Abstract
The lower jaws of archaeal RNA polymerase and eukaryotic RNA polymerase II include orthologous subunits H and Rpb5, respectively. The tertiary structure of H is very similar to the structure of the C-terminal domain of Rpb5, and both subunits are proximal to downstream DNA in pre-initiation complexes. Analyses of reconstituted euryarchaeal polymerase lacking subunit H revealed that H is important for open complex formation and initial transcription. Eukaryotic Rpb5 rescues activity of the ΔH enzyme indicating a strong conservation of function for this subunit from archaea to eukaryotes. Photochemical cross-linking in elongation complexes revealed a striking structural rearrangement of RNA polymerase, bringing subunit H near the transcribed DNA strand one helical turn downstream of the active center, in contrast to the positioning observed in preinitiation complexes. The rearrangement of subunits H and A′′ suggest a major conformational change in the archaeal RNAP lower jaw upon formation of the elongation complex.
INTRODUCTION
The transcriptional machinery of archaea is similar to, but far simpler than, the eukaryotic RNA polymerase II (polII) system. The archaeal RNA polymerase (RNAP) is recruited to the preinitiation complex (PIC) by association with promoter bound transcription factors TBP and TFB (1,2) that interact with the TATA-box and BRE element of promoter DNA. These archaeal factors are orthologs of the eukaryotic transcription factors TBP and TFIIB. Archaeal TFE is a polypeptide with sequence similarity to the N-terminal part of the α subunit of eukaryotic TFIIE (3–5). Like TFIIE, archaeal TFE interacts with the non-transcribed DNA strand upstream of the transcription start site, but in contrast to its eukaryotic counterpart, it interacts with and stabilizes the transcription bubble in elongation complexes (ECs; 6). The large subunits of archaeal RNA polymerases (B, which is similar in size to its eukaryotic ortholog Rpb2, and A′ and A′′, which correspond to consecutive parts of Rpb1) display higher sequence similarity with eukaryotic polII subunits than with the bacterial subunits β′ and β (7). The archaeal RNAP subunits F (conserved in eukaryotic RNAP II as Rpb4), H (Rpb5), E′ (Rpb7), N (Rpb10) and P (Rpb12) are only conserved between archaeal and eukaryotic RNAPs and have no orthologs in the multisubunit bacterial enzymes. Comparison of crystal structures of archaeal RNAP and eukyaryotic RNAP II revealed striking structural similarity (8,9). The major differences between archaeal RNAP and pol II are that in the archaea the homolog of Rpb1 is split into subunits A′ and A′′ and the smaller subunits F, P and H lack an N-terminal domain that is present in their eukaryotic counterparts Rpb 4, Rpb 12 and Rpb5, respectively (10). An Rpb8 ortholog is missing in euryarchaeota like Pyrococcus furiosus studied here, but was detected in the genomes of all hyperthermophilic crenarchaeota (11,12). Rpb8 was not found in crystals used for the first analysis of the structure of the Sulfolobus RNAP (9) but co-purified and crystallized with Sulfolobus RNAP characterized recently (13). Therefore, it seems to be a subunit that is easily lost during purification. An Rpb9 ortholog is not contained in purified archaeal RNAPs but the archaeal elongation factor TFS is clearly homologous to Rpb9 and to its Pol I and Pol II paralogs Rpa12 and Rpc11 and shares with Rpc11 the property to induce transcript cleavage (14,15). In spite of the missing N-terminal domain, the archaeal subunit P complements null mutants of yeast Rpb12 in vivo (16). This finding suggests a high degree of functional conservation between the two domains and indicates that the additional eukaryote-specific N-terminal domain is not required for basic RNAP functions.
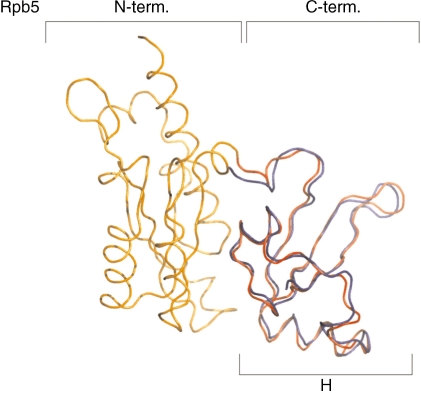
Analyses of the topography of PICs by photochemical cross-linking and molecular modeling on the basis of crystal structures revealed a highly conserved geometry of interaction of bacterial, eukaryotic and archaeal RNAPs with DNA in the PIC (17), e.g. the archaeal subunits A′ and A′′, the bacterial subunits β′ and eukaryotic Rpb1 are in close contact with DNA downstream of the transcription start site (17,18–21). Cross-linking studies with the archaeal PIC showed in addition close proximity of subunit H with downstream DNA from +10 to +20 (17). Eukaryotic Rpb5 has been cross-linked to DNA in initiation complexes from +5 to +15 (18,20), suggesting that both factors occupy similar locations in the archaeal and eukaryotic PIC. Eukaryotic Rpb5 shows a bipartite organization with an N-terminal domain that is unique to eukaryotic RNAPs, and a C-terminus that is highly conserved in eukaryotes and archaea (7). The C-terminus of Rpb5 is in close contact with subunit Rpb1 (8), and this feature is shared with the archaeal H, which is strongly associated with RNAP subunit A′′ (9,10,13). A superposition of Rpb5 and H reveals a very high similarity in tertiary structure of the C-terminal part of Rpb5 with H (Figure 1).
Figure 1.
The C-terminal domains of Rpb5 and RpoH are very similar. Superposition of the tertiary structures of subunits Rpb5 from yeast based on the RNAP II crystal structure (N-terminal domain coloured in orange and the C-terminal domain in red; PDB ID: 1I3Q; 8) and of the archaeal subunit H from S. solfataricus (coloured in blue; PDB ID: 2PMZ; 9) generated using DaliLite (45). The R.M.S. value of this alignment is 1.1 Å.
RNAPs from hyperthermophilic archaea can be reconstituted in vitro from bacterially expressed subunits (22,23). The reconstitution protocol allows investigation of functions of single subunits of RNAP by analysis of a reconstituted RNAP lacking a specific subunit. The small eukaryotic RNAP subunit Rpb12 can replace subunit P in archaeal RNAP without significant loss of function in assays covering most crucial steps of the transcription cycle (16). In this work, we focus on subunit H corresponding to the conserved C-terminal third of Rpb5. While the N-terminal domain of Rpb5 is involved in interactions with TFIIF and several regulators (24), the role of the conserved C-terminal domain in the transcription process is poorly understood. We show here that subunit H is required for early steps of initiation and—in striking contrast to Rpb5 in polII—undergoes an unexpected structural rearrangement in ECs.
MATERIALS AND METHODS
Purification of RNAPs
RNAP from P. furiosus and polII from yeast were purified as described (25,26). Wt Pyrococcus RNAP and ΔH RNAP were reconstituted from single subunits as described (24).
Cloning and purification of P. furiosus TFS
The gene encoding TFS was amplified by PCR from endogenous Pyrococcus DNA, cloned into the NdeI and EcoRI site of vectors pET-17b and over expressed in Escherichia coli BL21CP(DE3)-RIL. The crude extract was incubated for 30 min at 95°C and heat-soluble proteins in the supernatant were purified by gel filtration (Hi Load 16/60 Superdex 75 prep grade) and anion exchange chromatography (MonoQ 5/50 GL 1 ml). The ability of TFS to induce the nuclease activity of RNAP was assayed by addition of TFS to immobilized purified complexes stalled at +20, which were incubated in the absence of NTPs as indicated in Supplementary Figure S1. Under these conditions, the RNA in EC20 complexes was shortened in a TFS- and time-dependent manner.
Promoter-directed run-off in vitro transcription assay
In vitro transcription assays were performed as described (23).
KMnO4 footprinting of initiation complexes
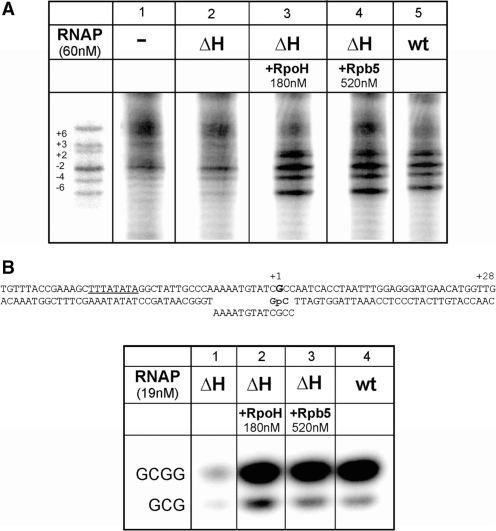
Footprinting experiments were performed essentially as described (16). Reconstituted wt RNAP or ΔH RNAP, respectively, was preincubated for 5 min at 70°C with gdh promoter DNA in reactions containing 500 nM TFE, 440 nM RpoE′-F complex, 60 nM RNAP, 60 nM TBP and 50 nM TFB. Subunits H (180 nM) and Rpb5 (520 nM) were added as indicated in Figure 3A.
