot Distiller 2.1 was used for raw data processing and for generating peak lists, essentially with standard settings for the Agilent ion trap. Mascot Server 2.2 was used for database searching with the following parameters: peptide mass tolerance: 1.1 Da, MS/MS mass tolerance: 0.3 Da, 13C: 1, enzyme: trypsin with max. two missed cleavage, variable modifications: Gln->pyroGlu (N-term. Q), oxidation (M) and carbamidomethyl (C).
The SwissProt 55.6 and WormPep 194 databases were used.
Microscopy
An Imager.Z1 microscope equipped with an AxioCamMRm CCD camera and the Axiovision software release 4.6 (all Carl Zeiss, Goettingen) were used for microscopic documentation of C. elegans. For cell culture observations, the Eclipse TS100 microscope (40× magnification) and the Digital Sight DS-2 MBW camera (both Nikon) were used.
FRET analysis
FRET microscopy of the strain BR5518 was performed using a Zeiss LSM510-META inverted confocal laser scanning microscope using the spectral detector in a virtual filter mode. Cerulean emission was detected from 471–503 nm, FRET-emission and Venus emission were detected from 524–567 nm. We used 457-nm excitation for Cerulean and FRET and 514-nm excitation for Venus in a two-track configuration. A C-Apochromat 63×/1.2W objective was used.
Spectral references for filter bleed-through were taken from myo-2::Cerulean and myo-2::Venus transgenic C. elegans strains, respectively. FRET was calculated with the PixFRET plugin (34) of ImageJ (35) using a Gaussian blur of 1.0 and a threshold of 0.8. BTdon (BTacc) was determined as 0.485 (0.742).
RESULTS
Identification and characterization of the SB1 binding epitope
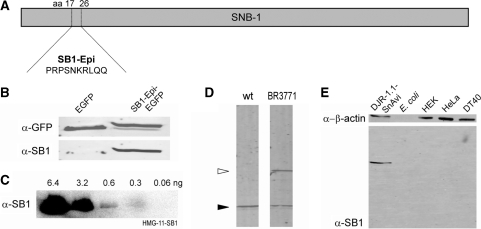
The SB1 mouse monoclonal antibody and the respective hybridoma cell line are publicly distributed by the Developmental Studies Hybridoma Bank (DSHB). Unlimited amounts of this antibody that targets the synaptobrevin protein (SNB-1, 36) of C. elegans (Figure 1D) can be produced with these hybridoma cells. Since this antibody did not show cross reactivity with other proteins we decided to identify the epitope in the SNB-1 sequence. For this purpose, several different fragments of the SNB-1 protein were expressed in HEK293T cells. The fragments that were recognized by the antibody were shortened in several rounds until a 10-amino-acid-long peptide stretch in the primary SNB-1 sequence was identified that is still recognized by the SB1 antibody. This peptide (SNB-1 17–26) has the amino acid sequence PRPSNKRLQQ and is recognized in synthetic protein fusions (Figure 1B). Notably, this antibody can also be used for protein detection in different vertebrate cell lines and in E. coli due to the fact that no or only very weak cross reactivity of the antibody with the respective proteomes is detectable (Figure 1E). The detection threshold of the antibody was determined in western blot analysis (Figure 1C). For detection, 0.3 ng of SB1 epitope tagged HMG-11 protein was sufficient. Interaction kinetics of the immobilized SB1 antibody with the epitope-tagged HMG-11 protein were further evaluated by SPR measurements. Interaction curves were fitted with a standard ‘A + B’ Langmuir model. In the most reliable measurement range of intermediate sample concentrations (e.g. 317 nM) the association rate constant was ka = 1.4 × 106 ± 2 × 104 M−1 s−1 and the dissociation rate constant kd = 3.4 × 10−3 ± 5 × 10−5 s−1 resulting in a dissociation constant of KD = 2.4 nM.
Figure 1.
