Abstract
A key step in meiotic recombination involves the nucleolytic resolution of Holliday junctions to generate crossovers. Although the enzyme that performs this function in human cells is presently unknown, recent studies led to the identification of the XPG-family endonuclease GEN1 that promotes Holliday junction resolution in vitro, suggesting that it may perform a related function in vivo. Here, we show that ectopic expression of GEN1 in fission yeast mus81Δ strains results in Holliday junction resolution and crossover formation during meiosis.
INTRODUCTION
Homologous recombination (HR) is a key process in chromosome biology. Its capacity to repair double strand breaks (DSBs) in DNA serves to maintain genome integrity, promote correct chromosome segregation during meiosis through the formation of chiasmata, and generate genetic diversity by creating new allelic combinations in the germline. The central intermediate of HR is a four-way DNA junction structure that physically connects the two recombining DNA molecules (1). These structures, known as Holliday junctions (HJs), have to be resolved so that the DNA molecules can segregate at cell division. Resolution is achieved by cleavage of a pair of strands at the junction centre, with the choice of strands determining whether crossover or non-crossover recombinants are made. Crossovers consist of a reciprocal exchange of DNA flanking the site of HJ resolution, and in meiosis, when the recombining molecules are the homologous chromosomes, give rise to chiasmata and allelic reassortment (2–5).
HJ resolution is catalysed by specialized nucleases, examples of which were first identified in bacteria and their phages (6–8). More recently, the Mus81-Eme1 complex, a member of the XPF-family of structure-specific endonucleases, was implicated in HJ resolution in eukaryotes (9,10). However, whilst Mus81-Eme1 can cleave HJs in vitro, the manner in which it does so is not representative of a canonical resolvase like Escherichia coli RuvC or RusA, which cleave HJs symmetrically to generate nicked linear duplex products that are repairable by DNA ligase (11–13). Moreover, Mus81-Eme1's preferred substrates are HJ precursors (displacement [D] loops and unligated HJs) rather than fully fledged HJs (14–16).
Most recently, two novel HJ resolvases have been identified in humans, which like RuvC and RusA, cleave synthetic HJs symmetrically. These are GEN1 (17,18) and SLX1-SLX4 (19–21). In the case of SLX1-SLX4 genetic data indicate that it plays an important role in promoting the repair of DSBs and interstrand crosslinks (ICLs) (19–21), and in Drosophila the orthologue of SLX4, MUS312, is critical for meiotic crossover formation (22). GEN1’s potential involvement in DNA repair and crossover formation has yet to be determined. However, genetic analysis of its orthologue in Saccharomyces cerevisiae Yen1 shows how at least one member of this family can play a redundant role with Mus81 in enabling the repair of ICLs and lesions that perturb replication fork progression (M.G. Blanco, J. Matos, U. Rass, S.C. Ip and S.C. West, manuscript submitted for publication). Here, we investigate whether human GEN1 has the ability in vivo to promote crossover formation by testing whether it can substitute for Mus81-Eme1 in promoting meiotic recombination in Schizosaccharomyces pombe.
MATERIALS AND METHODS
Yeast strains and plasmid construction
S. pombe strains used for this study are listed in Table 1. To express wild-type and mutated versions of GEN1 in fission yeast the following plasmids were constructed: From pDONR221-GEN1(1-527), pCMV6-GEN1(D30A) and pCMV6-GEN1(E134A/E136A), respectively (17), the open reading frames were amplified by PCR with Pfu polymerase (Stratagene, CA, USA) adding a NcoI-site upstream and a stop codon as well as a BamHI-site downstream of the gene (the FLAG-tags in the original constructs were omitted). These constructs were cloned into the pREP41-3HAN vector (23) (the NcoI–BamHI restriction endonuclease digest removes the N-terminal 3HA-tag entirely), to put GEN1 under the control of the thiamine-repressible nmt1-promotor. To express fission yeast Rqh1 from the nmt1-promotor the rqh1, open reading frame was amplified by PCR using Pfu polymerase from genomic DNA of the S. pombe strain MCW1221 adding SalI sites up and downstream of the gene and cloned into pREP41 (24). All resulting plasmids—pGEN1 (wild-type GEN1), pGEN1DA (GEN1 carrying the D30A point mutation), pGEN1EA/EA (GEN1 carrying the E134A and E136A point mutations) and pMW563 (pRqh1)—were sequenced to ensure that no mutations had been introduced.
