Abstract
We have developed an effective, easy-to-use two-step system for the site-directed insertion of large genetic constructs into arbitrary positions in the Escherichia coli chromosome. The system uses λ-Red mediated recombineering accompanied by the introduction of double-strand DNA breaks in the chromosome and a donor plasmid bearing the desired insertion fragment. Our method, in contrast to existing recombineering or phage-derived insertion methods, allows for the insertion of very large fragments into any desired location and in any orientation. We demonstrate this method by inserting a 7-kb fragment consisting of a venus-tagged lac repressor gene along with a target lacZ reporter into six unique sites distributed symmetrically about the chromosome. We also demonstrate the universality and repeatability of the method by separately inserting the lac repressor gene and the lacZ target into the chromosome at separate locations around the chromosome via repeated application of the protocol.
INTRODUCTION
The ability to engineer plasmid constructs containing genetic elements of arbitrary complexity has transformed biology. The use of engineered plasmids has become ubiquitous as a way to controllably express and study genes and gene networks, and has proven to be an indispensable tool for determining the function of many gene products. While proving extraordinarily useful, problems with copy number, DNA size and stability often arise. For example, plasmids and bacterial artificial chromosomes (BACs) are generally maintained in multiple copies, with copy numbers ranging over several orders of magnitude (1–1000), depending on the replication origin. Average plasmid copy numbers can also be affected by the growth state of the cell (1), and even when maintained at constant growth conditions, cell-to-cell plasmid copy number fluctuations can be substantial (2). While the systematic variation of average plasmid copy number and copy number fluctuations have been studied extensively for a few systems, for the majority of plasmid replication origins it is unknown how copy number depends on the growth state of the cell, and how much cell to cell variation there is. This can lead to problems with the interpretation of experimental data, for example, in measurements of noise in gene expression (3), because the magnitude and effects of these fluctuations are almost completely unknown.
Thus, it is advantageous to incorporate constructs directly into the chromosome where the construct can be stably maintained without the need for antibiotic selection. While the position of the insertion relative to the replication origin can still lead to cell-to-cell copy number variability because of multiple replication forks, this variability is systematic, well understood (4), and can be corrected for or exploited.
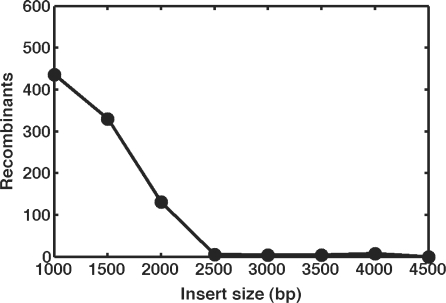
Unfortunately, it remains difficult to insert large DNA segments into the Escherichia coli chromosome. Currently, there are two main approaches to chromosomal integration: recombineering (5–11) and phage-derived methods (12). Recombineering is highly effective and easy to use, involving the expression of λ-Red enzymes in order to promote site-specific homologous recombination between the chromosome and a small linear polymerase chain reaction (PCR) fragment containing the desired sequence. By amplifying the linear fragment using primers, which contain 40–50-bp flanking regions homologous to the sequence of the desired insertion site, recombineering allows great flexibility in designing and choosing the chromosomal location and orientation. In addition, once the construct has been created, it can be inserted into various locations by designing new primers with the appropriate homology regions. Despite these advantages, recombineering in E. coli suffers from several shortcomings. For large fragments, it becomes increasingly difficult to generate PCR product in sufficient quantity, and the increased size of these fragments makes transformation and integration significantly less efficient. As a representative example, the number of recombinants we obtain when deleting lacZ with progressively larger PCR fragments bearing 50-bp homology extensions is illustrated in Figure 1 (6). Other laboratories have reported the successful and reliable integration of fragments up to ∼3.5 kb (13–16). In addition, integration efficiency can be enhanced by another order of magnitude by including homology regions 1 kb or larger (7). However, this generally requires the engineering of plasmid constructs bearing unique homology arms for each individual insertion fragment or location. Further restricting this approach is the general requirement to include an antibiotic marker on the inserted fragment to allow for the selection of successful integrants, occupying valuable real estate on the recombinant fragment. Because of these limitations, the insertion of large fragments into specific sites on the chromosome remains a non-trivial task.
Figure 1.