Mapping of the SB1 epitope and test of cross reactivity of SB1 in different expression systems. (A) The epitope that is recognized by the SB1 antibody was determined in the peptide sequence of the C. elegans protein SNB-1. (B) The SB1 antibody specifically detects EGFP when fused to the SB1 epitope (PRPSNKRLQQ, right lane). However, no signal is detectable in the negative control lacking the SB1 epitope (left lane). The upper blot is detected with α-GFP. The SB1 antibody is used for the detection of the lower blot. (C) The detection threshold of the SB1 antibody was determined in western blot analysis with SB1 epitope tagged HMG-11 protein as antigen and HRP-coupled secondary antibody. (D) The SB1 antibody specifically detects SNB-1 (filled arrow head) in C. elegans wild-type lysates and additionally in the lysate of the transgenic animal strain BR3771 also the protein fusion SNB-1::GFP (unfilled arrow head). Caenorhabditis elegans lysates were separated in a 12% SDS–PAGE. (E) Detection of the cross reactivity of the SB1 antibody with the lysates of different cell lines and E. coli. Equal amounts of proteins are separated in a 12% SDS–PAGE, blotted and detected with the SB1 or α–β-actin antibodies. DJR-1.1::SnAvi expressed in HEK293T cells is used as a positive control. Fluorescent secondary antibodies and the Odyssey scanner (LI-COR) were used for the detection of the western blots (B), (D) and (E).
The structural composition of the SnAvi-Tag allows a novel purification strategy
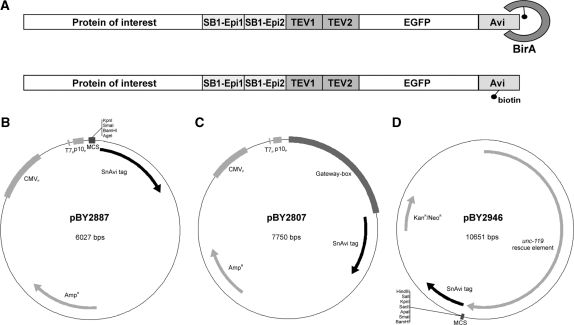
Since the cross-reactivity of the SB1 antibody with other proteins was low and the SB1 epitope maintained its reactivity in protein fusions, we selected the SB1 epitope as one component of our tandem tag. In order to gain avidity effects and to increase the chance of accessibility of the epitope in various protein fusions we included two copies of the SB1 epitope. These two SB1 epitopes were fused to the C-terminus of our target-proteins and can be used for both in vitro detection and purification. Furthermore, we incorporated the coding sequence for EGFP in our tandem tag to facilitate in vivo detection. Since the SB1 epitopes are separated from the EGFP by two TEV protease recognition motifs, the EGFP can be removed from the tagged protein during the purification procedure. As a last but very important feature, we also included an AviTag (15) in our system. This amino acid sequence is specifically biotinylated by the E. coli protein–biotin ligase birA and allows us to work with the biotin–(strept-)avidin interaction. This interaction is one of the strongest non-covalent interactions occurring in biochemistry and has a femtomolar dissociation constant. Using the AviTag for the first protein purification step allows us to purify also proteins of very low abundance. Furthermore, contaminating proteins can be removed in the first purification step by using stringent buffer conditions. A schematic view of the structure that we named SNB-1 Avi purification tag (SnAvi-Tag) is shown in Figure 2A.
Figure 2.
Composition of the SnAvi-Tag and SnAvi coding vectors. The SnAvi-Tag combines several peptide sequences with distinct features: two epitopes for the SB1 antibody (SB1-Epi1, SB1-Epi2), two recognition motifs for the TEV protease (TEV1, TEV2), the enhanced green fluorescent protein (EGFP) and the AviTag (Avidity). (A) The E. coli biotin ligase birA specifically ligates free D-biotin to the AviTag. (B)–(D) Vectors encoding the SnAvi-Tag. All vectors are available via www.addgene.org (B) The vector pBY2887 contains the coding sequence for the SnAvi-Tag under control of three different promoters (T7P, P10P and CMVP) allowing protein expression from this vector in E. coli, Sf9 and vertebrate cells. The tagged protein can be introduced in the multiple cloning site (MCS) by standard cloning procedures. (C) The vector pBY2807 shares the basic structure of pBY2887. Instead of a MCS it contains a cassette (‘Gateway-box’) that allows the introduction of a gene of interest using the Gateway-technology (Invitrogen). (D) The vector pBY2946 can be used for expression in C. elegans. It contains the coding sequence for the SnAvi-Tag and the rescue element for the unc-119(ed3) mutant as a genetic selection marker.
To make use of this tag in a variety of expression systems we generated several basic vectors in which any gene of interest can be introduced 5′ to the SnAvi coding sequence (Figure 2B–D). The plasmids pBY2887 and pBY2807 are derived from the pTriEx-5 vector (Novagen) that contain the CMV, T7 and P10 promoters and, thus, can be used for protein expression in either mammalian, E. coli or Sf9 cells. In pBY2887 a gene of interest can be introduced in a multiple cloning site (MCS) by classical cloning procedure. On the other hand, the vector pBY2807 gives the possibility to introduce a gene of interest by recombination with the help of the LR Clonase II enzyme (Invitrogen).