Table 1.
Schizosaccharomyces pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| MCW1019 | h+ rqh1Δ::kanMX6 ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| FO808 | h− ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1221 | h+ ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1237 | h− mus81Δ::kanMX6 ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW1238 | h+ mus81Δ::kanMX6 ura4-D18 leu1-32 his3-D1 arg3-D4 | Lab strain |
| MCW3200/ALP731 | h−smt0 his3+-aim ade6-L469 ura4-D18 his3-D1 arg3-D4 | This study |
| MCW3202/ALP733 | h+S ura4+-aim2 ade6-3083 ura4-D18 leu1-32 his3-D1 | This study |
| FO1267 | h− his3+-aim ade6-L469 ura4-D18 leu1-32 his3-D1 | Lab strain |
| MCW3514/ALP802 | h+S mus81Δ::kanMX6 ura4+-aim2 ade6-3083 ura4-D18 leu1-32 his3-D1 | This study |
| MCW3589/ALP822 | h−smt0 mus81Δ::kanMX6 his3+-aim ade6-L469 ura4-D18 his3-D1 arg3-D4 | This study |
| FO1260 | h− mus81Δ::kanMX6 his3+-aim ade6-L469 ura4-D18 leu1-32 his3-D1 | Lab strain |
Plasmids pREP41 (24), pRusA (pMW437, pREP1-NLS-rusA-GFP) (25), pRqh1 (pREP41-Rqh1), pMus81* (pMW592, pREP41-2myc6his-Mus81–Pk-Eme1) (15), pGEN1, pGEN1DA and pGEN1EA/EA were transformed into fission yeast strains MCW1019, FO808, MCW1221, MCW1237 and MCW1238, the resulting strains were used for spot assays and for spore viability testing. For the meiotic recombination assay, the empty vector pREP41 was transformed into the wild-type control strains MCW3202 and FO1267; the mus81Δ strains MCW3514 and FO1260 were transformed with pGEN1, pRusA and pMus81* (15).
Culture conditions and genetic methods
Yeast cells were cultured in YES broth and on YES plates, unless they contained plasmids, in which case the cells were grown in EMM2 broth and on EMM2 agar plates containing the required supplements and using the monosodium salt of l-glutamic acid as the nitrogen source instead of NH4Cl (http://www-rcf.usc.edu/∼forsburg/media.html). Sporulation of the crosses MCW3202 × MCW3200 and MCW3514 × MCW3589 were performed on ME agar and for all the crosses with strains containing plasmids on SPAS agar (26). Spot assays (25), determination of spore viability by random spore analysis (26) and the meiotic recombination assay (14,15) have been previously described in detail.
Protein extraction and western blotting
Whole-cell extracts were prepared as detailed before (27) and resolved on 12.5% SDS–PAGE gels, transferred to a Trans-Blot transfer medium membrane (Bio-Rad Laboratories, Hercules, CA, USA) and probed with an anti-GEN1 antibody (raised against the N-terminal 527AA of the protein; dilution 1:1000) or with anti-ß-actin (ab8224; Abcam plc, Cambridge, UK; dilution 1:2000) as a loading control. Blocking of membranes and antibody incubations using the anti-GEN1 antibody were performed in 1× PBS containing 5% skimmed milk, 2.5% BSA, 100 mM l-lysine and 0.1% Tween 20, all washes were done with 0.1% Tween 20 in 1× PBS. For the anti-β-actin immunoblotting 1× TBS containing 5% skimmed milk, 2.5% BSA, 100 mM l-lysine and 0.1% Tween 20 was used for blocking and 1× TBS containing 0.5% skimmed milk, 0.25% BSA, 10 mM L-lysine and 0.1% Tween 20 for the antibody incubations, all washes were done using 0.1% Tween 20 in 1× TBS. Following incubation with an appropriate secondary antibody conjugated to horse-radish peroxidase at a dilution of 1:20 000 the proteins were detected using Amersham ECL Western Blotting Detection Reagents (GE Healthcare UK Ltd., Little Chalfont, UK). Membranes were then exposed to Super RX medical X-ray film (Fujifilm, Tokyo, Japan).