Figure 3.
Subunit H is required for open complex formation and initial transcription. (A) RpoH and Rpb5 stimulate open complex formation. Promoter opening at 70°C was analyzed by KMnO4 footprinting as described (16,23). TBP, TFB, TFE and the RpoE′-F complex were present in all reactions. Additional components added to the reactions are indicated on top of the gel. (B) Abortive transcription from a preopened heteroduplex. The ability of ΔH and of wt RNAP to synthesize a 3 or 4 nt abortive transcript in the presence of TBP and TFB was analyzed on a preopened bubble mimicking an open complex (see figure on top of the gel). The reaction was dinucleotide (GpC) primed and was analyzed on a 24% PA gel. Subunits H and Rpb5 were added to transcription reactions as indicated.
Abortive transcription assays
The transcription reactions were performed similar to run-off transcription but with a −C/+20 cassette of the gdh promoter according to (27) but a heteroduplex template (Figure 3) was used to mimic an open complex. The transcription was dinucleotide primed with GpC (40 μM) and transcription was started by the addition of radiolabeled GTP (3.3 pmol). The 3-nt and 4-nt RNA products were analyzed on a 24% (w/v) polyacrylamide urea gel.
Immobilized in vitro transcription assay of stalled ternary complexes
Immobilized ternary complexes were stalled at position +20 relative to the transcription start site and were isolated according to (27). To remove unbound RNAP and TBP/TFB from promoter DNA, complexes were washed with transcription buffer containing 0.1% N-lauroylsarcosine (NLS). Then the isolated ternary complexes were resuspended in transcription buffer and chased by the addition of 4 NTPs (40 µM each) but no additional radioactivity in a total volume of 25 µl. Subunit H was added to the reaction as indicated on top of Figure 4.
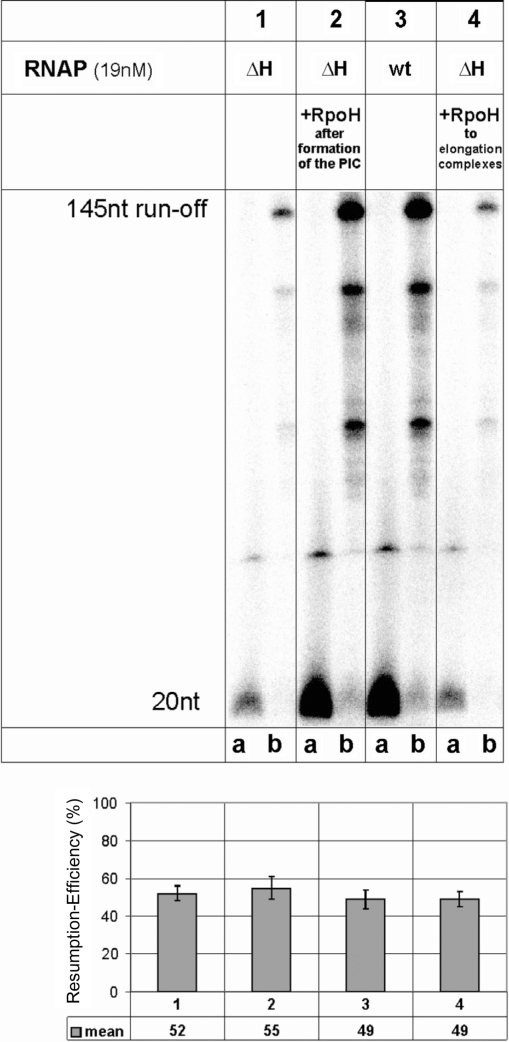
Figure 4.
Stalled ΔH RNAPs can efficiently elongate to the run-off. Ternary complexes were stalled at +20, washed and the reactions divided into two aliquots. One aliquot was untreated (lane a of each panel), the other was chased by the addition of a complete set of non-labeled NTPs for 3 min at 70°C (lane b in each panel), and both were then analyzed on a 20% polyacrylamide gel. In lanes 1, 2 and 4, the ability of the ΔH and wt enzyme to synthesize a 20-nt stalled transcript in the presence and absence of subunit H was analyzed. Reactions were chased by the addition of a complete set of NTPs not containing radioactivity and the synthesis of the 145-nt run-off product was analyzed. The RNA products of intermediate size are caused by specific pausing sites for RNAP. The quantification below shows the mean value of the transcriptional activity of the transcript in ternary complexes stalled at position +20 relative to the 145-nt run-off product.
Electrophoretic mobility shift assay
A DNA fragment spanning the gdh promoter region from −60 to +37 was used as probe as described earlier (10). The binding reactions contained 63 nM wt RNAP, 126 nM TBP, 106 nM TFB, 50 nM gdh promoter DNA and 1 μg of poly(dI–dC) as non-specific competitor DNA. Protein−DNA complexes were assembled in a 25 µl volume containing 40 mM Na-HEPES, pH 7.3, 250 mM NaCl, 25 mM MgCl2, 0.1 mM EDTA, 5 mM β–mercaptoethanol, 5 % (v/v) glycerol and 0.1 µg/ml bovine serum albumin.
Cloning and purification of the yeast RNAP subunit Rpb5
Genomic yeast DNA (Novagen) was used as template for PCR amplification of the gene encoding Rpb5 with help of the forward primer (5′-ATGGACCAAGAAAATGAAAG-3′) and the reverse primer (5′-CTACATACAGATTCTGTAAC-3′). The purified PCR product was cloned into pETBlue 1 vector (Novagen) and the DNA sequence was confirmed by sequencing.
Rpb5 was expressed in Tuner cells (Novagen). The protein was purified from the crude extract by a SP XL 5-ml column (GE Healthcare) and eluted with a linear salt gradient from 10 mM to 1 M NaCl in buffer containing 40 mM Na-HEPES pH 8, 10% (v/v) glycerol, 1 mM PMSF, 5 mM β-mercaptoethanol and complete protease inhibitor mix (Roche). Rpb5 eluted at 320 nM NaCl and was further purified by size exclusion chromatography on a Superdex 75 column.
Photochemical cross-linking of stalled ECs and of preinitiation complexes
Pyrococcus furiosus ECs were stalled essentially as described earlier (6): 2 nM of internally α-32P-NTP labeled gdh C-minus +20/+45 cassettes modified with photoactivatable azidophenacylated phosphorothioate at specific positions and prepared essentially as described (21) were incubated with 20 nM TBP, 60 nM TFB, 21 nM RNAP purified from Pyrococcus cells (Figures 6 and 7, lanes 2, 5 and 8) or reconstituted ΔH RNAP (Figure 7, lanes 3, 6 and 7), respectively. Rpb5 was added to 535 nM when indicated (Figure 7, lanes 4 and 7). Protein components and DNA were preincubated for 3 min at 70°C in 12.5 µl of transcription buffer containing 40 mM Na-HEPES, pH 7.3, 250 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, 600 ng of herring sperm DNA (acting as non-specific competitor DNA) and 40 µM each of adenosine triphosphate (ATP), guanosine triphosphate (GTP) and uridine triphosphate (UTP). Subsequently, gdh promoter DNA ranging from position −164 to +113 was added as specific competitor to a final concentration of 400 nM and incubation continued for 2 min at 70°C. Complexes were transferred to an ultraviolet (UV) chamber and UV-irradiated for 7 min at 70°C, followed by nuclease treatment (17). Cross-linked proteins were analyzed on 4–19% gradient PA-SDS gels and visualized using image plates and image analyzer (FLA-500, Fuji, Japan).
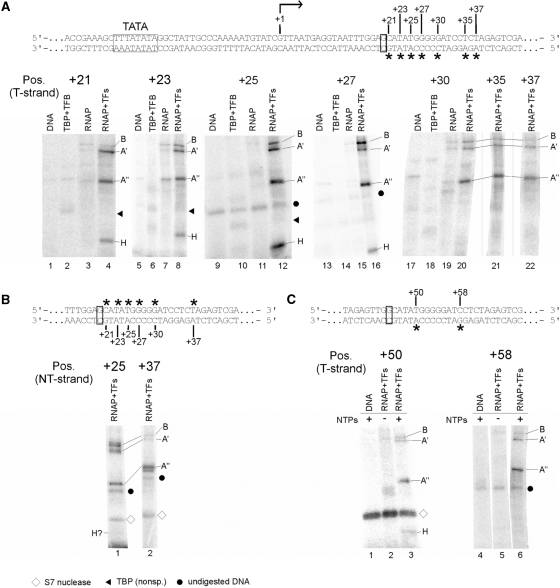
Figure 6.