The third basic vector we designed (pBY2946, Figure 2D) is suitable for gene expression in C. elegans. To simplify the selection of transgenic animals we included the rescue sequence for unc-119 mutants. This marker gene allows transgenic unc-119 mutants to enter the dauer larval stage (27). Thus, only transgenic animals can survive starvation periods. All described vectors are available via Addgene.
Expression and purification of SnAvi-tagged proteins in E. coli and HEK293T cells
Since we are interested in the function of the C. elegans orthologues of Parkinson’s-disease-related genes (37), we cloned the cDNAs of the C. elegans genes djr-1.1, pink-1 and lrk-1 in SnAvi-Tag expression vectors. Furthermore, the functionality of the SnAvi-Tag for the expression in different expression systems should be demonstrated with the C. elegans guanine nucleotide exchange factor rhgf-2.
For expression in HEK293T cells the respective vectors were co-transfected with the birA::mCherry encoding vector pBY2982. D-biotin (Sigma) was supplemented in a concentration of 100 µM in order to optimize the biotinylation efficiency (20).
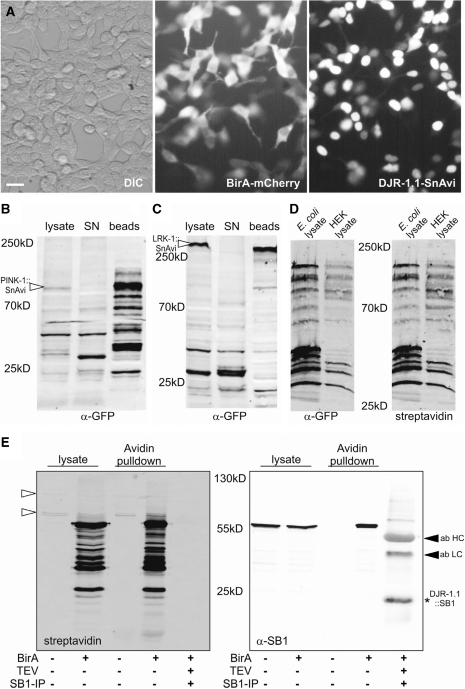
When djr-1.1::SnAvi and birA::mCherry were co-expressed, both proteins could be easily monitored in living cells. Using fluorescence microscopy we detected both protein fusions, birA::mCherry as well as DJR-1.1::SnAvi, independently from each other (Figure 3A). Only when the birA-encoding plasmid was co-transfected with djr-1.1::SnAvi in mammalian cells, the fusion protein could be pulled down from the cell lysates using avidin beads (Figure 3E). In order to test whether the TEV-cleavage sites can be recognized within the SnAvi-Tag a fraction of the avidin beads bound to DJR-1.1::SnAvi was incubated with the AcTEV-protease (Invitrogen). Subsequently, the AcTEV-eluate was incubated with SB1-bound Protein A/G Sepharose (Santa Cruz) to enrich DJR-1.1::SB1. The successful TEV-cleavage could be monitored due to the shift of the protein size after the removal of the EGFP-AviTag part of the protein fusion (Figure 3E). The size of the tagged DJR-1.1 protein was thereby reduced from 57 kDa (DJR-1.1::SnAvi) to 26 kDa (DJR-1.1::SB1, Figure 3E).
Figure 3.
The SnAvi-Tag can be used efficiently for protein expression and purification. (A) DJR-1.1::SnAvi and birA::mCherry can be detected in vivo. Recombinant protein fusions are visualized with the Eclipse TS100 microscope (40× magnification) and the Digital Sight DS-2 MBW camera (both Nikon). The scale bar represents 20 µm. PINK-1::SnAvi (B) and LRK-1::SnAvi (C) can be purified efficiently with the SB1-antibody cross-linked to Protein G Sepharose (Pierce). Most SnAvi-tagged proteins are cleared from the supernatant (SN) after incubation with the beads. (D) pBY3047 (rhgf-2::SnAvi in pTriEx) is used for expression of SnAvi-tagged proteins both E. coli as well as in HEK293T cells (in this case co-transfected with birA), biotinylated RHGF-2::SnAvi protein can be detected in the crude lysates of both expression systems. (E) Recombinant DJR-1.1::SnAvi expressed in HEK293T cells can only bind to avidin-beads when birA is co-transfected. Furthermore, DJR-1.1::SB1 (indicated by an asterisk) is detected after elution with the TEV-protease and enriched with SB1-coupled Protein A/G Sepharose (Santa Cruz). The heavy and light antibody chains are recognized by the secondary mouse antibody (ab HC and ab LC), indicated by filled arrow heads. Endogenously biotinylated proteins are indicated by unfilled arrow heads. In addition to the full-length proteins some partial or degraded protein products can be detected in (B)-(E). The detection for (B) and (C) was done with α-GFP antibodies (rabbit, Abcam, 1:2000), for (D) with α-GFP antibodies (mouse, Roche, 1:1000), and for (E) with the SB1 antibody (mouse, 4.6 µg/ml). Fluorescent secondary antibodies were used for detection in combination with the Odyssey (LI-COR). In (D) and (E) additionally Alexa 680 coupled streptavidin was used for detection.