Statistics
Statistical analysis for the recombination data was performed in Excel (Microsoft Office), in Matlab 7 (The MathWorks, UK) and on http://www.physics.csbsju.edu/stats/KS-test.html. First each data set was tested for normal distribution using a one-sample Kolmogorov–Smirnov goodness-to-fit test, rejecting the null hypothesis (H0; ‘data fits a normal distribution’) at an α-level of P < 0.05. Almost all the data were consistent with a normal distribution and were compared with the appropriate control experiment using a two-tailed, unpaired, two-sample t-test, assuming either equal (homoskedastic) or unequal variance (heteroskedastic) depending on the outcome of an F-test (H0 ‘data sets having an equal variance’ rejected at an α-level of P < 0.05). One data set did not follow a normal distribution and was therefore tested against the control using a two-sample Kolmogorov–Smirnov test, which is nonparametric and does not depend on the normal distribution of data sets. The P-values are presented in Supplementary Table S2, together with an indication of the type of test used for each comparison. H0 (‘data sets being similar’) was rejected at an α-level P < 0.01.
RESULTS
To determine whether GEN1 is capable of working in a meiotic in vivo environment, we made use of the fission yeast S. pombe. This is a useful eukaryotic system for validating HJ resolvases in vivo because the resolution of HJs, or their precursors, during meiotic recombination relies almost entirely on Mus81-Eme1 (28). Consequently, HJs accumulate in a mus81Δ mutant (28). Moreover, it has previously been shown that the ectopic expression of the bacterial HJ resolvase RusA can rescue vegetative and/or meiotic defects associated with HJ accumulation when Mus81-Eme1 or the RecQ-type helicase Rqh1 are deleted (9,15,25,29).
GEN1 can partially rescue the genotoxin sensitivities of rqh1Δ and mus81Δ mutants
Since RusA partially rescues different genotoxin sensitivities of vegetative yeast cells lacking Mus81 or Rqh1 (25,29), we first tested whether GEN1 could do likewise. Plasmid constructs carrying human GEN1 (amino acids 1-527), NLS-RusA-GFP (25), Rqh1 and 2myc6his-Mus81–Pk-Eme1 (15) under the thiamine-repressible nmt1-promotor were transformed into wild type, rqh1Δ and mus81Δ (see ‘Materials and Methods’ section). The resulting strains, carrying an empty vector or any of the above plasmids, were tested for sensitivities against the topoisomerase I poison camptothecin (CPT), the alkylating agent methyl methanesulfonate (MMS) and ultraviolet irradiation (UV).
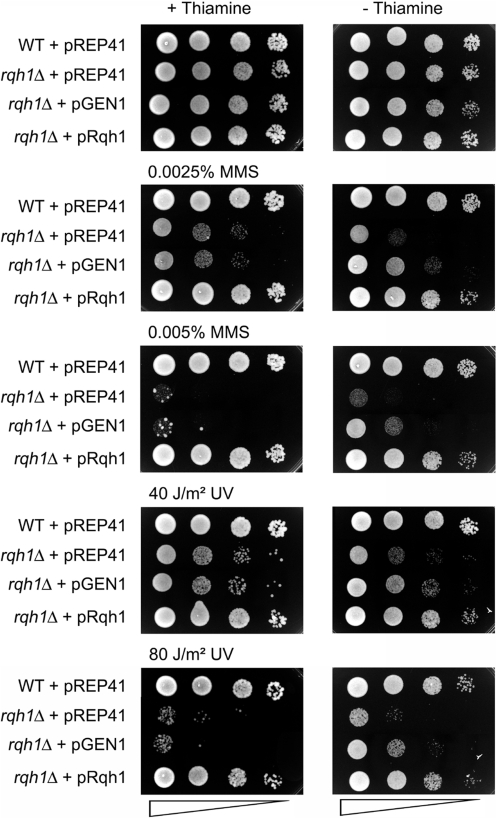