Mapping of RNAP subunits cross-linked to the gdh promoter DNA in stalled elongation complexes. (A) Processivity of ECs stalled at +20 is not affected by TFS. Increasing amounts of purified TFS were added to stalled ECs (lanes 4–9) and complexes were chased with a complete set of NTP after incubation in the absence (lane 3) and presence of TFS (lane 8). The amount of non-chaseable RNA in complexes is insignificant in both reactions and the amount of synthesized run-off transcript ∼ the same indicating that the stalled complexes were not backtracked in the absence of TFS. As expected, the 21-nt RNA most likely generated by misincorporation of an unpaired nt was removed upon TFS treatment. (B) Photocross-linking of proteins in a ternary complex stalled at position +20. The gdh promoter template spans bp −39 to +66 relative to the transcription start site (bent arrow). TATA box and the stalling site +20 are boxed. Locations of photoactivatable labels (4-azidophenacyl bromide coupled to a phosphorothioate modification in the DNA backbone) are indicated by asterisks. RNAP purified from Pyrococcus cells was used to stall elongation complexes at +20 as described earlier (6,27). Cross-linked subunits of the stalled complexes were analyzed via 4–20% SDS-PAGE. The position of the photoactive cross-linking site in the transcribed strand is indicated above the gels. Proteins present in the individual reactions are specified on top of the gels. Radiolabeled RNAP subunits were identified by their relative electrophoretic mobility and are indicated at the right-hand side of the gels. Dots indicate non-specific signals derived from undigested DNA, whereas triangles indicate non-specific TBP cross-links. (C) Cross-linking of RNAP subunits in stalled elongation complexes to the non-transcribed strand. Positions analyzed are indicated by asterisks above the sequence. No cross-linking of H was detectable at any of the tested positions except of position +25, where a very faint band suggests weak cross-linking of H. To exemplify this, positions +25 and +37 are shown. (D) H interacts with DNA close to the active center also in late elongation complexes. Mature ECs, stalled at position +45 were cross-linked to position +50 and +58 on the T-strand as described in (B). The diamond marks auto-radiolabeled S7 nuclease. Part of the DNA sequence is shown at the top of the panel.
Figure 7.
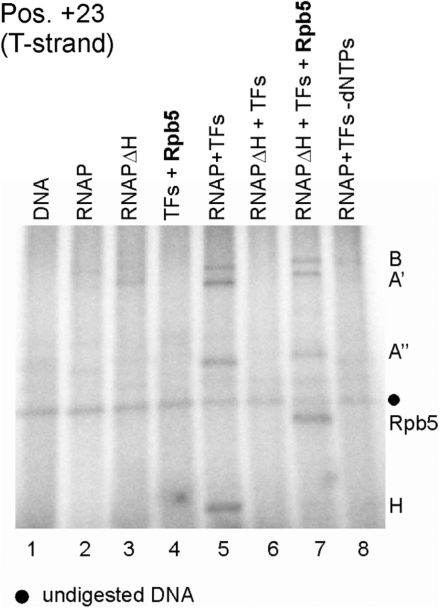
Rpb5 incorporated into the archaeal RNAP cross-links at the same position in elongation complexes as H. Archaeal ΔH RNAP complemented in transcription reactions with Rpb5 was cross-linked in ECs stalled at position +20 (lane 7). Reactions contained transcription factors TFB and TBP (TFs), RNAP purified from Pyrococcus cells (lanes 2, 5 and 8), reconstituted ΔH RNAP and subunit Rpb5 as indicated on top of the lanes. The template DNA was derivatized with APB at position +23. Note that the presence of Rpb5 is essentially required for the formation of cross-linkable stalled ECs by the ΔH RNAP (compare lanes 6 and 7). Subunits of reconstituted ΔH RNAP run at higher molecular weight because they are His-tagged.
Transcription of a derivatized template
Stalling of ECs on templates containing the photoactivateable azidophenacylated phosphorothioate at position 25 in the template strand (Figure 5A) was carried out as in photochemical cross-linking experiments, except few changes: the DNA template did not contain an internal radioactive label and stalled ECs were obtained by addition of 40 µM ATP, 40 µM GTP, 2.68 µM UTP and [α-32P]UTP with 0.15 MBq (110 tBq/mmol). NTPs (40 µM each) or 150 nM TFS to chase stalled complexes were added as indicated on top of the panel. After further incubation at 70°C for 2 min, reactions were stopped by addition of formamide loading buffer.
Figure 5.
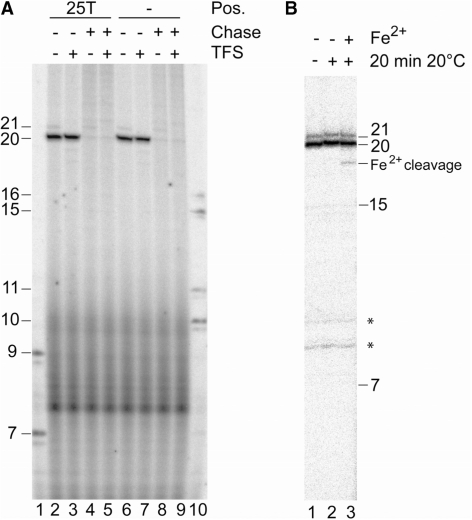
Resumption of stalled complexes and Fe2+ cleavage indicate that EC20 complexes are not backtracked. (A) Complexes stalled at the gdh promoter at position +20 on a template with and without aryl azide derivatization at position +25 of the template strand (25T; lanes 2, 3 and 6, 7) were challenged after incubation for 5 min at 70°C with a complete set of NTPs (chase) for 2 min in the presence and absence of TFS as indicated on top of the figure. Lanes 1 and 10 contained RNA size markers. (B) Stalled immobilized EC20 were extensively washed in the absence of Mg2+, as indicated in ‘Materials and Methods’ section, and incubated for 20 min at 70°C. Reactions were then cooled to 20°C, and Fe2+ and DTT were added to initiate the cleavage reaction. The ∼18-nt Fe2+ cleavage product contains a 3′ phosphate causing as slightly higher electrophoretic mobility (39). The weak 21-nt RNA band is due to misincorporation at the G-residue of template DNA.The asteriks indicate non-specific bands.
Fe2+-cleavage
ECs at position +20 (EC20) were stalled on immobilized templates as described (27), isolated and extensively washed with transcription buffer lacking MgCl2. Either immediately after washing or after further incubation for 15 min at 70°C the reaction was split in three parts: one-third was not further incubated, one-third was incubated for 20 min at 20°C without additives and one-third was treated with 3 mM (NH4)2Fe(SO4)2 and 30 mM DTT (Figure 5B). Reactions were analyzed on a 28% PA/urea gel. Incubation at 70°C after washing had no effect on Fe2+ cleavage (data not shown).
Kinetics of TFS induced RNA-cleavage
EC20 was stalled on immobilized templates according to (27). After 3 or 15 min, EC20 was isolated, washed three times and resuspended in transcription buffer without NTPs. TFS was added to a final concentration of 150 nM immediately or after 3 or 15 min further incubation at 70°C (Figure S1). Samples were taken 5 s, 45 s, 3 min and 15 min after TFS addition as indicated on top of each lane.
Cross-linking of PICs (Supplementary Figure S2) was performed essentially as described earlier, using α-32P-NTP labeled gdh C-minus +20 cassettes, derivatized with APB at various positions as indicated in Figure S1; 21 nM of native Pfu RNAP, 20 nM TBP and 60 nM TFB were incubated with 2 nM of photoactivatable template DNA. Buffer conditions resembled the conditions stated before, but NTPs were omitted from the reactions. Incubation of PICs, cross-linking, nuclease treatment and analysis of cross-linked proteins were performed as described in the previous section.
Chemical cleavage of cross-linked RNAP subunit H
The bands presumptively containing H were excised from the gel, eluted and concentrated as described previously (17). Cleavage by formic acid and cyanogen bromide (CNBr) was performed essentially as described previously (17), but the formic acid concentration was raised from 50 to 75% (v/v) and the reaction time was extended to 18 h at 37°C. The 2-nitro-thiocyanobenzoic acid (NTCB) cleavage reaction was conducted as described (28), with slight modification: NTCB cleavage at Cys residues was induced by treatment with 10 mM NTCB for 5 h at 37°C. The efficiency of CNBr and NTCB cleavage was tested by cleavage of cross-linked RNAP subunit B, which contains Cys and Met residues (data not shown). Following 4–12% VG/Mops gel electrophoresis, labeled proteins were visualized by autoradiography.