Representative recombineering efficiency as a function of insert size. 1000–4500-bp inserts containing the neo gene and bearing 50-bp flanking homology regions were inserted into the lacZ gene of strain K-12 MG1655 pTKRED via the method of Datsenko and Wanner (6). Cells were plated on LB agar + 25 μg/ml kanamycin, and the number of successful recombinants quantified as the number of resulting white colonies.
An alternative approach uses phage-integration systems to facilitate the insertion of synthetic constructs into the chromosome (12,17). Here, the donor plasmid contains a phage-specific attachment site (attP), which, when transformed into a host cell expressing the appropriate phage integrase enzyme, is integrated into complementary phage attB attachment sites in the chromosome. These phage-based systems have many advantages: they are highly efficient (17), and, in some instances, when the appropriate phage xis enzyme is expressed, the constructs can also be easily removed (12). Perhaps the greatest advantage of the phage-based systems is that there is effectively no limit to the size of the fragment that can be inserted at the attachment site. However, these approaches also have many disadvantages. Chief among these is the requirement for unique constructs to insert the same fragment into multiple different locations, since for each desired insertion location a new construct must be made bearing the required phage attP site. In addition, as the phage systems currently in widespread use in E. coli utilize endogenous chromosomal attachment sites, flexibility in choosing the insertion location is drastically reduced.
Recently, several groups have exploited the fact that double-strand DNA breaks stimulate in vivo recombination, thereby facilitating the high-throughput construction of plasmid libraries [MAGIC (18)], the subcloning of large fragments into BACs [ALFIRE (19)], or the recombination of short DNA fragments to introduce or repair mutations within the chromosome [gene gorging (20)]. These techniques utilize the yeast mitochondrial homing endonuclease I-SceI to introduce double strand breaks in the donor and/or recipient DNA molecule to enhance site-specific recombination. As the large 18-bp I-SceI recognition site does not exist naturally within the E. coli chromosome, introduction and cleavage of the recognition site at the desired location enhances site-specific recombination by several orders of magnitude (18) without any additional chromosomal damage.
Here, we describe a method for the chromosomal insertion of constructs that circumvents the limitations on insert size and location described above. To accomplish this, the cell is first transformed with a helper plasmid, pTKRED, harboring genes encoding the λ-Red enzymes, I-SceI endonuclease, and RecA. λ-Red enzymes expressed from the helper plasmid are used to recombineer a small (1.3 kb) ‘landing pad’, a tetracycline resistance gene (tetA) flanked by I-SceI recognition sites and 25-bp landing pad regions, into the desired location in the chromosome. After tetracycline selection for successful landing pad integrants, the cell is transformed with a donor plasmid carrying the desired insertion fragment; this fragment is excised by I-SceI and incorporated into the landing pad via recombination at the landing pad regions. In this manner, very large constructs can be inserted at any desired location within the chromosome. After successful integration, the I-SceI recognition sites in both the landing pad and the inserted fragment are eliminated, allowing successive applications of this protocol without modification of the landing pad regions. The entire procedure, from start to verified product, takes ∼1.5–2 weeks. This method has proven to be very easy to use and highly successful, allowing us to insert large (7 kb) fragments into several chromosomal locations without a single failure.
MATERIALS AND METHODS
Strains and Plasmids
Strains used were E. coli K-12 MG1655 (Coli Genetic Stock Center) in which the lac operon has been deleted by the method of Datsenko and Wanner (6) from the N terminal coding sequence of lacI to the C terminal coding sequence of lacA (henceforth denoted MG1655 Δlac, unless noted otherwise). Annotated sequences of pTKRED, pTKS/CS, and pTKIP versions are available as Genbank accession numbers GU327533, GU327534, GU327535, GU327536, GU327537, and GU327538 respectively).