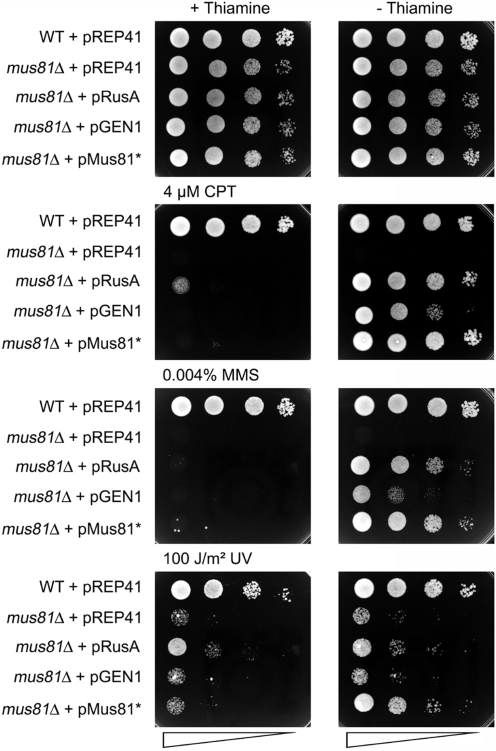
Expression of GEN1 partially rescued the sensitivities of rqh1Δ against UV and MMS (Figure 1) and mus81Δ against MMS and CPT (Figure 2, Supplementary Figure S1). However, it was unable to rescue the UV sensitivity of a mus81Δ mutant (Figures 2, Supplementary Figure S1). The degree of rescue by GEN1 was not as complete as by complementation of the mutants with plasmid-expressed Rqh1/Mus81-Eme1 (Figures 1 and 2, Supplementary Figure S1) and in the case of mus81Δ it was also not as great as achieved with RusA expression (Figure 2, Supplementary Figure S1). GEN1’s ability to only partially rescue the genotoxin sensitivities of mus81Δ and rqh1Δ is not a consequence of any overt toxicity associated with its expression, because in a wild-type it causes only a modest increase in UV sensitivity (Supplementary Figure S2). Overall, these data establish that GEN1 can at least partially substitute for Rqh1 and Mus81 in promoting resistance to genotoxins.
Figure 1.
Effect of GEN1 and Rqh1 (expressed from the thiamine-repressible nmt1-promotor in pREP41) on genotoxin sensitivities of a rqh1Δ (MCW1019) strain. The empty vector pREP41 in the wild-type strain MCW1221 and in MCW1019 serve as controls. The neat spot represents 105 cells.
Figure 2.
Effect of RusA (expressed from the thiamine-repressible nmt1-promotor in pREP1), GEN1 and Mus81-Eme1 (expressed from the thiamine-repressible nmt1-promotor in pREP41) on genotoxin sensitivities of a mus81Δ (MCW1238) strain. The empty vector pREP41 in the wild-type strain MCW1221 and in MCW1238 serve as controls. The neat spot represents 105 cells.
GEN1’s nuclease activity is required to rescue the genotoxin sensitivities of rqh1Δ and mus81Δ mutants
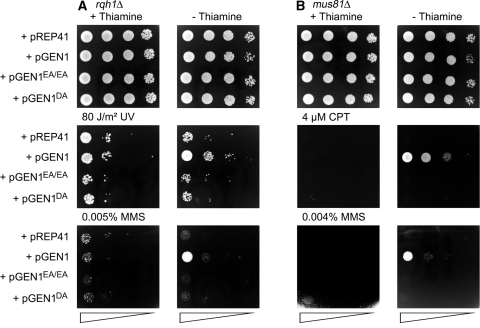
To determine whether GEN1’s catalytic activity is needed for its rescue of mus81Δ and rqh1Δ genotoxin sensitivities, we repeated the above experiments using plasmids that express nuclease-dead mutants of GEN1 (GEN1E134A/E136A and GEN1D30A). As shown in Figure 3, neither mutant protein can rescue the genotoxin sensitivities of rqh1Δ or mus81Δ strain. These data suggest that it is GEN1’s HJ resolution activity that is critical for its ability to rescue mus81Δ and rqh1Δ, which in turn is consistent with the idea that unresolved HJs accumulate in these mutants.
Figure 3.
Effect of wild-type and nuclease-defective GEN1 (expressed from the thiamine-repressible nmt1-promotor in pREP41) on genotoxin sensitivities of (A) rqh1Δ (MCW1019) and (B) mus81Δ (MCW1238) strains. pREP41 serves as the empty vector control. The neat spot represents 105 cells.