Cross-linking of archaeal RNAP and of polII to elongation scaffolds
The assembly of the elongation scaffolds was performed essentially as described in (29), but azidophenacylated phosphorothioate with adjacent internal radiolabel were introduced at positions +6 or +15 of the T-strand, respectively. To anneal RNA with the T strand, hybridization reactions were started by heating the oligonucleotides for 3 min at 90°C. The tubes were transferred to a heating block kept at 70°C, which was switched off after the transfer. After an incubation period of 15 min, the tubes were transferred to a heating block kept at 40°C which was again immediately switched off after transfer and cooled down to 20°C. This DNA–RNA hybrid (2 nM) was incubated with increasing amounts of polII (36 nM, 72 nM or 110 nM) at 20°C in 500 mM potassium acetate, pH 7.6, 100 mM HEPES, pH 7.6, 5 mM EDTA, 25 mM MgOAc (30), 5 % glycerol (v/v) and heparin 50 ng/µl. The NT-strand was added after 5 min to a final concentration of 0.5 nM and reactions were further incubated at 20°C. After 10 min, reactions were transferred to a 30°C heating block and preincubated for 5 min and then the open tubes were UV254 irradiated for 10 min at 30°C. Photoactivatable elongation scaffolds for cross-linking analyses of RNAP purified from Pyrococcus cells contained 2 nM of the T-strand DNA/RNA hybrid, 46 nM RNAP and 380 nM TFE as indicated in Figure 6b. Hybridization reactions were incubated for 5 min at 20°C prior to the addition of the NT-strand. Ten minutes after the addition of the NT-strand, reactions were preincubated for 5 min at 70°C, followed by UV254 irradiation for 10 min at 70°C. After UV irradiation, reactions were nuclease treated as described (6), denatured and analyzed by 4–19% SDS-PAGE.
Transcription on assembled elongation scaffolds
Activity of polIIs on the elongation scaffold containing the cross-linker was tested under essentially the same conditions as described in the previous section but reactions contained 0.4 µM unlabeled DNA/RNA hybrid and NT-strand, respectively. Radiolabeled nucleotides were added prior to incubation for 30 min at 30°C.
RESULTS
Eukaryotic Rpb5 can functionally replace H in the archaeal RNAP
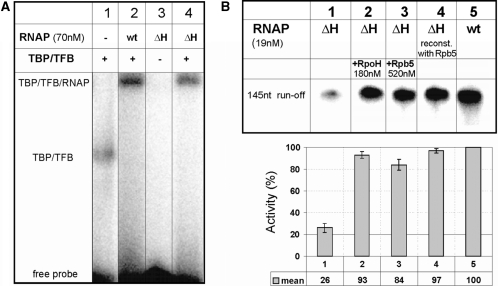
The high structural similarity between H and Rpb5 suggests conserved function for these RNAP subunits in archaea and eukaryotes. To test this, we compared activity of reconstituted archaeal RNAP with or without H, or with Rpb5 replacing H. In electrophoretic mobility shift assays containing gdh promoter DNA and the general transcription factors TFB and TBP, RNAP reconstituted without subunit H (ΔH RNAP) formed stable PICs. The efficiency of PIC formation was ∼75% when compared to the reconstituted RNAP containing all subunits (wt RNAP; see lanes 2 and 4 in Figure 2A). This suggests that subunit H is not essential for PIC formation.
Figure 2.
Characterization of promoter recruitment and of run-off transcripts of ΔH RNAP. (A) ΔH RNAP forms stable preinitiation complexes. EMSAs with a probe containing the Pyrococcus gdh promoter (10) were conducted in the presence and absence of TBP, TFB and reconstituted wt or ΔH RNAP as indicated on top of the lanes. The position of the TBP–TFB and TBP–TFB–RNAP complexes are indicated. (B) Eucaryotic Rpb5 can functionally replace RpoH in an archaeal RNAP. The synthesis of a 145 nt run-off transcript from the Pyrococcus gdh promoter was analyzed in standard multiple round transcription assays (‘Materials and Methods’ section) and RNA products were analyzed on 6% denaturing PA gels. Lane 4 shows transcription products synthesized by ΔH RNAP reconstituted with Rpb5. In lanes 2 and 3 subunit H or Rpb 5 were added to transcription reactions. The diagram below the gel shows the mean value of the transcriptional activity for each lane. The quantification was done with the Aida image analyzer software version 3.28.
To analyze the ability of the ΔH RNAP to initiate RNA synthesis in vitro, we performed specific run-off assays on linearized DNA containing the gdh promoter (31). H or its eukaryotic homolog Rpb5 were added to reconstitution reactions involving denaturation and renaturation of all subunits (23), or were added after reconstitution of ΔH RNAP as indicated on top of the lanes. As reported earlier (22,32), the ΔH RNAP showed a dramatic defect in transcription activity (Figure 2B, lane 1) when compared to the reconstituted wild-type RNAP (Figure 2B, lane 5). An archaeal RNAP reconstituted in the presence of the eukaryotic RNAP subunit Rpb5 showed the same transcriptional activity as the archaeal RNAP reconstituted with H (Figure 2B, lanes 4 and 5), indicating that the N-terminal part of Rpb5 does not disturb the proper folding of the archaeal enzyme during reconstitution. Furthermore, when recombinant Rpb5 was added to transcription reactions with ΔH RNAP the activity was ∼80% of wt levels (Figure 2B, lane 3). The addition of archaeal H to transcription reactions also rescued transcription (Figure 2B, lane 2). These findings suggest that both the archaeal subunit H and eukaryotic Rpb5 can be easily and rapidly incorporated into the reconstituted ΔH enzyme. Furthermore, these data provide evidence that function of H and the C-terminal part of Rpb5 are conserved.
Subunit H is required for efficient open complex formation and initial transcription
Defects in transcription can be caused at several stages of the transcription process. After formation of the PIC, the next step is opening of the DNA around the transcription start site. To analyze the ability of the ΔH enzyme to form open complexes, we performed potassium permanganate footprinting experiments as described previously (6). No reactivity of T-residues in complexes formed by the ΔH RNAP could be detected with the exception of a weak signal at the T-residue at −6 (Figure 3A, compare lanes 1 and 2), marking the upstream boundary of the archaeal open complex (27). Wild-type levels of reactivity of T-residues were observed when H or Rpb5 were added to footprinting reactions (Figure 3A, lanes 3–5). These results indicate that subunit H is required for the formation of stable and full-length open complexes. To investigate initiation from a preformed open complex, a heteroduplex primed by a dinucleotide was used as template, as previously described (Figure 3B; 11). The ΔH RNAP formed only small amounts of tri-and tetranucleotide transcript in this assay (Figure 3B, lane 1), but could be rescued by the addition of H or of Rpb 5 to transcription reactions (Figure 3B, lanes 2 and 3). These findings demonstrate that formation of the stable open complex is not the only effect of the H subunit. Transcription on a DNA template containing a preformed transcription bubble shows that H is also essential for formation of the first phosphodiester bonds during the abortive phase of transcription.
A small fraction of ΔH RNAP is able to synthesize full-length transcripts
To investigate the ability of the ΔH enzyme to perform later steps in transcription, RNAP was stalled at position +20 on an immobilized template (6,27) and resumption of transcription was analyzed in the presence and absence of H. The ΔH RNAP formed fewer ternary complexes stalled at position +20 compared to complete RNAP (Figure 4, compare lanes 1a and 3a), but was rescued when H was added to PICs before initiation (Figure 4, lane 2a). However, the ternary complexes formed by the ΔH enzyme elongated, synthesizing run-off transcripts of 145 nt as efficiently as complete RNAP (Figure 4, lanes1a and b). The ratio of RNA in stalled ECs to run-off RNA formed after addition of NTPs to stalled complexes was the same for ΔH RNAP and complete RNAP (Figure 4 lanes a and b, and see quantification following the gel). Furthermore, the addition of H to stalled complexes of the ΔH enzyme did not increase resumption of elongation by the ΔH enzyme (Figure 4, lane 4). This finding suggests that subunit H contributes significantly to RNAP activity during early steps of transcription and probably also to the transition from initiation to elongation, but once the EC is formed, it is stable and equally active in the absence of H.
