Abstract
Despite recent advances in sequencing, complete finishing of large genomes and analysis of novel proteins they encode typically require cloning of specific regions. However, many of these fragments are extremely difficult to clone in current vectors. Superhelical stress in circular plasmids can generate secondary structures that are substrates for deletion, particularly in regions that contain numerous tandem or inverted repeats. Common vectors also induce transcription and translation of inserted fragments, which can select against recombinant clones containing open reading frames or repetitive DNA. Conversely, transcription from cloned promoters can interfere with plasmid stability. We have therefore developed a novel Escherichia coli cloning vector (termed ‘pJAZZ’ vector) that is maintained as a linear plasmid. Further, it contains transcriptional terminators on both sides of the cloning site to minimize transcriptional interference between vector and insert. We show that this vector stably maintains a variety of inserts that were unclonable in conventional plasmids. These targets include short nucleotide repeats, such as those of the expanded Fragile X locus, and large AT—rich inserts, such as 20-kb segments of genomic DNA from Pneumocystis, Plasmodium, Oxytricha or Tetrahymena. The pJAZZ vector shows decreased size bias in cloning, allowing more uniform representation of larger fragments in libraries.
INTRODUCTION
Nearly complete genomic sequence is available for over 1300 eukaryotic and 4400 prokaryotic organisms (1) (http://www.genomesonline.org/). ‘Next-generation’ sequencing technologies are rapidly expanding this database, without the need to construct genomic DNA libraries (2). However, cloning and library construction are still critical for de novo assembly of novel genomes and for gap closure of larger genomes (3). Accurate cloning is also essential for functional analysis of entirely novel genes.
Unfortunately, many of the sequenced coding regions as well as gaps in the assemblies are difficult to capture in conventional circular plasmids. For example, the ‘finished’ human genome contained at least 241 gaps within the euchromatic sequence, which were not detected in BAC, phage P1, fosmid or cosmid libraries (4,5). Many of the uncloned regions were also missing from ‘next-generation’ sequence assemblies, presumably due to their highly repetitive nature (5–7). Significantly, rapidly evolving genes specific to humans can be found in some of these regions (8,9). Skewed sequence composition also can impede cloning in E. coli. Very AT—rich genomes are notoriously difficult to clone, such as that of the slime mold Dictyostelium discoideum (10), the malaria parasite Plasmodium (11–13) (80–85% AT) and the fungal pathogen Pneumocystis carinii (75% AT) (http://pgp.cchmc.org/html/genome_pro_clonelib.html). Similar difficulties are likely for the genomes of other parasitic organisms, as they are often AT—rich (14,15).
Difficulties in cloning consequently prevent in vitro expression and functional analysis of newly discovered Open Reading Frames. The Mammalian Gene Collection (MGC) was a high-throughput endeavor by the National Institutes of Health to clone and express ∼22 000 putative protein-coding cDNAs from the human genome (16) (http://mgc.nci.nih.gov/, 2009). Years of effort yielded ∼18 000 unique clones. The remaining genes were specifically targeted by PCR or gene synthesis, yet nearly 1600 of the genes were never recovered (16). Included among these are genes for over 100 G protein-coupled receptors, a family of proteins that are involved in a wide range of cell signal transduction processes. Possible explanations for the inability to capture the cDNAs are that they were toxic to bacteria, contained repetitive sequences or were longer than 4–6 kb (17).
Instability of cloned fragments is exacerbated by features of typical cloning vectors (18). Transcription of the cloned DNA may result in selection against open reading frames encoding toxic proteins or it may cause rearrangement or deletion of repetitive sequences, such as di- or tri-nucleotide repeats or poly-T tracts (19–21). In addition, active promoters within the insert can transcribe into the vector backbone, interfering with plasmid replication or expression of the selectable marker (20,22). High copy number of the vector can reduce the ability to clone large fragments (e.g. >8 kb) or regions with strong secondary structure (23,24). Plasmid supercoiling can induce cruciforms and other secondary structures, favoring deletions or rearrangements (25,26).
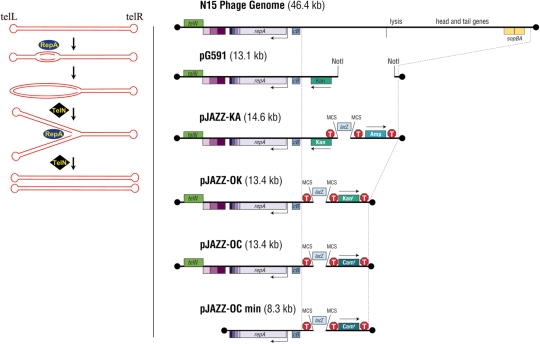
To circumvent some of these issues, the first generation linear vectors ‘pG591’ and ‘pN15L’ were constructed (27,28). These vectors were based upon the coliphage N15, which has a linear, dsDNA genome. Upon infection, the N15 phage DNA normally becomes circularized via its cohesive ends. A unique phage enzyme termed protelomerase (TelN) cuts at a specific site, telRL, linearizing the phage DNA. TelN subsequently seals each newly cut end, generating covalently closed ‘hairpin’ ends (29).