GEN1 can rescue the meiotic defects of a mus81Δ mutant
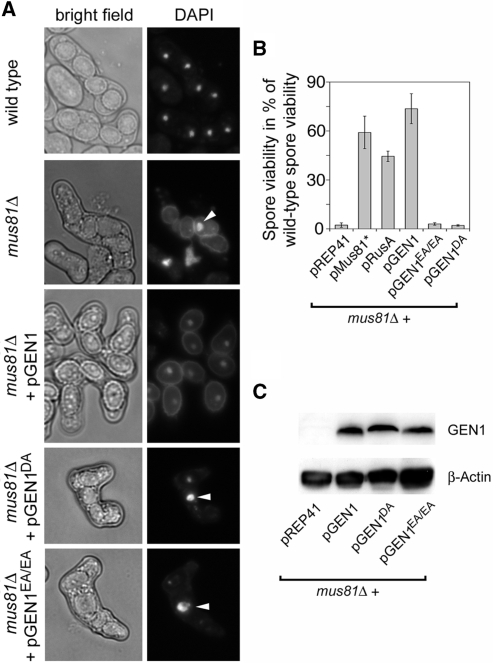
Having established that GEN1 can partially compensate for the lack of Rqh1 and Mus81 in the tolerance/repair of DNA damage, we next looked to see whether it could rescue the meiotic defects of a mus81Δ mutant. In S. pombe, the deletion of mus81 causes a very severe meiotic phenotype, where the meiotic nuclear division cannot proceed due to unresolved HJs (9,14,28). The resulting asci, despite holding up to four spores, very often contain only a single DNA mass, and therefore many of the spores are completely devoid of DNA (Figure 4A). Consequently spore viability is very low (Figure 4B). Expression of GEN1 from the nmt1-promotor ameliorates both the chromosome segregation defect and the poor spore viability of a mus81Δ mutant just as well as does RusA or Mus81-Eme1 (Figure 4, Supplementary Table S1). As with its rescue of genotoxin sensitivities, suppression of mus81Δ's meiotic defects depends on GEN1’s nuclease activity because neither GEN1E134A/E136A nor GEN1D30A expression provided any amelioration (Figure 4, Supplementary Table S1).
Figure 4.
Effect of expression of RusA, as well as wild-type and nuclease-defective GEN1 on mus81Δ during meiosis. (A) Typical examples of asci from crosses of wild-type and mus81Δ strains carrying plasmids as indicated. The DNA is stained with DAPI and arrowheads indicate non-segregated DNA. (B) Restoration of spore viability in a mus81Δ mutant by pMus81*, pRusA and pGEN1. See also Supplementary Table S1. (C) Western blot showing protein levels of wild-type and nuclease-defective GEN1 in cultures of mus81Δ strains carrying the respective plasmids, as indicated, grown for 20 h without thiamine. β-actin serves as loading control.
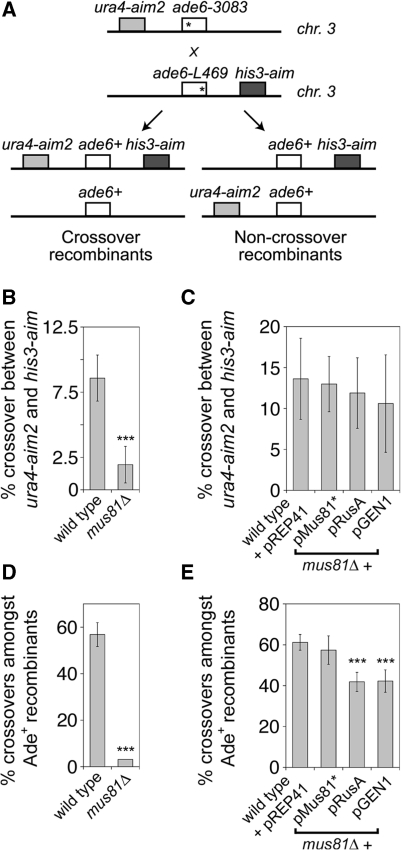
The resolution of HJs and/or their precursors by Mus81-Eme1 generates crossover recombinants during meiosis. To see if GEN1 is capable of producing crossovers as well, the wild-type pGEN1 construct was transformed into mus81Δ strains carrying a meiotic recombination reporter substrate in which gene conversion is measured at the ade6 locus using the ade6-3083 hotspot (30) and crossing over is assessed by two flanking auxotrophic markers, ura4-aim2 and his3-aim (Figure 5A). Similar to previous data at ade6-M26, loss of mus81 reduces crossover formation between ura4-aim2 and his3-aim by at least 4-fold (Figure 5B) (14). This can be rescued back to wild-type levels by expression of GEN1, RusA or Mus81-Eme1 (Figure 5C). In wild-type fission yeast, there is a general bias to resolve meiotic gene conversion events into crossover recombinants. As such >55% of gene conversions at ade6 are associated with a crossover of the flanking markers (Figure 5D). Importantly, unlike the wild type, or the mus81Δ mutant complemented by Mus81-Eme1 expressed from a plasmid, expression of GEN1 or RusA results in crossing over in only ∼40% of gene conversion events (Figure 5E), which is essentially the same as that determined previously for RusA at the ade6-M26 hotspot (15). Presumably, both GEN1 and RusA resolve the HJs that accumulate in a mus81Δ mutant without bias for either crossover or non-crossover recombinant, whereas Mus81-Eme1’s ability to act on HJ precursors enables it to establish a crossover bias (3). The reason that ∼40% of gene conversion events are associated with a crossover rather than 50% is probably because ∼20% of gene conversion events occur via synthesis-dependent strand annealing (SDSA) (see ‘Discussion’ section). These data highlight an important difference between canonical HJ resolvases, like RusA and GEN1, and Mus81-Eme1 in dealing with recombination junctions.