Characterization of DNA–protein contacts in stalled ECs
ECs stalled at position +20 of the gdh promoter do not undergo backtracking
For analysis of protein–DNA contacts in stalled ECs by photochemical cross-linking, it is essential that the 3′-end of RNA is located in the polymerase active site. In backtracked complexes, the active site is relocated to an internal position of RNA (33–37). Previously published data provide evidence that complexes stalled on the immobilized Pyrococcus gdh promoter at position +20 show exoIII digestion patterns and resumption after challenge with NTPs (27) that are typical for active complexes in which the active site of the RNAP has not moved to an internal position within the transcript. To provide additional biochemical evidence that the complexes stalled on the template derivatized with aryl azide at position +25 (Figure 5A, lanes 2–5) are not backtracked, transcription and resumption of complexes stalled at the template derivatized with the cross-linker at position +25 (Figure 5A, lanes 2–5) were compared with the complexes stalled on an identical but underivatized DNA template (Figure 5A, lanes 6–9). On both templates, the 20-nt RNA was the dominant transcript and it was completely chaseable to a longer run-off transcript after challenge with a complete set of NTPs (Figure 5A). The same resumption was observed when stalled ECs were challenged with NTPs in the presence of TFS (Figure 5A, lanes 5 and 9), which is known to induce a cleavage activity in the archaeal (and eukaryotic) RNAP (15). Since TFS rescues backtracked, RNAP in ECs by inducing RNA cleavage, the finding that resumption is essentially 100% in the absence of TFS, and is therefore not affected by TFS, also argues against backtracking of stalled RNAP.
Analysis of the kinetics of TFS induced cleavage activity on these complexes confirmed this conclusion. When EC20 complexes were incubated with TFS for 5 s in the absence of NTPs, an 18-nt RNA was observed as first cleavage product (Supplementary Data Figure S1, lanes 3, 13, 18 and 22). In backtracked complexes TFIIS induced cleavage released RNA fragments of 7–14 nt (35). Therefore, cleavage of two nucleotides in the archaeal EC20 indicates that the active site is close to the 3′-end of RNA as predicted for an active complex. When stalled complexes were formed with 3- or 15-min incubation times and then challenged after removal of NTPs with TFS for 5–15 min, the TFS-induced cleavage patterns were almost identical (Supplementary Figure S1, compare e.g. lanes 4 and 14 or 6 and 16). The patterns still remained the same when the RNAP was incubated after removal of NTPs for 3 or 15 min prior to TFS addition (Supplementary Figure S1, lanes 17–26). If backtracking were occurring, longer incubation would be expected to result in increased backtracking, causing a different TFS-dependent cleavage pattern. Since no changes in the pattern occurred, this indicates that all of the RNA cleavage was TFS-dependent and independent of backtracking. Taken together, these findings suggest that significant backtracking does not occur even after 15 min of incubation of EC20 at 70°C, while the cross-linked complexes in Figure 6 were formed for 5 min prior to UV radiation (‘Materials and Methods’ section).
Fe2+ substituting for Mg2+ in the RNAP active site was previously shown to cause highly localized cleavage of RNA (38). In ECs that do not backtrack, the 3′-end of the RNA stays close to the active site and Fe2+ therefore cleaves close to the 3′-end. Repositioning of the active site in backtracked complexes can be detected as an upstream shift of Fe2+-induced cleavage. Therefore, cleavage close to the active center is characteristic of active complexes where the 3′-end of RNA is still close to the bivalent cation. When Mg2+ was substituted by Fe2+ in complexes stalled at +20 an RNA fragment of ∼18 nt was formed (Figure 5b, lane 3) and therefore cleavage occurred close to the 3′-end of the transcript. This finding provides direct evidence that the complexes used for cross-linking experiments are not backtracked.
Subunit H cross-links to the DNA of the transcribed strand immediately downstream of the active center in elongation complexes
Previous cross-linking experiments provided evidence that subunit H, located in the lower jaw of RNAP, guides downstream DNA toward the active site (9,17). Our finding that H is required for early steps of transcription suggests a more important role of H during the transition from initiation to elongation. To investigate this in more detail the DNA–RNAP contacts were analyzed in ECs stalled at the gdh gene at position +20 and +45. For photochemical cross-linking of stalled complexes, the RNAP purified from Pyrococcus cells (nat RNAP) was used.
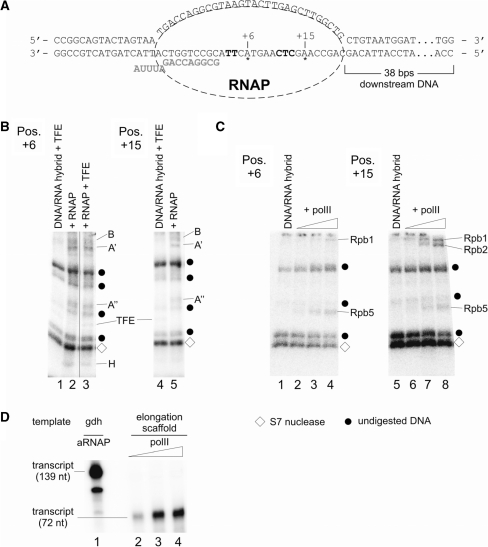
In the PIC, subunit H cross-links to the non-transcribed strand from position +14 to +20 and to the transcribed strand from position +12 to +20, as previously shown (17) and confirmed here using the same conditions used for the cross-linking of ECs (Supplementary Figure S2). If the geometry of RNAP–DNA interaction of the PIC is conserved in ECs, H would be predicted to interact with DNA from +30 to +40 in an EC stalled at +20. We analyzed cross-linking of stalled RNAP to DNA derivatized with APB (p-azidophenacyl bromide) on the transcribed (T) and non-transcribed (NT) DNA strands at various positions downstream of the transcription halt site at +20 (Figure 6A). When cross-links in the EC stalled at +20 were analyzed we observed prominent cross-links to subunits A′ and A′′ and weaker cross-links to subunit B at positions +30, +35 and +37 on the T strand (Figure 6A). No cross-link to subunit H was observed at either of these positions. However, when DNA derivatized at positions +21 to +30 was analyzed, a cross-link to a protein of ∼9–10 kDa was observed, most prominently at position +25. On the NT strand strong cross-links to B, A′ and A′ were observed at position +25, while position +37 showed strong cross-links to A′′ and weaker cross-links to B and A′ (Figure 6C). Surprisingly, only very faint cross-linking to H was observed at position +25NT (Figure 6C, lane 1). It is noteworthy that H cross-linking is most prominent at position +25 on the T strand as well (Figure 6B lane 12). At position +37 (Figure 6C, lane 2) and at all other positions tested between +21 and +35 on the NT strand, no cross-link of subunit H could be detected (data not shown). These findings suggest that H cross-links mainly to the transcribed strand immediately downstream of the active center in elongation complexes.
To identify the cross-linked 9–10-kDa protein in ECs as subunit H we used specific chemical cleavage of this particular cross-linked RNAP subunit essentially as described previously (17). RNAP subunit H does not contain cysteine (Cys) and methionine (Met) residues, but does contain an aspartic acid-proline motif (9,10). The 9–10-kDa protein cross-linked at +25T was excised from the gel and treated with cyanogen bromide (CNBr; cleaving at Met residues), 2-nitro-thiocyanobenzoic acid (NTCB; cleaving at Cys residues) and formic acid (cleaving at Asp-Pro motifs), respectively. No cleavage was detectable in the cleavage reactions containing CNBr or NTCB, indicating that the cross-linked protein did not contain Cys or Met residues (Supplementary Figure S3, lanes 1–4), whereas cleavage of the protein occurred when treated with formic acid (Supplementary Figure S3, lanes 5 and 6). This finding provided evidence that the protein cross-linking to DNA in close proximity of the active center in ECs contains an Asp-Pro motif, and thus it can be identified as RNAP subunit H. The cleavage activity of CNBr and NTCB was successfully tested in reactions with the largest subunit B, which contains Met and Cys residues (data not shown).
Subunit H cross-links at identical sites in a late EC
Several lines of evidence indicate that polII ECs stalled at position +45 are stable and have a low tendency for backtracking (39,40). To study DNA–RNAP interactions in a highly processive form of an EC, RNAP was stalled at position +45 and cross-linking to DNA derivatized at positions +50 and +58 was analyzed. Subunit H was cross-linked at position +50, which is 5 nt downstream of the active center, consistent with the results seen for the +20 stalled complex. At position +58, no interactions of H with the DNA were detectable (Figure 6C). Therefore, the location of H in late elongation complexes matches the position of H in RNAP complexes stalled at position +20. This finding suggests that the conformation of archaeal elongation complexes is the same whether the transcript is 20 or 45 nt long.