In a next step, we tested the possibility of using the SB1-epitope within the SnAvi-Tag for immunoprecipitation. LRK-1::SnAvi as well as PINK-1::SnAvi were expressed and purified from HEK293T cells. For this purpose, the SB1 antibody was cross-linked to Protein G agarose (Pierce). The SB1-cross-linked beads were incubated with the respective lysates and PINK-1::SnAvi as well as LRK-1::SnAvi were purified together with these beads (Figure 3B or C, respectively), despite of the huge protein size of LRK-1::SnAvi (>300 kDa). Notably, all purification steps described for DJR-1.1::SnAvi worked as well for both PINK-1::SnAvi and LRK-1::SnAvi (data not shown).
rhgf-2::SnAvi was used for expression in mammalian cells and in the Rosetta E. coli strain. For this purpose, we used the plasmid pBY3047 that contains promoters for both expression systems. Due to the fact that birA is a natural E. coli protein recombinantly expressed birA was not required in this case. RHGF-2::SnAvi could be detected in both the lysates of the pBY3047 transformed Rosetta E. coli strain as well as in lysates of pBY3047 transfected HEK293T cells (Figure 3D). In addition, the biotinylation of RHGF-2::SnAvi was confirmed when Alexa 680 coupled streptavidin was used for detection (Figure 3D).
Expression and purification of SnAvi-tagged proteins from C. elegans
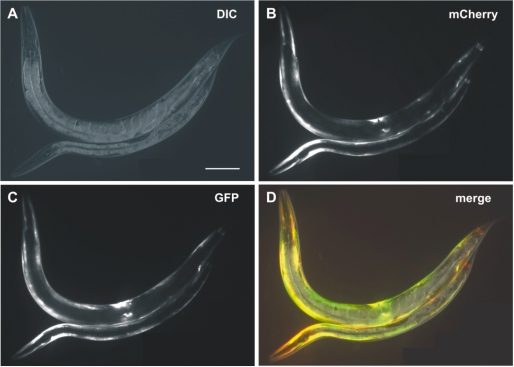
Since the SnAvi-Tag was proven to be an effective tool for protein purifications in HEK293T cells and in E. coli, we wanted to test it in a system in which tandem affinity purification has not yet been established as a canonical method. Therefore, we decided to purify SnAvi-tagged proteins and protein complexes from C. elegans lysates. For this purpose, we cloned the chn-1 coding sequence as fusion to the SnAvi-Tag coding sequence. To restrict the expression of CHN-1::SnAvi to the body wall muscle of C. elegans the myo-3 promoter was chosen. The same promoter was used to express the birA::mCherry fusion. Both plasmids that additionally carried an element to rescue the unc-119 mutant phenotype were mixed and injected (33) in unc-119(ed3) to yield the transgenic C. elegans strain BR5072. As shown in Figure 4 both fusion proteins can be detected in the body wall muscle of transgenic animals.
Figure 4.
Expression of myo-3::birA::mCherry and myo-3::chn-1::SnAvi in C. elegans. The worm strain BR5072 carries the transgene byEx715. This transgenic array allows expression of both birA::mCherry and CHN-1::SnAvi in the body wall muscle of C. elegans. The upper animal is an adult hermaphrodite, whereas the lower is a L4-larva. For both animals anterior is left. Scale bar: 100 µm. (A) Differential interference contrast image: (B) mCherry-fluorescence; (C) GFP-fluorescence; and (D) merge.