Figure 5.
Effect of expression of RusA and wild-type GEN1 on meiotic recombination in a mus81Δ background. (A) Schematic representation of the meiotic recombination assay and the possible recombinants associated with gene conversions at ade6 (note that crossover recombinants in the ura4-aim2–his3-aim interval can also arise without an associated gene conversion event at ade6). Asterisks indicate the position of point mutations in the ade6− alleles. (B and C) Percentage of crossovers between ura4-aim2 and his3-aim in the indicated strains. (D and E) Percentage of crossovers between ura4-aim2 and his3-aim associated with a gene conversion at ade6-3083 that produces an Ade+ prototroph. Data in (B–E) are mean values from at least 10 independent crosses with error bars being standard deviations about the mean. Statistical significance at ***P < 0.001 against the wild-type control within each chart (for details see Supplementary Table S2).
DISCUSSION
We have investigated whether GEN1 has the ability to resolve HJs in vivo by testing it in a heterologous system under three different conditions where HJs are expected, or have been shown, to accumulate. In the first case, we asked whether GEN1 could rescue the hypersensitivity of a rqh1Δ mutant to two different genotoxins, UV and MMS, both of which are known to cause lesions in DNA that perturb replication fork progression and promote Rad51-dependent recombination (31–33). In S. cerevisiae, the RecQ-type helicase Sgs1 has been implicated in processing Rad51-dependent DNA junctions that can form between sister chromatids following replication fork perturbation (31,32,34,35). Such processing is thought to include HJ branch migration, which when combined with topoisomerase III(α) activity can result in the dissolution of double HJs (36). In the absence of this activity, HJs can accumulate and impede chromosome segregation (34). This may account for the high frequency of aberrant mitoses in a rqh1Δ mutant following replication fork stalling (37). Importantly, RusA partially rescues this chromosome segregation defect in S. pombe, together with associated genotoxin sensitivities, presumably by resolving HJs that would otherwise physically prevent sister chromatid separation (25). We suspect that the amelioration of rqh1Δ's; UV and MMS sensitivities by GEN1 expression is similarly associated with the resolution of HJs that would otherwise impede chromosome segregation and cause cell death.
In the second case, we showed that, like RusA, GEN1 can partially rescue the hypersensitivity of a mus81Δ mutant to MMS and CPT. Mus81-Eme1 can cleave quite a wide range of substrates in vitro, including 3′ flaps, forked DNAs, and four-way DNA junctions including fully ligated HJs (9,14–16,29,38). Exactly which of these substrates is targeted in vivo to promote the repair and/or tolerance of DNA damage is still a matter of conjecture (39). However, at least in the case of replication fork breakage induced by CPT, a strong argument can be made for Mus81-Eme1 being required for resolving the single four-way DNA junction (D-loop/unligated HJ/ligated HJ) that is formed during the repair of the broken fork by Rad51 (29,39,40). Moreover, the fact that both RusA and GEN1 have a preference for cleaving HJs in vitro (17,41), makes it likely that their partial suppression of mus81Δ mutant genotoxin sensitivity is directly as a result of this ability in vivo.