Construction of the helper plasmid pTKRED
All PCRs were performed using Phusion Hi-Fi master mix (Finnzymes) and the sequences for all primers are listed in Supplementary Table SI. The helper plasmid pTKRED was constructed using the MAGIC plasmid pML104, the kind gift of Dr Steven Elledge (18), which contains the λ-Red enzymes under the control of a LacI regulated promoter, as well as constitutively expressed RecA. The I-SceI gene was amplified from the MAGIC strain BUN21 (18) and purified. An araC-ParaC-ParaBAD fragment was amplified from the plasmid pKD46 (6) using a 3′ primer with an extension homologous to the 5′ sequence of the I-SceI gene. This araC fragment was purified and fused to the I-SceI gene in a fusion PCR reaction. The resulting araC-ParaC-ParaBAD-I-SceI fragment was gel-purifed (using QIAEX II) and ligated into the SphI site of pML104, to create pTKRED-I. The lac repressor, along with its native promoter PlacI, was amplified from K-12 MG1655 and ligated into the NheI site of pTKRED-I, creating pTKRED.
Dependence of recombineering efficiency on insert size
The size-dependent efficiency of linear PCR fragment recombineering was assayed using the recombineering template pL451 (9), from which 1000–4500-bp fragments varying in length by 500-bp increments and containing the neo gene were prepared by PCR. These fragments were then used to transform competent MG1655 pTKRED, according to the method of Datsenko and Wanner (6). Competent cells were prepared in super optimal broth (SOB) medium supplemented with 0.5% w/v glucose and 100 μg/ml spectinomycin; 2 mM IPTG was added with the initial inoculum to induce expression of λ-Red enzymes. An identical culture was prepared in parallel without isopropyl β-d-1-thiogalactopyranoside (IPTG) to serve as a negative control. Once the OD (λ = 600 nm) of the culture reached ∼0.5–0.6, the cells were placed on ice and washed 3× with ice-cold 10% v/v glycerol. One-hundred microliters of the resulting competent cells was added to ∼100 ng of purified PCR fragment and the resulting mixture was electroporated in 0.1-cm-gap cuvettes (USA Scientific) at 2.0 kV, 25 uF, 200 Ω in a Gene Pulser electroporation apparatus (BioRad). Cells were resuspended in 1 ml SOC medium. The cells were allowed to recover at room temperature for 24 h, and then plated on LB agar plates with 25 μg/ml kanamycin, 0.1% X-Gal and 2 mM IPTG and incubated overnight at 37°C. The number of successful recombinants was the number of white colonies per plate.
Construction of the donor plasmid pTKIP
Two unique, random 25-bp sequences with ∼50% GC content were generated using a random sequence generator in MATLAB (MathWorks). The resulting sequences were BLASTed against the E. coli genome to ensure their uniqueness. The sequences obtained in this way and used throughout this communication were landing pad region 1: 5′-TACGGCCCCAAGGTCCAAACGGTGA-3′; landing pad region 2: 5′-GATGGCGCCTCATCCCTGAAGCCAA-3′. To generate the donor plasmid pTKIP, primers containing I-SceI recognition sites were used to amplify the pBR322 backbone containing the bla ampicillin resistance gene and the pMB1 replication origin. Primers containing I-SceI sites, as well as the 25-bp landing pad regions given above, were used to amplify the multiple cloning site (MCS) and neo resistance gene from the recombineering plasmid pL451 (9). Both the backbone and fragment were digested with I-SceI (New England Biosciences), gel purified and ligated together. The resulting insertion platform includes a bla ampicillin resistance marker in the backbone for simplified screening against clones retaining pTKIP after insertion.
Several versions of the pTKIP plasmid were generated containing antibiotic resistance genes within the insertion fragment: neo (kanamycin resistance from pL451), cat [chloramphenicol resistance; amplified from pZA31-luc (21)], dhfr [trimethoprim resistance; amplified from pAH145 (12)] or hph [hygromycin B resistance; amplified from p220KattBfull (17)], the kind gift of Dr Michele Calos]. These alternate versions of pTKIP were generated using recombineering to exactly replace the neo gene of pTKIP-neo in SW105 (11). These plasmids contain the indicated antibiotic resistance genes flanked by flippase recognition target (FRT) sites. After successful integration, the resistance genes can be eliminated via expression of flippase recombination enzyme (FLP) recombinase from e.g. pCP20 (6).