To purify SnAvi-tagged CHN-1 transgenic animals were grown in liquid culture. Protein lysates were incubated with avidin beads to bind CHN-1::SnAvi. The beads were washed and then used for the TEV-cleavage procedure. For the last step of the purification SB1 antibody covalently coupled to Protein G agarose was used to bind CHN-1::SB1 along with its complex partners. The bound proteins were eluted from the beads and completely used for analysis with two-dimensional (2D) LC-MS/MS tandem MS.
Several proteins were identified by MS (Table 1). Proteins that were also identified in CHN-1 unrelated experiments were considered as unspecific and subtracted. In these control experiments, the only interaction partner that was specifically purified with the SnAvi tag was the C. elegans protein NAS-37 that thus was included in our background subtraction list (data not shown). Among the remaining identified proteins, CHN-1 was abundant. In addition to CHN-1, HSP-1 was identified as complex partner. Interestingly, the human CHN-1 orthologue CHIP was originally described as interaction partner of Hsp70 (38). CHIP interacts with the C-terminus of Hsp70 and was even named after this interaction (C terminus of Hsp70 interacting protein). For instance, CHIP and Hsp70 are known to act together in a complex with the chaperon protein Csp to mediate the ubiquitylation and degradation of the unfolded cystic fibrosis transmembrane conductance regulator protein CFTR (39), thus validating a physiological relevance of this interaction.
Table 1.
Identification of CHN-1 interactors with 2D-LC-MS/MS
| Caenorhabditis elegans protein | Protein mass (KDa) | Amino acids | Amino acids identified by MS-analysis | Peptide coverage, n % |
|---|---|---|---|---|
| CHN-1 | 31.1 | 266 | 73 | 27.4 |
| HSP-1 | 69.7 | 640 | 54 | 8.4 |
| UBC-25 | 44.1 | 387 | 16 | 4.1 |
CHN-1 complexes were purified by tandem purification using the SnAvi-Tag and SB1-coupled protein G agarose. Purified proteins were analysed by 2D LC-MS/MS. Proteins that were also recognized in other CHN-1 unrelated experiments were considered as unspecific and thus subtracted.
Since we also identified the Hsp70 homologue CeHSP-1 as interactor for the C. elegans CHIP orthologue CHN-1 we conclude that this interaction is important and conserved among species.
Furthermore, we also identified the E2 ubiquitin-ligase UBC-25 which plausibly could form a functional complex with the E3/E4 ubiquitin transferase CHN-1. Interestingly, this interaction was also found in the vice versa experiment in which the purification procedure was performed with UBC-25::SnAvi. Here, CHN-1 was identified with MS as the most prominent co-purified protein (data not shown).
Investigation of the CHN-1 and UBC-25 interaction in vivo
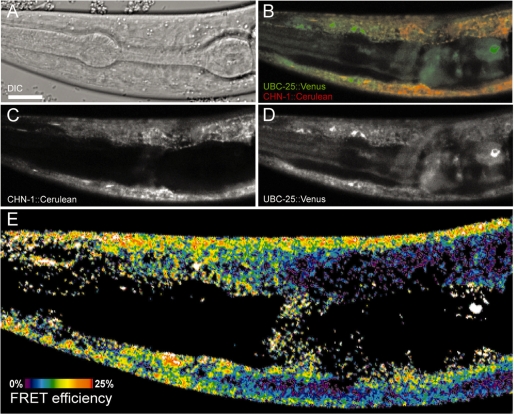
To verify the interaction of CHN-1 and UBC-25 with a SnAvi-tag independent method the C. elegans strain BR5518 was generated and used for microscopic analysis. This strain carries both the translational fusion chn-1::Cerulean under control of the myo-3 promoter as well as the ubc-25::Venus translational fusion under control of the ubc-25 promoter. In optical sections both protein fusions clearly co-localized in not further characterized cytosolic structures of the body wall muscle cells (Figure 5B). In addition, UBC-25::Venus was detected in the nucleus as described previously (31). Moreover, we were able to measure FRET signals in regions where co-localization of UBC-25 and CHN-1 occurred (Figure 5E). These results suggest that the distance between Cerulean and Venus does not exceed more than 7–10 nm, indicating a molecular interaction.
Figure 5.
UBC-25 and CHN-1 are interacting in C. elegans in vivo. The worm strain BR5518 carries the transgene byEx780. This enables the expression of UBC-25::Venus under control of its natural promoter and CHN-1::Cerulean expression in the body wall muscle of the animals. A L3-stage larva is shown. Optical sections of the animal were analysed with a Zeiss LSM510-META inverted confocal laser scanning microscope. Scale bar: 10 µm. (A) Differential interference contrast image. (B) The merged image of UBC-25::Venus (green) and CHN-1::Cerulean (red) shows a partial co-localization of both proteins. (C) CHN-1::Cerulean fluorescence. (D) UBC::25::Venus fluorescence. (E) FRET efficiency colour encoded from 0% (dark blue) to 25% (red).