During lysogeny, the prophage replicates as a linear dsDNA molecule, maintaining the covalently closed ends (Figure 1) (27,30). Priming of N15 replication is carried out by the N15 RepA protein (31) and the phage genome is replicated by E. coli DNA polymerase. After replication of the ends, TelN cleaves the duplicated palindromic site created at each telomere. It again seals the cut ends to recreate the characteristic closed hairpin structure (30). pG591 and pN15L utilize the replication and partitioning system of N15, but they lack the structural and lytic genes.
Figure 1.
Replication and structure of phage N15 and linear pJAZZ vectors. Left panel: Replication of phage N15 and the pJAZZ vectors requires the phage proteins RepA and TelN. Bidirectional replication is initiated by RepA within the repA gene. TelN cleaves the newly replicated telomeres to re-create the closed hairpin ends. Right panel: The lysis and structural genes of N15 were deleted to create pG591. The vector pJAZZ-KA was derived from pG591 by insertion of a lacZ-alpha ‘stuffer’ fragment, situated between a pair of identical multiple cloning sites. A drug resistance gene was added to the right arm. The vectors pJAZZ-OK, pJAZZ-OC and pJAZZ-OCmin were derived from the pJAZZ-KA vector. The lacZ-alpha stuffer is removed before ligation to target DNAs. Transcription is indicated by arrows and transcriptional terminators by ‘T’. Dark balls represent the hairpin telomeres; cB, regulator of replication; MCS, Multiple Cloning Site (SmaI, NotI, AflII … LacZ-alpha … AflII, NotI, SmaI).
Here we describe a second-generation linear cloning system, the ‘pJAZZ’ series of transcription-free, linear cloning vectors. We have also generated a host cell strain, E. coli ‘TSA’, that provides efficient transformation with the pJAZZ vectors. We show that the pJAZZ vectors in the E. coli TSA strain can stably maintain templates that are difficult or impossible to clone in circular vectors, including AT-rich inserts of up to 30 kb and short tandem repeats of up to 2 kb.
MATERIALS AND METHODS
Construction of the E. coli TSA host strain
The telN gene from phage N15 was amplified by PCR and cloned into the expression vector pGZ119EH (32), under control of isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible Ptac promoter. The Ptac-telN fragment was excised and subcloned into the chromosome-integration vector pJW22 (33). A fragment containing the sop operon of phage N15 under control of its own promoter and the antA antirepressor gene under control of the arabinose-inducible araPBAD promoter were excised from the plasmid pCD31sop (34). The sop-antA fragment was sub-cloned into the integration plasmid with the Ptac-telN fragment.
The resulting plasmid was partially digested with NotI to excise a fragment containing telN-sopAB-antA and the vector’s β-lactamase gene (bla; ampR). The fragment was purified by gel-electrophoresis, circularized by self-ligation and transformed into E. cloni® 10G cells (Lucigen, Middleton, WI, USA), which had been transformed previously with the lambda integrase-producing plasmid pJW289t (33). Colonies that contained an integrated telN-sopAB-antA-bla fragment and which had lost the integration plasmid, were selected. The resulting ampicillin resistant strain is designated E. coli TSA.
Construction of the pJAZZ vectors
A cloning cassette was constructed containing the lacZ-alpha coding region, bounded on each side by a transcriptional terminator (phage T7 and E. coli rrnB terminator on the left and right, respectively), a unique primer binding site and a multiple cloning site. This cassette was inserted into the NotI cloning site of pG591, along with a β-lactamase gene (encoding ampicillin resistance) followed by the TonB terminator. The resulting linear plasmid is the pJAZZ-KA vector (Figure 1).
To create a vector with only a single drug-resistance marker, the kanamycin resistance gene was deleted, leaving the origin of replication as a selectable marker for the left arm. The ampicillin resistance gene was replaced by chloramphenicol resistance to select for the right arm. The resulting vector is pJAZZ-OC. Replacing the chloramphenicol resistance gene with the kanamycin resistance gene generated the pJAZZ-OK vector.
The pJAZZ-KA, -OC and -OK vectors contain several N15 genes that are not essential in cis. To construct a minimal linear vector, telN and several genes of the antirepressor operon (N15 genes 30–32) were deleted from the vector. The minimal vector backbone was excised from the pJAZZ-OC vector as an 8.2-kb AgeI – BglII fragment, comprising the repA gene and origin of replication, the cloning region and the chloramphenicol resistance gene. It was ligated to a 0.4-kb PCR product containing the telRL region of N15, which was amplified from the N15 phage genome with primers tosLOC (5′-GAGAGATCTCTATCTCTTCCGTCTC) and tosROC-2 (5′-TCTACCGGTGTCTCTGGATATCGTAACAC). The circular ligation products were transformed into the E. coli TSA strain. In vivo cutting at the telRL site by TelN produced the 8.6-kb linear plasmid, pJAZZ-OCmin (Figure 1). TelN is provided by inducing expression of the chromosomally integrated copy of the gene in the E. coli TSA cells.