Rpb5 cross-links like H in an EC formed by an archaeal hybrid RNAP containing Rpb5
The high activity of ΔH RNAP in a complex with Rpb5 prompted us to investigate cross-linking of the hybrid enzyme containing the eukaryotic subunit. Since the ΔH enzyme is not able to initiate transcription efficiently in the absence of H or Rbp5, respectively, no detectable cross-links were formed by the ΔH RNAP (Figure 7, lane 6). When the hybrid enzyme containing the eukaryotic subunit was analyzed, a prominent cross-link of Rbp5 to position +23 was observed (Figure 7, lane 7), analogous to the cross-links observed to H at the same promoter position (Figures 6A and 7, lane 5). The appearance of B, A′ and A′′ cross-links in reactions containing the ΔH enzyme and Rpb5 give evidence that cross-linked Rpb5 essentially is incorporated into the stalled hybrid enzyme (Figure 7, compare lanes 6 and 7). According to this finding, Rbp5 not only rescues the transcriptional activity of the ΔH enzyme, but also seems to be positioned similarly to H in the archaeal elongation complex.
The cross-linking patterns of subunit Rpb5 are similar in an RNA polymerase II PIC and elongation complex
In the light of the high similarities between archaeal RNAPs and polII, along with the unexpected finding that subunit H or Rpb5 change positions relative to the template DNA in the archaeal EC compared to the PIC, we asked whether this structural rearrangement also happens with polII. To investigate this, the location of subunit H in the archaeal enzyme and of Rpb5 in polII was analyzed in a promoter independent assembly of an EC (29; Figure 8A, scaffold a) assembled as described in ‘Materials and Methods’ section. This scaffold is a good template for archaeal RNAP (16) even if it contains the APB cross-linking labels (data not shown). To investigate the position of H and Rpb5, the transcribed strand was derivatized with APB at positions located 6 and 15 nt downstream of the active center, respectively. As expected, subunit H was cross-linked to DNA derivatized at 6 nt downstream (Figure 8B, lanes 2 and 3) but was not cross-linked to the derivatized position 15 nt downstream of the active site (Figure 8B, lanes 4 and 5). At position 6 nt downstream, cross-linking of A′ and A′′ and weak cross-linking of B occurred. At position 15 nt downstream, cross-linking of B, A′ and A′′ was detected. TFE was cross-linked non-specifically to DNA at both positions (Figure 8B, lanes 1, 3 and 4) as it binds to single stranded DNA (6). As expected, TFE did not influence cross-linking of subunits B, A′, A′′ and H since it is not able to functionally interact with already formed ECs (6; Figure 8B, compare lanes 2 and 3).
Figure 8.
Mapping of archaeal RNAP and eukaryotic polII subunits cross-linked to an elongation scaffold. The templates for this assay were assembled essentially as described (29) but contained an azidophenacylated phosphorothioate substitution at positions +6 or +15, respectively, with adjacent internal radiolabel. (A) Sequence of the template in the final assembly. RNA is marked in bold gray letters, position of the cross-linker is indicated with asterisks, and internal radiolabels are highlighted in bold letters. (B) Cross-linking of archaeal RNAP subunits to positions +6 (left) and +15 (right). ECs were formed as described in ‘Materials and Methods’ section in the presence of heparin as non-specific competitor. Lanes 1, 3 and 4 contained TFE, which cross-linked non-specifically to both templates as described (6). Radiolabeled RNAP subunits and TFE were identified by their relative electrophoretic mobility and are indicated at the right-hand side of the gels. Note that subunit H cross-links only to position +6 and not to position +15. (C) Cross-linking of polII subunits to positions +6 (left) and +15 (right) on an elongation scaffold. Control lanes 1 and 5 contained the template without polII. Triangles above lanes 2–4 and 6–8 indicate an increase of polII from 36 to 110 nM polII in the reaction. As described in (B), po lII subunits were identified by their mobility by 4–19 % SDS-PAGE. As in (B), diamonds mark auto-radiolabeled S7 nuclease, while dots specify undigested DNA. (D) Eukaryotic polII is active on elongation scaffolds. Lane 1 shows RNA products synthesized in a standard in vitro transcription assay containing 46 nM archaeal nat RNAP and Pyrococcus gdh promoter DNA. Reactions analyzed in lanes 2–4 contained increasing amounts (36–110 nM) of polII and the elongation scaffold shown in (A) as template (for details see ‘Materials and Methods’ section).
In the eukaryotic PIC polII cross-links to positions +5 to +15 (20), but cross-linking of RNAP subunits with DNA in polII ECs have not yet been studied. To investigate the location of Rbp5 in the eukaryotic EC, polII was cross-linked to the elongation scaffold shown in Figure 8A. This elongation scaffold was transcribed with high efficiency by polII (Figure 8D, lanes 2–4; lane 1 is a run-off transcript synthesized by the archaeal RNAP shown for comparison) as described (29). When cross-linking of the complex formed by polII was analyzed at position 6 nt downstream of the active site, Rpb5 was clearly cross-linked (Figure 8C, lanes 2–4) and a weak cross-linking signal for Rpb1 was also detected. When the derivatized position was located 15 nt downstream of the active site, a clear cross-linking signal for Rpb5 and two signals corresponding to Rpb1 and Rpb2 were found (Figure 8C, lanes 6–8). These findings are consistent with the idea that the position of Rpb5 in the polII EC resembles the position of Rpb5 in RNAP II pre-initiation complex.
DISCUSSION
The position of RNAP subunit H changes during the transition from initiation to elongation
The cross-linking data of archaeal ECs presented in this study indicate a novel rearrangement of the RNAP’s lower jaw, consisting of the subunits A′, A′′ and H (Figure 9C). Cross-links to subunits B and A′, to DNA in the EC, or in the PIC (17 and Figure 6A) did not change relative to the active center (Figure 9A and B). However, unexpected changes in cross-linking patterns were observed with subunit H. While H marks the downstream boundary of protein–DNA contacts in the PIC at the T strand, cross-linking to positions +12 to +21 (17 and Figure 6A), no cross-links of H to the T strand were detectable 10–17 nt downstream of the active center in stalled ECs, but instead were observed on the T strand 1–7 nt immediately downstream of the active center (Figure 6A). The cross-linking pattern of H to the DNA of the NT strand also revealed dramatic changes of protein–DNA interactions in the EC compared to the PIC. While H cross-linked to NT strand DNA 14–20 nt downstream of the active center in the PIC, no H interactions with the NT strand were observed 17 nt downstream of the active center in the EC (Figure 6C) and at almost all other positions tested (data not shown). Only at position +25 on the NT strand a very faint signal suggests weak contacts of H to the DNA of the NT strand (Figure 6B). These findings indicate rearrangement of subunit H in ECs. Our lab previously reported cross-linking results for the +20 stalled complexes with cross-linkers incorporated into DNA upstream of the transcription halt site. We note that the upstream boundary of A′′ cross-linking was dramatically extended by more than one helical turn of DNA in ECs. The 5′-end of the A′′ cross-linking sites was 11 nt upstream of the active center in the EC, while it was 1 nt upstream of the active site in the PIC (6 and Figure 9). The downstream boundary of A′′–DNA cross-linking sites was not exactly determined in ECs, but extended at least to 17 nt downstream of the active site (Figure 6A) which is consistent with the DNA–A′′ cross-links observed in the PIC (17 and Supplementary Figure S2 and Figure 9B). The possibility that the novel-upstream cross-links to A′′ in the EC are caused by RNAP molecules paused close to the transcription start site due to APB derivatization is extremely unlikely, since all cross-linking sites observed for subunits B and A′ are located at identical positions relative to the active center in the PIC and EC. Inhibition of transcription read through due to APB derivatization can be excluded a priori for the cross-linking sites of H, since all APB derivatization sites cross-linked to H are located downstream of the active center (Figure 6A).
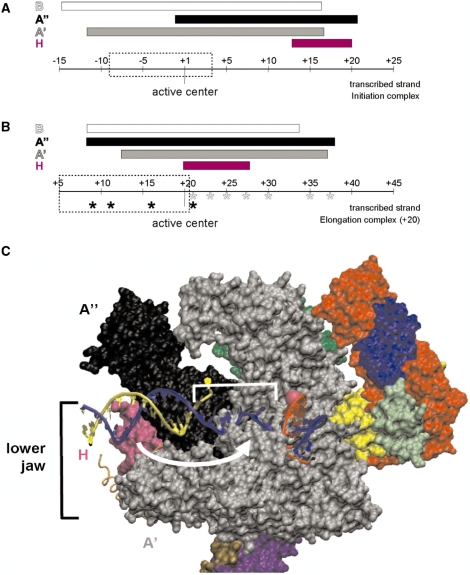
Figure 9.