DISCUSSION
Many proteins exert their function within protein complexes. Thus, the isolation and identification of components of protein complexes may extend our understanding of their physiological role. However, to purify a tagged protein requires reagents with a high binding affinity to the target protein as well as low cross-reactivity with other unrelated proteins. One possible approach to increase sample purity is the use of TAP strategies, most prominent among them the original TAP-tag (2). This tag is composed of an IgG-binding domain and of the CBP. It was used very successfully for the purification of protein complexes from yeast, but it unfortunately has several drawbacks for the work with other cell types. We recognized in our initial experiments that the CBP domain did not bind to the calmodulin affinity matrix if it was expressed in C. elegans. A possible reason for this could be a strong binding of endogenous proteins of this species directly to the tag structure. Here, we demonstrate that our newly designed tandem tag, the SnAvi-Tag, is able to overcome these problems. The AviTag (15) is used for the first step of the purification that is based on the high affinity interaction of biotin to (strept-)avidin. It has previously been employed for chromatin purification and allows the use of 10 times less amount of protein compared to the classical antibody-based chromatin immunoprecipitation (ChIP) experiment (40). Furthermore, it enables the application of more stringent washing conditions in order to improve purity. Therefore, the SnAvi-Tag should be applicable for ChIP-like approaches as well as for the identification of low-abundant protein complexes. The latter is of special importance because the mass action law sets a defined limit to the application of all other affinity tags, whose KD is many orders of magnitudes higher.
In addition to the AviTag the SnAvi-Tag encodes EGFP for the visualization of protein fusions in vivo. This allows the assessment of the expression and functionality of protein fusions. Only if green fluorescence is visible and is localized to the expected sub-cellular compartments the fusion protein can be expected to achieve the proper physiological function.
Another advantage of the AviTag for the work with multicellular model organisms is the requirement of an enzymatic activation of this tag. Since we can express the E. coli protein biotin ligase birA in a tissue specific manner, we can selectively purify proteins from a target cell type, regardless of the expression pattern of the SnAvi-tagged protein. This is of special importance for small model organisms, such as C. elegans, in which cell fractionation of post-embryonic stages is impossible. Until now, it was only possible to purify proteins from a specific subset of C. elegans cells when tissue specific promoters were used to express a tagged protein. Often, it is not an easy task to find a promoter that results in appropriate expression levels in the required tissue. With our tag system, it is now possible to use the endogenous promoter. The tissue specificity is obtained by the cell-specific expression of birA, since biotinylated SnAvi-tagged protein is produced only in those cells in which both promoters are active. The AviTag is not biotinylated by the birA C. elegans orthologue BPL-1 which was tested in preliminary experiments (data not shown). Compared to other enzymatically activated tag-systems, such as the Halo-Tag (41) or the SNAP-Tag (42), the SnAvi-Tag has one major advantage: its co-factor D-biotin is an essential vitamin and must be taken up by every cell to ensure its survival. Therefore, it is available in every cell of the organism. In contrast to this, the SNAP-tag and the Halo-tag need the presence of artificial small molecules to label the tagged protein. Thus, these tags can only be used if all cells internalize the required compounds. This sets serious limitations to the application of these tags in multicellular model organisms.
A relevant disadvantage of the AviTag is the presence of endogenously biotinylated proteins, such as carboxylases in all eukaryotic cells that will be co-purified with (strept-) avidin in the first affinity purification. However, a specific elution of the tagged protein from the first affinity matrix is already achieved by TEV-protease cleavage. This allows the elution of the tagged protein from the (strept-)avidin column material using non-denaturing conditions, thus preserving protein complexes.
In a second step, the tagged protein and its complex binding partners are further purified using the SB1 antibody. The SB1-producing hybridoma cell line is publicly shared and distributed. In this study, we mapped the epitope recognized by this antibody and introduced it as a part of the SnAvi-Tag. We could show that this antibody specifically recognizes the SB1-epitope without cross reactivity in diverse expression systems including E. coli, vertebrate cell culture, and C. elegans.
One major concern is steric hindering of native protein complex formation by tag structures. We propose that the long unfolded amino acid stretch in the SnAvi-Tag composed of two SB1 and two TEV-recognition sites should reduce such effects.