The GenBank Accession numbers of the linear vectors are as follows: pJAZZ-KA (DQ391279); pJAZZ-OC (EF583812); pJAZZ-OK (FJ160465) and pJAZZ-OCmin (GQ469986).
Construction of libraries in the pJAZZ vectors
The pJAZZ vectors were digested with SmaI (blunt) or AhdI (3'C; overhang) to excise the lacZ-alpha stuffer fragment, which separates the vector arms. The digests were dephosphorylated using calf intestinal alkaline phosphatase (Promega, Madison, WI, USA) and purified with Qiaquick® PCR Cleanup columns (Qiagen, Valencia, CA, USA).
Genomic DNA was mechanically sheared to the desired size range (HydroShear® device, Genomic Solutions, Ann Arbor, MI, USA) and end-repaired with the DNATerminator® Kit (Lucigen, Middleton, WI, USA). The DNA was size fractionated by electrophoresis, excised, purified (Gel Extraction Kit, Qiagen) and ligated to a SmaI digest of a pJAZZ vector. Alternately, a single 3'G; tail was added to the blunt DNA with PyroPhage™ 3173 DNA polymerase (Lucigen). G-tailed DNA was ligated to an AhdI digest of a pJAZZ vector.
Ligation reactions were performed with the CloneDirect® ligation kit (Lucigen), using 50–100 ng of digested linear vector and 100–300 ng of prepared insert DNA. After heat inactivation, ligation reactions were electroporated into TSA cells. Recombinants were selected on YT agar plates containing X-GAL (Amresco, Solon, OH, USA) plus the appropriate antibiotic (12.5 µg/ml chloramphenicol for pJAZZ-OC; 30 µg/ml kanamycin for pJAZZ-OK; 100 µg/ml carbenicillin plus 20 µg/ml kanamycin for pJAZZ-KA).
Ligations were also carried out in parallel with the circular high copy plasmids pUC19 and the transcription-free vector pSMART®-HCKan, which contains transcriptional terminators on each side of the cloning site; the low—copy circular vector pSMART-LCKan, which is transcription-free and maintained at ∼15–20 copies per cell and a single-copy, transcription-free vector (pSMART-BAC, Lucigen) (18).
Single colonies were grown in TB medium containing the appropriate antibiotic. l-arabinose was added to 0.01% (w/v) at the time of inoculation to induce the copy number. Cultures of 1.5 ml typically yielded ∼5–20 µg of linear plasmid DNA, using standard alkaline lysis methods. Miniprep DNA was digested with NotI to excise inserts for gel analysis. Dye terminator cycle sequencing was performed using 150–250 ng of miniprep DNA, according to the manufacturer’s instructions (Amersham, Piscataway, NJ, USA; or Applied Biosystems, Inc., Foster City, CA, USA).
RESULTS
Vector construction
The vector pG591 (Figure 1) is capable of cloning large palindromes or regions with abnormal structures [(28) and data not shown]. However, the digested vector preparations generated aberrant clones consisting of circular permutations or dimers of the left arm alone (data not shown).
To create the pJAZZ vectors, the parental plasmid pG591 was modified by addition of: (i) a multiple cloning site, (ii) a lacZ-alpha ‘stuffer’ fragment in the cloning site to allow blue/white screening against uncut vector, (iii) transcriptional terminators on each side of the cloning site to prevent transcription from the insert into the vector, (iv) a transcriptional terminator in the vector backbone to prevent transcription from the vector into the insert and (v) a selectable marker on the right arm of the vector to select for recombinants containing both arms of the vector (Figure 1; ‘Materials and Methods’ section).
The first of the new series was the vector pJAZZ-KA, which contains a kanamycin resistance marker on the left arm and an ampicillin resistance marker on the right arm. Selection for both arms ensures that only recombinants with the correct structure are recovered. Subsequent testing indicated that no antibiotic resistance gene was required on the left arm of the vector, as the origin of replication provides selection. We therefore created the vectors pJAZZ—OK, which is resistant to kanamycin, and pJAZZ-OC, encoding resistance to chloramphenicol (Figure 1). Except for their drug resistance, they showed the same behavior and functionality as the pJAZZ-KA parent (data not shown).
A truncated version, pJAZZ—OCmin (Figure 1), lacks telN and genes #30–32 of the N15 antirepressor operon. The truncated vector is propagated in the same way as the full-length version, with one notable exception. Upon induction of the vector copy number, there appears to be insufficient basal expression of TelN from the chromosomal gene driven by the lac promoter in TSA cells. As a result, a small but detectable fraction of molecules are not cleaved by the pro-telomerase, generating tail-to-tail dimers (data not shown). This situation is remedied by IPTG induction to increase expression TelN. Induction of TelN is not needed for the full-length versions of the pJAZZ vectors, which carry their own copy of the telN gene, as its gene dosage is increased along with the copy number of the vector. Efficient transformation with the full-length vector nonetheless requires TSA cells, which provide an endogenous supply of TelN prior to transformation to protect the ends of the incoming linear DNA.