Schematic summary of the repositioning of H and A′′ in elongation complexes. The range of protein/DNA interaction of RNAP subunits H (magenta), A′ (black), A′ (gray) and B (white) is symbolized by bars. The position of the RNAP’s active center is highlighted and the extension of the transcription bubble (27) is marked by dashed boxes. (A) Interactions of archaeal RNAP subunits with the transcribed DNA-strand in the PIC. H marks the far downstream end of the complex (based on the results published by ref. 17). (B) Schematic representation of cross-links of archaeal RNAP in a ternary complex stalled at +20. Cross-linking from position +9 to +21 was analyzed previously (6), and those derivatized sites are labeled by black asterisks. Cross-linkers from position +21 to +37 from this study are indicated with gray asterisks. Note that H is localized in close proximity to the active center and that the upstream boundary of A′′ is extended by ∼10 nt in stalled ECs. (C) Model for the path of DNA relative to RNAP in the archaeal elongation complex. The S. shibatae RNAP structure (PDB ID: 2WAQ; 13) was aligned with eukaryotic RNAP II in an EC (PDB ID: 2E2H; 35) using C-alpha coordinate ‘Iterative Magic Fit’ from Swiss PDBViewer 4.01 (54). R.M.S. deviation of the aligned structures was 1.55 Å for 1786 atoms. The RNAP II EC DNA and RNA coordinates were then merged with the archaeal RNAP structure coordinates and rendered concurrently using VMD 1.8.6 (54), revealing a good fit with few clashes. RNAP subunit colors are as used in (9), except for subunits G (darker green, obscured) and Rpo13 (orange trace) that were not part of the structure solved by (9). Subunit B was removed to allow visualization of the nucleic acids. The DNA transcribed and non-transcribed strands are blue and yellow, respectively, and the RNA is red. The active site is indicated by a pink sphere, and represents the +1 position in the EC. +1 to +10 of the T strand is bracketed, and the white arrow suggests the conformational change that subunit H (in magenta) would need to make to be cross-linked by aryl azide derivatizations within this region.
One possible explanation for shifted H and A′′ contacts is that the transcription EC may have backtracked when deprived of NTPs (39,40). However, several pieces of data argue against this. First, the exoIII border of Pfu EC20 is at +30 and therefore about 10 nt downstream of the last transcribed DNA base at the RNAP active center (27), whereas the last transcribed base is close to the downstream exoIII footprint edge when backtracking has occurred (35,41). Backtracked complexes show a low propensity to be elongated after challenge with NTPs and this ability is restored by TFS/TFIIS induced cleavage (41,42). The archaeal EC20 complex could be easily chased to the run-off and TFS did not affect this property (Figure 5A) indicating again that the 3′-end of the RNA was located close to the active site in the complex analyzed in cross-linking experiments. Furthermore, analysis of the kinetics of TFS induced cleavage did not reveal any evidence that backtracking occurred during longer incubation times (Supplementary Figure S1). When Mg2+ in the active center is exchanged by Fe2+, cleavage of RNA occurs through a radical mechanism. Fe2+-induced cleavage of RNA was observed in the internal part of RNA in backtracked complexes formed by E. coli RNAP but close to the 3′-end of RNA in active complexes (38). RNA cleavage in the archaeal EC20 complex containing Fe2+ led to a ∼18-nt RNA cleavage product (Figure 5B), indicating that the RNA 3′ terminus was close to the active site in these complexes. Taken together, these data provide strong evidence that the unexpected cross-linking patterns of subunit H reported here do not correlate with backtracking of RNAP to an internal position of RNA, and are therefore due to a conformational change of RNAP in the EC. Further analyses of a late archaeal EC stalled at position +45 (Figure 6c), and of EC complexes of the archaeal RNAP assembled in a quite different manner on an elongation scaffold (Figure 8) revealed basically the same cross-linking pattern and thus confirmed this conclusion.
Our findings that both A′′ and H interactions with DNA were considerably shifted in +20-stalled complexes suggest a major conformational change of the RNAP lower jaw in ECs, which is unexpected in the light of EC structural data (18, 43–45). To visualize the structural changes that must occur during transition from initiation to elongation by archaeal RNAP, DNA and RNA from a polII EC crystal structure (46) was modeled into the recently solved Sulfolobus shibatae RNAP crystal structure (13) by alignment of the RNAP subunits (Figure 9c). The nucleic acids fit in the archaeal RNAP structure with essentially no clashes, and the proximity of DNA to RNAP subunits is consistent with cross-links seen in the initiation complex (17). Downstream DNA is to the left of the active site (indicated by the pink sphere), with the RNA/DNA duplex extending out of the plane of the paper to the right of the active site. To explain the cross-links observed in the archaeal EC, a rearrangement in the lower jaw would be needed to bring H in close proximity to DNA near +3 relative to the active site (suggested by the white arrow), and also to bring A′′ close to transcribed strand DNA in the RNA/DNA duplex.
A conformational change in ECs may be a common property of the archaeal RNAP and the phylogenetically unrelated T7-RNAP
Darst and co-workers (47) reported a striking conformational change in the E. coli RNAP compared to the high-resolution crystal structure of the Taq RNAP and this conformational change was dominated by a 20° rotation of a region partially analogous to the lower jaw domain of the archaeal RNAP (47). This rotation results in the opening of the downstream RNAP channel by nearly 25 Å, suggesting a role in the formation of transcriptional active complexes. The flexibility of the region comprising structural components of the β and β′ subunits was further confirmed in single-pair fluorescence resonance energy transfer assays (48).
Structural studies revealed that the N-terminal domain of the single subunit RNAP from bacteriophage T7 undergoes substantial changes in conformation between initiation and elongation. A transcription mechanism involving two stages has been described for the T7 enzyme (49) and seems to operate also in multisubunit RNAPs (50), including the archaeal polymerase (27). Synthesis of the first 8 nt does not involve major conformational changes, the second stage occurring between synthesis of 9 and 14 nt includes a major conformational change of the T7-RNAP observed in processive ECs (51). Analysis of T7-RNAP bound to promoter DNA containing an 8-nt RNA revealed rotation of the promoter bound domain in the N-terminus and the bound promoter by ∼45°, allowing the active site to accommodate a growing heteroduplex as required in processive elongation complexes (52). It is tempting to speculate that the major transition involving repositioning of subunit H in the archaeal enzyme and establishing the processive conformation of the enzyme encountered in ECs occurs also during the transition from the first to the second stage in archaea.
Our finding that Rpb5 cross-links to the transcribed strand in an EC both at +5 and +15 suggests that a rearrangement involving subunit Rpb5 is unlikely in polII ECs. All the structural data available indicate that no significant structural change occurs in the lower jaw region during the transition from initiation to elongation in crystal structures of free forms of the eukaryotic polymerase with various elongation scaffolds (43–45). The differences in the molecular interactions with RNAP and DNA between the PIC and ECs reported here could be a specific property of the archaeal RNAP. Since the archaeal RNAP does not contain a stably bound Rpb9-like subunit (10,13), stabilization of the polII active center region mediated by Rbp9-Rpb1 contacts (53) and the presence of the eukaryotic specific N-terminal domain in Rpb5 could explain why a rearrangement of the lower jaw region in ECs might not be shared by the archaeal enzyme and polII.
The data presented here suggest a large rearrangement of the archaeal RNAP polymerase lower jaw during the transcription cycle. Future experiments will be necessary to determine at which point during early elongation the rearrangement of the RNAP lower jaw occurs, and whether subunit H assists or is passive in the rearrangement. In addition, the flexibility of the lower jaw is likely to be subject to modulation by external factors, and identification of such factors could identify regulators that control gene expression by influencing the structure of the transcription elongation complex. During termination, there is likely to be a rearrangement of the lower jaw to its pre-elongation state, and the timing and control of this rearrangement could also be subject to regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Priority program of the Deutsche Forschungsgemeinschaft (DFG) ‘genome function and gene regulation in archaea’; Forschergruppe FOR 1068/1 (Th422/11-1) established at the University of Regensburg (to S.G. and M.T.); National Institute of Health (NIH to M.S.B.); National Institute of General Medical Sciences (NIGMS) Academic Research Enhancement Award (AREA-1R15GM083306). Funding for open access charge: Funds of the University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The advice of Winfried Hausner for the experiments with TFS is appreciated. We thank Patrick Cramer and Herbert Tschochner for providing RNA polymerase II.