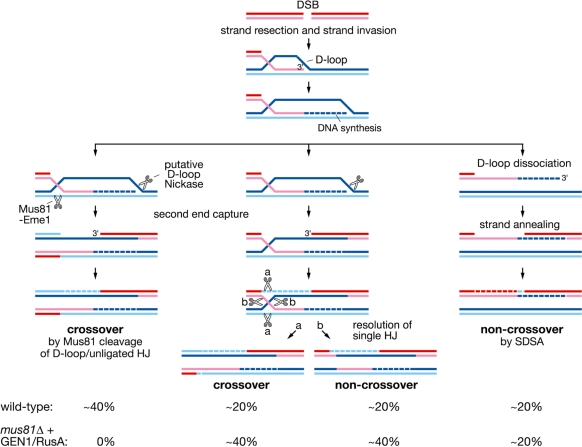
The third, and most compelling, case for GEN1 being able to resolve HJs in vivo is its ability to promote crossover formation during meiosis in a mus81Δ mutant. In S. pombe, there is strong evidence that Mus81-Eme1 is the principal enzyme responsible for resolving meiotic recombination intermediates (9,14,28,42). Moreover, it resolves these intermediates with a marked bias for generating crossover recombinants. HJs are essentially symmetrical structures, and can therefore be cleaved by HJ resolvases in either orientation to generate equal numbers of crossover and non-crossover products. This lack of cleavage bias was recently shown to hold true in E. coli where each of the known resolution systems (i.e. RecG-, RuvABC- and RusA-dependent) produced approximately equal proportions of crossovers and non-crossovers during DSB repair (43). What guides Mus81-Eme1 to deliver junction resolution with a bias for crossover recombinants is not certain; however, one simple model is that it resolves D-loops and unligated HJs, which are the precursors of fully ligated HJs (14). These structures contain a 5′ DNA end at or close to the junction centre that directs Mus81 cleavage to a specific strand, which, in the context of DSB repair, generates only crossovers (14) (Figure 6). Clearly not all gene conversion events in S. pombe are associated with a crossover, which implies that resolution does not always proceed via Mus81-Eme1-mediated cleavage of D-loops/unligated HJs. It is likely that some DSB repair occurs via SDSA, which is a form of HR that does not necessitate HJ formation and resolution and generates only non-crossovers (Figure 6). It is also possible that some D-loops/unligated HJs evade early processing and mature into fully fledged HJs that are cleaved by Mus81-Eme1 without a bias for generating crossovers (15) (Figure 6).
Figure 6.
Pathways of meiotic interhomologue DSB repair in S. pombe. Here, we have invoked the existence of a D-loop nickase to account for the fact that mostly single HJs accumulate in a mus81Δ mutant (28). However, in the absence of such an activity, Mus81-Eme1 could perform essentially the same cleavage following second end capture (14). Regardless of whether single and/or double HJs accumulate in a mus81Δ mutant, both GEN1 and RusA appear to be able to resolve them efficiently into either crossover or non-crossover products. The bottom panel shows the percentage of interhomologue DSB repair events at the ade6 locus that could proceed via each of the postulated pathways to account for the observed crossover values in wild-type and GEN1/RusA-rescued mus81Δ mutant cells.
In contrast to Mus81-Eme1, RusA does not utilize a DNA end to guide strand cleavage of four-way DNA junctions, and therefore appears to have no intrinsic mechanism for generating a crossover bias (14,15). We suspect the same is true for GEN1. Moreover these enzymes exhibit preferences for specific nucleotide sequences being present at the junction centre for efficient strand cleavage (12,17). This property makes it more likely that the four-way junction has migrated and become a fully fledged HJ before being resolved by them (6). As such both RusA and GEN1 should deliver equal numbers of crossover and non-crossover recombinants in a mus81Δ mutant. Therefore, the fact that both of them produce ∼40% crossovers amongst Ade+ convertants, suggests that overall ∼80% of interhomologue recombination events at this locus proceed via junction resolution (Figure 6). We presume that the other ∼20% are SDSA events. These data also imply that in a wild-type Mus81-Eme1 is responsible for processing ∼80% of interhomologue recombination intermediates. So the fact that it generates only ∼60% of crossovers amongst Ade+ convertants suggests that ∼40% of events may involve HJ cleavage generating equal numbers of crossovers and non-crossovers as proposed previously (15) (Figure 6).