Host strain construction
The pJAZZ vectors may be transformed into any laboratory strain of E. coli, but three problems must be addressed. First, cellular nucleases of E. coli degrade the hairpin telomeres of incoming linear molecules (35). Transformation with pJAZZ vectors therefore is about two orders of magnitude less efficient than with circular DNA of the same size. Second, due to the absence of a partitioning system, the low copy number vector is spontaneously lost. Third, the low yield of DNA is inconvenient for subsequent plasmid manipulation.
These problems were solved by incorporation of three genetic elements of phage N15 (telN, sopAB and antA) into the chromosome of the E. coli host strain. Degradation of incoming linear DNA is prevented by binding of TelN to hairpin telomeres (35). The sopAB operon ensures stable inheritance of plasmids containing the N15 centromere site, which lies within repA. The rate of loss is <0.1% per generation (36). In addition, the copy number of the plasmids can be elevated to ∼50/cell from ∼5/cell by expression of the antirepressor AntA (31). The efficiency of transformation of the resulting E. coli ‘TSA’ strain with linear pJAZZ DNA is about the same as with circular DNA of the same size.
Utility of the linear cloning vector
Cloning AT-rich DNA
AT-rich inserts are often very unstable in conventional circular vectors. Several lines of evidence suggest that the instability is a function of both the copy number of the vector and its level of transcription. Circular plasmids were used to clone fragments of various sizes from a number of AT—rich genomes, including those of Lactobacillus helveticus (67% AT), the ciliated protozoans Tetrahymena and Oxytricha (∼75% AT) and the fungal pathogen Pneumocystis carinii (∼75% AT). AT-rich inserts as small as 1–3 kb were unstable in pUC19, which has a high copy number and transcribes actively into the cloning site due to the blue/white screen (Figure 2A). However, these inserts were stable in the high-copy, transcription-free vector pSMART-HCKan, which contains transcriptional terminators on each side of the cloning site (18; data not shown).
Figure 2.
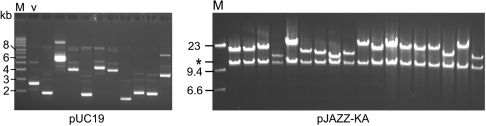
Increased stability of AT-rich DNA in the transcription-free linear vectors. Left panel: Genomic DNA from L. helveticus was sheared to 1–3 kb and cloned into pUC19. Recombinants frequently were deleted, yielding clones smaller than the empty vector control (lane ‘V’). Uncut plasmid DNAs are shown. Right panel: Genomic DNA from L. helveticus was sheared to 10–20 kb, end-repaired and cloned into the pJAZZ—KA vector. Inserts were excised with NotI. Left arm of linear vector is 12 kb (asterisk); the 2-kb right arm ran off the gel. M, lambda/HindIII size marker.
Larger fragments could only be stably maintained in plasmids with lower copy number. AT-rich inserts of 4–6 kb required the use of the lower—copy circular vector pSMART-LCKan, which is transcription-free and maintained at ∼15–20 copies per cell. Fragments of 10–20 kb from the Tetrahymena or Oxytricha genomes were only recovered in the single-copy, transcription-free circular vector (pSMART-BAC, Lucigen) (18). However, even in the single-copy vectors the large insert libraries yielded very few clones, and these clones typically grew poorly.
In contrast, 10–20 kb genomic DNA fragments from these organisms were stably maintained in both pJAZZ-OC and pJAZZ-KA. Ligation reactions containing 100–300 ng of prepared DNA typically yielded ∼10 000 cfu. Recombinants grew well on plates and in liquid culture, and inserts were stably maintained (Figure 2B). To our knowledge, these are the only libraries of Oxytricha or Tetrahymena DNA of >10 kb to be successfully generated, despite numerous attempts in our lab and in others using traditional cloning systems. The pJAZZ-OC vector was also used for constructing a genomic library from Bifidobacterium animalis (65% GC). Stable inserts of up to 30 kb were obtained, confirming that this system is suitable for cloning GC-rich fragments as well (data not shown).
Cloning repetitive DNA
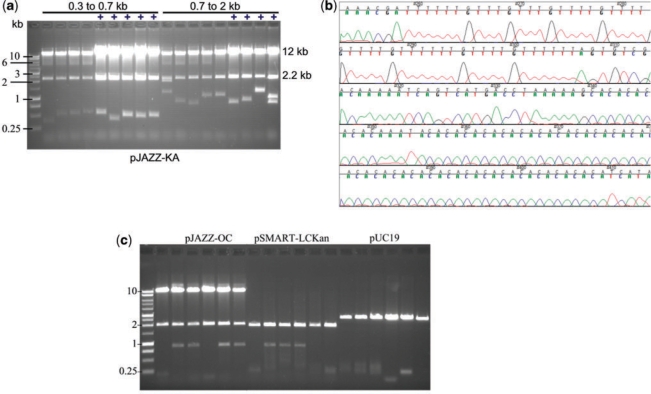
cDNA fragments from a marine mollusk were size-selected into two fractions of 0.3–0.7 kb and 0.7–2.0 kb. The fractions were cloned into pUC19 and into the transcription-free vectors pSMART-HCKan or pSMART-LCKan (Lucigen Corp., Middleton, WI, USA). All inserts recovered from these vectors were <100 bp, and many were <10 bp (data not shown). In contrast, cloning the same cDNA fractions into the pJAZZ—OC linear vector resulted in clones that were all within the expected size ranges (Figure 3A). Sequence analysis confirmed that many of these clones consisted of di-, tri- and tetra-nucleotide repeats (Figure 3B).