In order to demonstrate the efficacy of the SnAvi-Tag in C. elegans, we investigated the E3/E4 ubiquitin-protein ligase CHN-1 in body-wall muscle cells. These ligases typically function in the context of heterologous multiprotein complexes (43). Since we are interested in disease relevant functions of CHN-1 and other members of the ubiquitylation machinery (44–46) we selected this protein as a demonstration target. Together with CHN-1::SnAvi we could co-purify known as well as new complex components, including UBC-25. Vice versa, CHN-1 was co-precipitated using UBC-25::SnAvi (data not shown). Moreover, we were able to proof the interaction of UBC-25 and CHN-1 with a SnAvi-tag independent method in vivo. For this purpose, transgenic animals carrying CHN-1::Cerulean and UBC-25::Venus were generated and analysed in FRET experiments. Taken together, we have evidence of CHN-1 being indeed an interaction partner of UBC-25, demonstrating that the SnAvi-Tag is a suitable tool for protein complex purifications even from whole animals.
In summary, we could show that the SnAvi-Tag is efficient in a broad range of organisms in five basic applications: (i) for in vivo imaging of protein fusions; (ii) for immunoprecipitation with the SB1 antibody; (iii) for single-step affinity purification using (strept-)avidin; (iv) for tandem affinity purification employing in vivo biotinylation, streptavidin binding, mild TEV-protease elution and final purification using the SB1 antibody; and (5) for cell-type-specific protein interaction studies in small multicellular model organisms.
FUNDING
The German Research Foundation (grants SFB780 to R.B. and GRK843 to U.S.). R.B. was additionally supported by the Excellence Initiative of the German Federal and State Governments (EXC294 and FRIAS), and by Deutsche Forschungsgemeinschaft (CRC746, CRC780). K.M.M. was supported by the Excellence Initiative of the German Federal and State Governments (EXC 294). Funding for open access charge: Institutional funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Antje Thien, Meta Rath, Caroline Scherzinger, Birte Manßhard, Caroline Keck, Gregor Bochenek, Erika v. Gromoff, and Birgit Holzwarth for experimental contributions to this project, and Bettina Schulze and Julia Sämann for critically reading the manuscript. Plasmids encoding mCherry, Cerulean and Venus proteins were kindly provided by R. Tsien, D. W. Piston and A. Miyawaki, respectively. The SB1 hybridoma cell line was developed by Michael Nonet and Gayla Hadwiger. We obtained it from the Developmental Studies Hybridoma Bank, which was developed under the auspices of the NICHD and is maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. Some strains in this study were obtained from the Caenorhabditis Genetics Center (CGC), which is funded by the NIH National Centre for Research Resources (NCRR). We thank Agilent Technologies for supporting us with instrumentation.
REFERENCES
- 1.Polanowska J, Martin JS, Fisher R, Scopa T, Rae I, Boulton SJ. Tandem immunoaffinity purification of protein complexes from Caenorhabditis elegans. Biotechniques. 2004;36:778–780, 782. doi: 10.2144/04365BM05. [DOI] [PubMed] [Google Scholar]
- 2.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 3.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- 4.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates J.R., III, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 6.Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 7.Schimanski B, Nguyen TN, Gunzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai A, Carstens RP. An optimized protocol for protein purification in cultured mammalian cells using a tandem affinity purification approach. Nat. Protoc. 2006;1:2820–2827. doi: 10.1038/nprot.2006.371. [DOI] [PubMed] [Google Scholar]
- 9.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 10.Connelly PR, Varadarajan R, Sturtevant JM, Richards FM. Thermodynamics of protein–peptide interactions in the ribonuclease S system studied by titration calorimetry. Biochemistry. 1990;29:6108–6114. doi: 10.1021/bi00477a031. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T, Morone N, Kashiyama T, Oyamada H, Kurebayashi N, Murayama T. Engineering a novel multifunctional green fluorescent protein tag for a wide variety of protein research. PLoS ONE. 2008;3:e3822. doi: 10.1371/journal.pone.0003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laitinen OH, Hytonen VP, Nordlund HR, Kulomaa MS. Genetically engineered avidins and streptavidins. Cell. Mol. Life Sci. 2006;63:2992–3017. doi: 10.1007/s00018-006-6288-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane MD, Rominger KL, Young DL, Lynen F. The Enzymatic synthesis of holotranscarboxylase from apotranscarboxylase and (+)-biotin. Ii. Investigation of the reaction mechanism. J. Biol. Chem. 1964;239:2865–2871. [PubMed] [Google Scholar]