REFERENCES
- 1.Hausner W, Wettach J, Hethke C, Thomm M. Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J. Biol. Chem. 1996;271:30144–30148. doi: 10.1074/jbc.271.47.30144. [DOI] [PubMed] [Google Scholar]
- 2.Bell SD, Jackson P. The role of transcription factor B in transcription initiation and promoter clearance in the Archaeon Sulfolobus acidocaldarius. J. Biol. Chem. 2000;275:12934–12940. doi: 10.1074/jbc.275.17.12934. [DOI] [PubMed] [Google Scholar]
- 3.Hanzelka BL, Darcy TJ, Reeve JN. TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEalpha. J. Bacteriol. 2001;183:1813–1818. doi: 10.1128/JB.183.5.1813-1818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell SD, Brinkman AB, van der Oost J, Jackson SP. The archaeal TFIIEalpha homologue facilitates transcription initiation by enhancing TATA-Box recognition. EMBO Rep. 2001;2:133–138. doi: 10.1093/embo-reports/kve021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meinhart AJ, Blobel J, Cramer P. An extended winged helix domain in general transcription factor E/IIEalpha. J. Biol. Chem. 2003;278:48267–48274. doi: 10.1074/jbc.M307874200. [DOI] [PubMed] [Google Scholar]
- 6.Grünberg S, Bartlett MS, Naji S, Thomm M. Transcription factor E is a part of transcription elongaton complexes. J. Biol. Chem. 2007;282:35482–35490. doi: 10.1074/jbc.M707371200. [DOI] [PubMed] [Google Scholar]
- 7.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in Archaea. Similarity to that in Eukarya. Proc. Natl Acad. Sci. USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Ångstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 9.Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008;451:851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goede B, Naji S, von Kampen O, Ilg K, Thomm M. Protein-protein interactions in the archaeal transcriptional machinery. J. Biol. Chem. 2006;281:30581–30592. doi: 10.1074/jbc.M605209200. [DOI] [PubMed] [Google Scholar]
- 11.Koonin EV, Makarova KS, Elkins JG. Orthologs of the small RPB8 subunit of the eukaryotic RNA polymerases are conserved in hyperthermophilic Crenarchaeota and “Korarchaeota”. Biol. Dir. 2007;2:38. doi: 10.1186/1745-6150-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwapisz M, Beckouët F, Thuriaux P. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends Genet. 2008;24:211–215. doi: 10.1016/j.tig.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Korkhin Y, Unligil UM, Littlefield O, Nelson PJ, Stuart DI, Sigler PB, Bell SD, Abrescia N.GA. Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol. 2009;7:e102. doi: 10.1371/journal.pbio.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chédin S, Riva M, Schultz P, Sentenac A, Carles Ch. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hausner W, Lange U, Musfeldt M. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J. Biol. Chem. 2000;275:12393–12399. doi: 10.1074/jbc.275.17.12393. [DOI] [PubMed] [Google Scholar]
- 16.Reich C, Zeller M, Milkereit P, Hausner W, Cramer P, Tschochner H, Thomm M. The archaeal RNA polymerase subunit P and the eukaryotic polymerase subunit Rpb12 are interchangeable in vivo and in vitro. Mol. Microbiol. 2009;71:989–1002. doi: 10.1111/j.1365-2958.2008.06577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartlett MS, Thomm M, Geiduschek EP. Topography of the euryarchaeal transcription initiation complex. J. Biol. Chem. 2004;279:5894–5903. doi: 10.1074/jbc.M311429200. [DOI] [PubMed] [Google Scholar]
- 18.Kim TK, Lagrange T, Wang YH, Griffith JD, Reinberg D, Ebright RH. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc. Natl Acad. Sci. USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 20.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1421. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 21.Bartlett MS, Thomm M, Geiduschek EP. The orientation of DNA in an archaeal transcription initiation complex. Nat. Struct. Biol. 2000;7:782–785. doi: 10.1038/79020. [DOI] [PubMed] [Google Scholar]
- 22.Werner F, Weinzierl R.OJ. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 23.Naji S, Grünberg S, Thomm M. The RPB7 orthologue E’ is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J. Biol. Chem. 2007;282:11047–11057. doi: 10.1074/jbc.M611674200. [DOI] [PubMed] [Google Scholar]
- 24.Le TTT, Zhang S, Hayashi N, Yasukawa M, Delgermaa L, Murakami S. Mutational analysis of human RNA polymerase II subunit 5 (RPB5): The residues critical for interactions with TFIIF subunit RAP30 and Hepatitis B virus X protein. J. Biochem. 2005;238:215–224. doi: 10.1093/jb/mvi119. [DOI] [PubMed] [Google Scholar]
- 25.Kusser AG, Bertero MG, Naji S, Becker T, Thomm M, Beckmann R, Cramer P. Structure of an archaeal RNA polymerase. J. Mol. Biol. 2008;376:303–307. doi: 10.1016/j.jmb.2007.08.066. [DOI] [PubMed] [Google Scholar]
- 26.Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl Acad. Sci. USA. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spitalny P, Thomm M. Analysis of the open region and of DNA-protein contacts of archael RNA polymerase transcription complexes during transition from initiation to elongation. J. Biol. Chem. 2003;278:30497–30505. doi: 10.1074/jbc.M303633200. [DOI] [PubMed] [Google Scholar]
- 28.Shah SM, Kumar A, Geiduschek EP, Kassavetis GA. Alignment of the B′′ subunit of RNA polymerase III transcription factor IIIB in its promoter complex. J. Biol. Chem. 1999;274:28736–28744. doi: 10.1074/jbc.274.40.28736. [DOI] [PubMed] [Google Scholar]
- 29.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 30.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hethke C, Geerling A.CM, Hausner W, de Vos W, Thomm M. A cell-free transcription system for the hyperthermophilic Archaeon Pyrococcus furiosus. Nucleic Acids Res. 1996;24:2369–2376. doi: 10.1093/nar/24.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouhammouch ME, Werner F, Weinzierl R.OJ, Geiduschek P. A fully recombinant system for activator-dependent archaeal transcription. J. Biol. Chem. 2004;279:51719–51721. doi: 10.1074/jbc.C400446200. [DOI] [PubMed] [Google Scholar]
- 33.Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in 3′-5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 34.Izban MG, Luse DS. The increment of SII-facilitated transcript cleavage varies dramatically between elongation competent and incompetent RNA polymerase II ternary complexes. J. Biol. Chem. 1993;268:12874–12885. [PubMed] [Google Scholar]
- 35.Nudler E, Goldfarb A, Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- 36.Rudd MD, Izban MG, Luse DS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc. Natl Acad. Sci. USA. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl Acad. Sci. USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 39.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 40.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samkurashvili I, Luse DS. Structural changes in the RNA polymerase II transcription complex during transition from initiation to elongation. Mol. Cell. Biol. 1998;18:5343–5354. doi: 10.1128/mcb.18.9.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wind M, Reines D. Transcription elongation factor SII. BioEssays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 44.Westover DD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 45.Kettenberger H, Armache K.-J, Cramer P. Complete RNA polymerase II elongation complex structure and ist interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;12:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W. Conformational flexibility of bacterial RNA polymerase. Proc. Natl Acad. Sci. USA. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coban O, Lamb DC, Zaychikov E, Heumann H, Nienhaus GU. Conformational heterogeneity in RNA polymerase observed by single-pair FRET microscopy. Biophys. J. 2006;90:4605–4617. doi: 10.1529/biophysj.105.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steitz TA. Visualizing polynucleotide polymerase machines at work. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal M, Luse DS. The initiation-elongation transition: lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23. Proc. Natl Acad. Sci. USA. 2003;100:5700–5705. doi: 10.1073/pnas.1037057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandwar RP, Ma N, Emanuel SA, Anikin M, Vassylyev DG, Patel SS, McAllister WT. The transition to an elongation complex by T7 RNA polymerase is a multistep process. J. Biol. Chem. 2007;282:22879–22886. doi: 10.1074/jbc.M702589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durniak KJ, Bailey S, Steitz TA. The structure of a transcribing T7 RNA polymerase in transition form initiation to elongation. Science. 2008;322:553–557. doi: 10.1126/science.1163433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaros C, Briand J.-F, Boulard Y, Labarre-Mariotte S, Garcia-Lopez MC, Thuriaux P, Navarro F. Functional organization of the Rpb5 subunit shared by the three yeast RNA polymerases. Nucleic Acids Res. 2007;35:634–647. doi: 10.1093/nar/gkl686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.