Figure 3.
Cloning repetitive DNA in the pJAZZ vectors. Mollusk cDNA fractions of 0.3–0.7 kb and 0.7–2.0 kb were cloned into the pJAZZ-KA vector. (A) DNA was isolated from randomly selected clones from each library and digested with NotI to excise the insert. Nine of the samples were subject to copy number amplification during growth (indicated by ‘+’). All clones contained inserts of the expected sizes. (B) Chromatogram from one of the recovered mollusk cDNA clones, showing repetitive sequence. (C) A PCR fragment containing ∼220 copies of the CGG repeat from the Fragile X locus was cloned into the pJAZZ-OC linear vector or into circular vectors. DNA was isolated from randomly selected colonies and digested to excise the insert from the vector. No intact CGG fragment was obtained in pUC19.
Equally improved stability was observed with a DNA fragment containing 220 CGG•CCG repeats, representing an allele that gives rise to Fragile X syndrome, a human repeat expansion disorder. A PCR product containing these repeats was generated from an expanded allele from a Fragile X mouse model (37) and cloned into the linear or circular vectors. Full-length inserts were recovered from approximately half the clones in the pJAZZ-OC linear vector and in the transcription-free pSMART-LCKan circular vector (Figure 3C). The remaining clones in these vectors appeared to contain PCR artifacts or primer dimers. The vector pUC19 yielded no clones with intact inserts.
Serial culturing showed that the CGG inserts were stable in the pJAZZ vector, using conditions that were designed to maximize the chance of rearrangements or deletions. Subcultures were grown overnight to saturation in 1000 ml of medium at 37°C in the presence of arabinose, which increases the plasmid copy number. The following day they were diluted 1: 1000 into fresh medium and re-grown overnight. After three rounds of subculturing, the plasmid DNA was analyzed. No apparent change in the fraction of intact CGG insert was detectable, although in all cultures there appeared to be a fraction of vectors lacking the insert (data not shown). In contrast, the inserts were lost upon the first attempt to re-grow small cultures of the pSMART—LCKan clones, confirming the instability in supercoiled plasmids (data not shown).
Assembly of genomic libraries
As a broad assessment of cloning bias, the pJAZZ-OC vector was used to clone and sequence the genome of Flavobacterium columnare (69% AT, 3.1 Mb). Genomic DNA was randomly sheared, end-repaired and size-selected to 2–6 kb, 6–10 kb and 10–12 kb. The fractions were cloned separately into three libraries in the pJAZZ-KA vector. A total of 88 Mb was cloned and 33 Mb was sequenced (∼10X coverage). Automated assembly followed the expected Lander–Waterman curve (Figure 4). Initial assembly after 8X genome coverage yielded 21 major contigs of >40 kb, 15 contigs of 2–40 kb, plus 21 small contigs or singletons. Importantly, the gaps between contigs appeared to be short sequencing gaps of <10 kb, rather than physical clone gaps.
Figure 4.
Assembly of the Flavobacterium genome (69% AT) in the pJAZZ vector. Fragments of 2–6 kb, 6–10 kb and 10–12 kb were cloned into the pJAZZ-KA vector. Clones were end-sequenced to provide ∼10X genomic coverage. Assembly of pJAZZ libraries of this AT-rich genome closely approximated the predicted Lander–Waterman curve.
Reduced size bias
Ligation of a fragment into a circular vector involves an intermolecular ligation reaction to join one end of the insert to the vector, followed by intramolecular ligation of the recombinant molecule into a circular form. The efficiency of circularization depends directly on the size of the fragments, decreasing dramatically as the length of the vector or insert is increased. The effect is evident even among inserts in a relatively small size range, causing the well-known bias against larger inserts. On the other hand, formation of a viable linear plasmid from two separate arms of the vector involves two independent ligation events. Ligation of a vector arm to each end of the target DNA is not affected by the size of the fragments. The linear vector was therefore expected to show minimal bias for cloning fragments of various sizes.
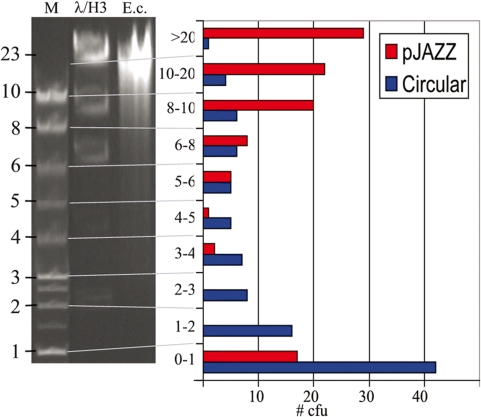
To test this notion, E. coli DNA was randomly sheared to ∼8–20 kb, purified without size selection and cloned into the pJAZZ-OC vector or into the circular vector pUC19. Another aliquot of the DNA was run on a gel to visualize the size range of the fragments. The size of the inserts in the linear vector reflected the size range of the input DNA. In contrast, the inserts in the circular vector were strongly biased toward smaller clones. (Figure 5).