- 14.Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO, Templin MF, Pawlita M. Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins. Clin. Chem. 2005;51:1845–1853. doi: 10.1373/clinchem.2005.052381. [DOI] [PubMed] [Google Scholar]
- 15.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (NY) 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 17.Ashraf SS, Benson RE, Payne ES, Halbleib CM, Gron H. A novel multi-affinity tag system to produce high levels of soluble and biotinylated proteins in Escherichia coli. Protein Expr. Purif. 2004;33:238–245. doi: 10.1016/j.pep.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 18.Athavankar S, Peterson BR. Control of gene expression with small molecules: biotin-mediated acylation of targeted lysine residues in recombinant yeast. Chem. Biol. 2003;10:1245–1253. doi: 10.1016/j.chembiol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Duffy S, Tsao KL, Waugh DS. Site-specific, enzymatic biotinylation of recombinant proteins in Spodoptera frugiperda cells using biotin acceptor peptides. Anal. Biochem. 1998;262:122–128. doi: 10.1006/abio.1998.2770. [DOI] [PubMed] [Google Scholar]
- 20.Penalva LO, Keene JD. Biotinylated tags for recovery and characterization of ribonucleoprotein complexes. Biotechniques. 2004;37:604, 606, 608–610. doi: 10.2144/04374ST05. [DOI] [PubMed] [Google Scholar]
- 21.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Audhya A, Desai A. Proteomics in Caenorhabditis elegans. Brief Funct. Genomic Proteomic. 2008;7:205–210. doi: 10.1093/bfgp/eln014. [DOI] [PubMed] [Google Scholar]
- 23.Gottschalk A, Almedom RB, Schedletzky T, Anderson SD, Yates J.R., III, Schafer WR. Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. EMBO J. 2005;24:2566–2578. doi: 10.1038/sj.emboj.7600741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 2006;25:2178–2188. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D., Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 26.Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 27.Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 29.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 30.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 31.Schulze E, Altmann ME, Adham IM, Schulze B, Frode S, Engel W. The maintenance of neuromuscular function requires UBC-25 in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2003;305:691–699. doi: 10.1016/s0006-291x(03)00824-6. [DOI] [PubMed] [Google Scholar]
- 32.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mello C, Fire A. DNA transformation. Methods Cell. Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 34.Feige JN, Sage D, Wahli W, Desvergne B, Gelman L. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 2005;68:51–58. doi: 10.1002/jemt.20215. [DOI] [PubMed] [Google Scholar]
- 35.Rasband WS. (1997–2005) Image J. http://rsb.info.nih.gov/ij/, U. S. National Institutes of Health, Bethesda, MD, USA. [Google Scholar]
- 36.Nonet ML, Saifee O, Zhao H, Rand JB, Wei L. Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J. Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sämann J, Hegermann J, Gromoff EV, Eimer S, Baumeister R, Schmidt E. Caenorhabditis elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J. Biol. Chem. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt BZ, Watts RJ, Aridor M, Frizzell RA. Cysteine string protein promotes proteasomal degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) by increasing its interaction with the C terminus of Hsp70-interacting protein and promoting CFTR ubiquitylation. J. Biol. Chem. 2009;284:4168–4178. doi: 10.1074/jbc.M806485200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viens A, Mechold U, Lehrmann H, Harel-Bellan A, Ogryzko V. Use of protein biotinylation in vivo for chromatin immunoprecipitation. Anal. Biochem. 2004;325:68–76. doi: 10.1016/j.ab.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 42.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 43.Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size' doesn't; fit all. Trends Biochem. Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Nyamsuren O, Faggionato D, Loch W, Schulze E, Baumeister R. A mutation in CHN-1/CHIP suppresses muscle degeneration in Caenorhabditis elegans. Dev. Biol. 2007;312:193–202. doi: 10.1016/j.ydbio.2007.09.033. [DOI] [PubMed] [Google Scholar]
- 45.Hoppe T, Cassata G, Barral JM, Springer W, Hutagalung AH, Epstein HF, Baumeister R. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell. 2004;118:337–349. doi: 10.1016/j.cell.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Springer W, Hoppe T, Schmidt E, Baumeister R. A Caenorhabditis elegans Parkin mutant with altered solubility couples alpha-synuclein aggregation to proteotoxic stress. Hum. Mol. Genet. 2005;14:3407–3423. doi: 10.1093/hmg/ddi371. [DOI] [PubMed] [Google Scholar]