In summary, we have provided evidence that GEN1 can work as a HJ resolvase in vivo. Moreover, we have shown that in a meiotic environment it can generate crossovers, and thereby is a potential candidate for performing this function in humans. However, like RusA, and other canonical HJ resolvases, GEN1 does not appear to have an intrinsic mechanism for producing a crossover bias during DSB repair. So if HJs in humans are resolved predominantly into crossover recombinants, like in S. cerevisiae (44,45), additional factors would be needed to ensure that GEN1 delivers such a bias.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Senior Research Fellowship from the Wellcome Trust (to M.C.W.); Erwin Schrödinger-Fellowship from the Austrian Science Fund (FWF; to A.L.) in part. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank F. Osman and W. W. Steiner for providing strains.
REFERENCES
- 1.Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 2.Lorenz A, Whitby MC. Crossover promotion and prevention. Biochem. Soc. Trans. 2006;34:537–541. doi: 10.1042/BST0340537. [DOI] [PubMed] [Google Scholar]
- 3.Whitby MC. Making crossovers during meiosis. Biochem. Soc. Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 4.Mézard C, Vignard J, Drouaud J, Mercier R. The road to crossovers: plants have their say. Trends Genet. 2007;23:91–99. doi: 10.1016/j.tig.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 6.Whitby MC. Holliday junction resolution. In: Aguilera A, Rothstein RJ, editors. Molecular Genetics of Recombination. Topics in Current Genetics. Vol. 17. Berlin - Heidelberg, Germany: Springer; 2007. pp. 169–199. [Google Scholar]
- 7.Déclais AC, Lilley D.MJ. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr. Opin. Struct. Biol. 2008;18:86–95. doi: 10.1016/j.sbi.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Lilley DMJ, White MF. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2001;2:433–443. doi: 10.1038/35073057. [DOI] [PubMed] [Google Scholar]
- 9.Boddy MN, Gaillard P.-HL, McDonald WH, Shanahan P, Yates J.R., III, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 10.Chen X.-B, Melchionna R, Denis C.-M, Gaillard P.-HL, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 11.Connolly B, Parsons CA, Benson FE, Dunderdale HJ, Sharples GJ, Lloyd RG, West SC. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc. Natl Acad. Sci. USA. 1991;88:6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SN, Harris L, Bolt EL, Whitby MC, Lloyd RG. Sequence specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J. Biol. Chem. 1997;272:14873–14882. doi: 10.1074/jbc.272.23.14873. [DOI] [PubMed] [Google Scholar]
- 13.Dunderdale HJ, Benson FE, Parsons CA, Sharples GJ, Lloyd RG, West SC. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- 14.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 15.Gaskell LJ, Osman F, Gilbert R.JC, Whitby MC. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillard PHL, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves holliday junctions by a nick and counternick mechanism. Mol. Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 17.Ip SCY, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 18.West SC. The search for a human Holliday junction resolvase. Biochem. Soc. Trans. 2009;37:519–526. doi: 10.1042/BST0370519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates J.R., III, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Svendsen JM, Smogorzewska A, Sowa ME, O'C;onnell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craven RA, Griffiths D.JF, Sheldrick KS, Randall RE, Hagan IM, Carr AM. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- 24.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 25.Doe CL, Dixon J, Osman F, Whitby MC. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuo Y, Asakawa K, Toda T, Katayama S. A rapid method for protein extraction from fission yeast. Biosci. Biotechnol. Biochem. 2006;70:1992–1994. doi: 10.1271/bbb.60087. [DOI] [PubMed] [Google Scholar]
- 28.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 30.Steiner WW, Smith GR. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2005;169:1973–1983. doi: 10.1534/genetics.104.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicolli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mankouri HW, Ngo H.-P, Hickson ID. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell. 2007;18:4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osman F, Adriance M, McCready S. The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 2000;38:113–125. doi: 10.1007/s002940000145. [DOI] [PubMed] [Google Scholar]
- 34.Mankouri HW, Ngo HP, Hickson ID. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009;20:1683–1694. doi: 10.1091/mbc.E08-08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, Hickson ID. The Bloom's; syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 37.Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom's; and Werner's; syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 39.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolt EL, Lloyd RG. Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell. 2002;10:187–198. doi: 10.1016/s1097-2765(02)00560-9. [DOI] [PubMed] [Google Scholar]
- 42.Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grove JI, Harris L, Buckman C, Lloyd RG. DNA double strand break repair and crossing over mediated by RuvABC resolvase and RecG translocase. DNA Repair. 2008;7:1517–1530. doi: 10.1016/j.dnarep.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 44.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 45.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.