Figure 5.
Minimal size bias in the pJAZZ vector. HMW E. coli genomic DNA was hydrodynamically sheared to ∼8–20 kb. One aliquot was visualized on a gel (lane ‘E.c.’), along with a 1-kb size marker (‘M’) and a HindIII digest of lambda DNA (‘λ/H3’). Another aliquot was cloned without size selection into the pJAZZ-OC vector or pUC19. Clones from each library were randomly isolated for analysis of insert size. The bar graph depicts the number of inserts of each size. The size distribution of pJAZZ clones corresponds to that of the input DNA.
Full-length versus minimal pJAZZ vector
To determine the minimal construct needed for a functional linear vector, several regions of the pJAZZ-OC vector, including the telN gene, were removed to create the vector pJAZZ-OCmin (8.3 kb; Figure 1). The pJAZZ-OCmin vector was able to transform TSA cells efficiently, and it was stably maintained in culture.
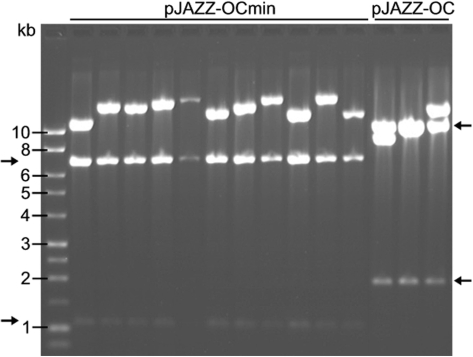
Several libraries were constructed in parallel using the pJAZZ-OC and -OCmin vectors. Unexpectedly, the truncated pJAZZ-OCmin vector consistently yielded 4–10-fold more colonies than the parental vector. The difference was observed with a variety of targets, but it was most pronounced with large, AT-rich inserts, e.g. 10–20 kb fragments from Tetrahymena (75% AT) (Figure 6).
Figure 6.
AT-rich libraries in the pJAZZ-OCmin vector. Tetrahymena genomic DNA was sheared to ∼10–20 kb, end-repaired, and cloned into the pJAZZ-OCmin or pJAZZ-OC vectors. DNA inserts from randomly selected clones were excised by digestion with NotI. The minimal vector yielded ∼10-fold more clones than the parental vector and all clones analyzed had inserts of >10. The arms of each vector are marked with arrows.
Size limit of inserts in the pJAZZ vectors
Although derived from the N15 phage genome, the pJAZZ vectors are never packaged into phage particles. Therefore, the size of recombinant molecules is not constrained by packaging. To assess the maximum size that can be cloned, several sources of large fragments were ligated into the pJAZZ-OC vector. For example, genomic DNA from the nematode Meloidogyne spp. (68% AT) was randomly sheared, size selected to 30 kb and cloned into the vector pJAZZ-OCmin. Approximately 1800 clones were recovered from 60 ng of input DNA. Clones had inserts in the expected size range of 30 kb (Figure 7). Additional sources of large fragments were also tested, but inserts of >30 kb were not recovered.
Figure 7.
The 30-kb inserts of Nematode DNA. HMW genomic DNA from nematodes was randomly sheared to 30 kb, end-repaired and cloned into the pJAZZ-OCmin vector. Inserts were excised by NotI digestion. Average insert size is 30 kb. Size markers ‘M’: HindIII digest of phage lambda DNA plus full-length concatamers.
DISCUSSION
While most DNA fragments can be cloned successfully into supercoiled circular vectors, there has been no satisfactory system for cloning regions whose biological properties or encoded proteins make them prone to deletion and rearrangement. Instability is often regarded as an intrinsic property of the DNA being cloned, whereas the effect of the cloning vector itself has received relatively little scrutiny.
We have previously shown that the stability of inserts in circular vectors can be increased by the use of transcriptional terminators to block transcription into and out of the cloning region (18). We show here that many types of DNA sequences can be more stably propagated by cloning into a linear vector that is also transcription-free. The most notable examples were provided by AT—rich inserts of >10 kb and repetitive DNAs of up to 2 kb.
CGG•CCG-repeats as well as other short tandem repeats are notoriously unstable in bacteria (21). Our previous efforts to clone fragments with <100 CGG repeats into circular vectors were successful only when cells were grown for short periods, at low temperatures, at low densities (avoiding saturation), and in specific E. coli strains; even then deletions were common. However, with the linear plasmid, CGG inserts more than twice this size were reproducibly cloned and stably maintained with standard protocols.
To our knowledge, this is the first report of the successful stable cloning of such a large number of CGG repeats. Significantly, tracts of expanded CGG-repeats are responsible for a number of human genetic diseases, including the Fragile X-related disorders (38). The ability to stably propagate constructs with large numbers of repeats may be useful for generating cell lines and animals for studying this disorder and others belonging to the Repeat Expansion Disease family. In the past, the generation of suitable animal and cell culture models for these diseases has been limited by the elaborate, inefficient and time-consuming procedures required to generate long repeat tracts in vitro (37,39,40).
The pJAZZ system currently is the only method of cloning and maintaining DNAs exclusively as linear plasmid molecules in E. coli. Although bacteriophage lambda vectors are ligated into linear concatamers for packaging, the molecule is re-circularized for replication. The instability caused by supercoiling is therefore likely to affect inserts in lambda clones.
The linear cloning system has other applications as well. The ability of the pJAZZ vectors to maintain fragments of 30–40 kb, even if they are rich in repetitive DNA, may be useful to close genome gaps resulting from Sanger or ‘next-generation’ sequencing. The pJAZZ system also can be used as a simple alternative to fosmid cloning, without the packaging restrictions on the insert size. The structure or topology does not seem to limit the size of linear molecules, as shown by the stability of a linear BAC of 100 kb with N15 telomeres (41). In fact, the E. coli chromosome itself has been linearized through the addition N15 telomeres, yielding viable clones with stable, linear genomes (42). Whether there is an upper limit on insert size in the pJAZZ vectors remains unclear.
The absence of transcriptional interference allows cloning of large cDNAs or operons. Further, the linear vector can be transfected efficiently into mammalian cells (data not shown). Future plans include addition of a mammalian expression signals to the pJAZZ constructs. These linear shuttle vectors will allow the mammalian expression and analysis of genes that cannot be readily cloned in conventional plasmids.
ACCESSION NUMBER
FUNDING
An SBIR grant awarded to Lucigen by the National Institutes of Health (grant no. 5R44 HG003076-03). Funding for open access charge: Page charges will be funded from Lucigen’s general budget. The source of funds is from commercial sales of products.
Conflict of interest statement. R.G., D.M., V.D. and C.W. are employees of Lucigen Corporation. The pJAZZ vectors and TSA cell line described in this article are currently or in the future may be made available for commercial sale by Lucigen.
REFERENCES
- 1.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat. Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 4.Kouprina N, Leem SH, Solomon G, Ly A, Koriabine M, Otstot J, Pak E, Dutra A, Zhao S, Barrett JC, et al. Segments missing from the draft human genome sequence can be isolated by transformation-associated recombination cloning in yeast. EMBO Rep. 2003;4:257–262. doi: 10.1038/sj.embor.embor766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:RESEARCH0079. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.She X, Jiang Z, Clark RA, Liu G, Cheng Z, Tuzun E, Church DM, Sutton G, Halpern AL, Eichler EE. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- 7.Garber M, Zody MC, Arachchi HM, Berlin A, Gnerre S, Green LM, Lennon N, Nusbaum C. Closing gaps in the human genome using sequencing by synthesis. Genome Biol. 2009;10:R60. doi: 10.1186/gb-2009-10-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 9.Paulding CA, Ruvolo M, Haber DA. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc. Natl Acad. Sci. USA. 2003;100:2507–2511. doi: 10.1073/pnas.0437015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glockner G, Eichinger L, Szafranski K, Pachebat JA, Bankier AT, Dear PH, Lehmann R, Baumgart C, Parra G, Abril JF, et al. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature. 2002;418:79–85. doi: 10.1038/nature00847. [DOI] [PubMed] [Google Scholar]
- 11.Gardner MJ. A status report on the sequencing and annotation of the P. falciparum genome. Mol. Biochem. Parasitol. 2001;118:133–138. doi: 10.1016/s0166-6851(01)00390-5. [DOI] [PubMed] [Google Scholar]
- 12.Vaughan A, Chiu SY, Ramasamy G, Li L, Gardner MJ, Tarun AS, Kappe SH, Peng X. Assessment and improvement of the Plasmodium yoelii yoelii genome annotation through comparative analysis. Bioinformatics. 2008;24:i383–389. doi: 10.1093/bioinformatics/btn140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 15.Moran NA. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr. Opin. Microbiol. 2003;6:512–518. doi: 10.1016/j.mib.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Temple GF. The completion of the Mammalian Gene Collection (MGC) Genome Res. 2009 doi: 10.1101/gr.095976.109. Oct 28 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Temple G, Lamesch P, Milstein S, Hill DE, Wagner L, Moore T, Vidal M. From genome to proteome: developing expression clone resources for the human genome. Hum. Mol. Genet. 2006;15(Spec No 1):R31–43. doi: 10.1093/hmg/ddl048. [DOI] [PubMed] [Google Scholar]
- 18.Godiska R. Beyond pUC: Vectors for Cloning Unstable DNA. In: Kieleczawa J, editor. DNA Sequencing: Optimizing the Process and Analysis. 1st edn. Vol. 1. Sudbury, Massachusetts: Jones and Bartlett Publishers; 2005. pp. 55–76. [Google Scholar]
- 19.Kiyama R, Oishi M. Instability of plasmid DNA maintenance caused by transcription of poly(dT)-containing sequences in Escherichia coli. Gene. 1994;150:57–61. doi: 10.1016/0378-1119(94)90857-5. [DOI] [PubMed] [Google Scholar]
- 20.Adhya S, Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982;29:939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- 21.Bowater RP, Wells RD. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:159–202. doi: 10.1016/s0079-6603(00)66029-4. [DOI] [PubMed] [Google Scholar]
- 22.Stueber D, Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1:1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham TP, Montelaro RC, Rushlow KE. Lentivirus envelope sequences and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vectors. Gene. 1993;124:93–98. doi: 10.1016/0378-1119(93)90766-v. [DOI] [PubMed] [Google Scholar]
- 24.Feng T, Li Z, Jiang W, Breyer B, Zhou L, Cheng H, Haydon RC, Ishikawa A, Joudeh MA, He TC. Increased efficiency of cloning large DNA fragments using a lower copy number plasmid. Biotechniques. 2002;32:992–998. doi: 10.2144/02325bm03. [DOI] [PubMed] [Google Scholar]
- 25.Leach D, Lindsey J. In vivo loss of supercoiled DNA carrying a palindromic sequence. Mol. Gen. Genet. 1986;204:322–327. doi: 10.1007/BF00425517. [DOI] [PubMed] [Google Scholar]
- 26.Malagon F, Aguilera A. Genetic stability and DNA rearrangements associated with a 2 x 1.1-Kb perfect palindrome in Escherichia coli. Mol. Gen. Genet. 1998;259:639–644. doi: 10.1007/s004380050858. [DOI] [PubMed] [Google Scholar]
- 27.Ravin NV, Kuprianov VV, Gilcrease EB, Casjens SR. Bidirectional replication from an internal ori site of the linear N15 plasmid prophage. Nucleic Acids Res. 2003;31:6552–6560. doi: 10.1093/nar/gkg856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravin NV, Ravin VK. Use of a linear multicopy vector based on the mini-replicon of temperate coliphage N15 for cloning DNA with abnormal secondary structures. Nucleic Acids Res. 1999;27:e13. doi: 10.1093/nar/27.17.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deneke J, Ziegelin G, Lurz R, Lanka E. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl Acad. Sci. USA. 2000;97:7721–7726. doi: 10.1073/pnas.97.14.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravin NV. Mechanisms of replication and telomere resolution of the linear plasmid prophage N15. FEMS Microbiol. Lett. 2003;221:1–6. doi: 10.1016/S0378-1097(03)00125-3. [DOI] [PubMed] [Google Scholar]
- 31.Mardanov AV, Ravin NV. Functional characterization of the repA replication gene of linear plasmid prophage N15. Res. Microbiol. 2006;157:176–183. doi: 10.1016/j.resmic.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Lessl M, Balzer D, Lurz R, Waters VL, Guiney DG, Lanka E. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wild J, Hradecna Z, Szybalski W. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 2002;12:1434–1444. doi: 10.1101/gr.130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mardanov AV, Strakhova TS, Smagin VA, Ravin NV. Tightly regulated, high-level expression from controlled copy number vectors based on the replicon of temperate phage N15. Gene. 2007;395:15–21. doi: 10.1016/j.gene.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 35.Dorokhov BD, Strakhova TS, Ravin NV. [Expression regulation of the protelomerase gene of the bacteriophage N15] Mol. Gen. Mikrobiol. Virusol. 2004;2:28–32. [PubMed] [Google Scholar]
- 36.Ravin N, Lane D. Partition of the linear plasmid N15: interactions of N15 partition functions with the sop locus of the F plasmid. J. Bacteriol. 1999;181:6898–6906. doi: 10.1128/jb.181.22.6898-6906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Entezam A, Biacsi R, Orrison B, Saha T, Hoffman GE, Grabczyk E, Nussbaum RL, Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oostra BA, Willemsen R. FMR1: a gene with three faces. Biochim. Biophys. Acta. 2009;1790:467–477. doi: 10.1016/j.bbagen.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grabczyk E, Usdin K. Generation of microgram quantities of trinucleotide repeat tracts of defined length, interspersion pattern, and orientation. Anal. Biochem. 1999;267:241–243. doi: 10.1006/abio.1998.2962. [DOI] [PubMed] [Google Scholar]
- 40.Osborne RJ, Thornton CA. Cell-free cloning of highly expanded CTG repeats by amplification of dimerized expanded repeats. Nucleic Acids Res. 2008;36:e24. doi: 10.1093/nar/gkn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ooi YS, Warburton PE, Ravin NV, Narayanan K. Recombineering linear DNA that replicate stably in E. coli. Plasmid. 2008;59:63–71. doi: 10.1016/j.plasmid.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Cui T, Moro-oka N, Ohsumi K, Kodama K, Ohshima T, Ogasawara N, Mori H, Wanner B, Niki H, Horiuchi T. Escherichia coli with a linear genome. EMBO Rep. 2007;8:181–187. doi: 10.1038/sj.embor.7400880. [DOI] [PMC free article] [PubMed] [Google Scholar]