Construction of insertion fragment
A 7-kb regulatory unit consisting of a lacI:venus translational fusion and a lacZ reporter gene was constructed to test the efficacy of the insertion method. A T1 Rho-independent terminator was amplified from pZA31-luc (21) and purified. A PlacI-lacI:venus fusion was then amplified from strain JE13, the kind gift of Dr Sunney Xie (22), using primers with an overlap extension homologous to the previously amplified T1 terminator. lacI:venus and the T1 terminator were then coupled together in a fusion PCR reaction and inserted into the KpnI and SalI sites of pTKIP-neo, yielding pTKIP-IvT. The insertion fragment is referred to throughout as the IvT-neo cassette. Primers including a PLlacO1 promoter (21) were then used to amplify lacZ from MG1655, which was subsequently inserted into the HindIII and NheI sites of pTKIP-IvT to yield pTKIP-IvT-O1Z. This large fragment is referred to throughout as the IvT-O1Z-neo cassette. To test the repeatability of the protocol, an additional construct was made carrying only PLlacO1-lacZ on plasmid pTKIP-cat without the lacI:venus:T1 regulator. This insertion fragment is referred to as the O1Z-cat cassette.
Construction of the landing pad plasmid pTKS/CS
pTKS/CS was generated using the technique described above for pTKIP. A backbone including the cat chloramphenicol resistance gene and a p15A replication origin was amplified from pZA31-luc (21) using primers containing the 25-bp landing pad regions as well as I-SceI recognition sites. Primers containing I-SceI recognition sites and the promoter PlacIQ1 (23) were used to amplify tetA from the CRIM plasmid pAH162 (12). These fragments were digested with I-SceI and ligated together, yielding pTKS/CS. This plasmid contains the tetA tetracycline resistance gene flanked by the 25-bp landing pad regions and I-SceI recognition sites, and is used as a PCR template for the preparation of linear landing pad fragments for recombineering. The cat gene in the backbone can be used to screen successful landing pad integrants from cells transformed by undigested pTKS/CS.
Landing pad integration
pTKS/CS was used as a PCR template to amplify landing pad fragments using the landing pad regions as standardized priming sites. The primers included 50-bp sequence homology for the desired insertion location in the chromosome. PCR conditions were as follows: 95°C for 30 s, followed by 35 cycles of 98°C for 15 s, 55°C for 15 s and 72°C for 30 s. The resulting PCR reactions were digested with 1 μl DpnI per 50 μl PCR reaction for at least 2 h at 37°C and purified using a QIAquick spin column. Cells containing pTKRED were prepared and electroporated with ∼100 ng of purified landing pad fragment as detailed earlier. After electroporation, 1 ml SOC medium was added. After recovery for at least 2 h, 500 μl was plated on LB agar plates containing 10 μg/ml tetracycline, 100 μg/ml spectinomycin and 0.5% glucose and grown overnight at 30°C. The remaining ∼500 μl was allowed to recover overnight and plated the next day. Potential integrants were picked onto LB plates supplemented with 34 μg/ml chloramphenicol to screen against colonies which had been transformed with undigested pTKS/CS plasmid. The high level of expression from the PlacIQ1 promoter combined with the significantly decreased growth rate of cells expressing tetA (24) makes it easy to distinguish landing pad integrants from higher copy number pTKS/CS transformants, and, consequently, all potential integrants passed this screen. We also verified that chromosomal landing pad integrants cannot grow on low nutrient LB agar plates supplemented with fusaric acid (25). Samples were verified by colony PCR across the desired insertion junctions.
Fragment insertion
Individual colonies were inoculated into 5 ml of EZ-Rich Defined Medium (RDM; Teknova) +0.5% glycerol, 2 mM IPTG, and 0.2% w/v l-arabinose. After growing at 37°C for 1 h in a shaking water bath (New Brunswick Scientific), 100 μg/ml spectinomycin was added to the culture and the tubes were transferred to a 30°C shaking water bath for 4 h. The appropriate antibiotic for the given insertion fragment was then added (25 μg/ml kanamycin, 34 μg/ml chloramphenicol, 100 μg/ml hygromycin or 300 μg/ml trimethoprim), and the cultures were grown overnight. The next day, samples were diluted 105× and 100 μl was plated on LB plates with the appropriate antibiotic and grown at 37°C. Potential integrants were picked and screened on LB plates containing 100 μg/ml ampicillin or 10 μg/ml tetracycline to verify the loss of the landing pad and donor plasmid. Clones passing this screen were again verified by colony PCR across the integration junction, and the resulting PCR fragments were sequenced (Genewiz).
Curing of pTKRED
Verified clones containing the desired fragment were picked into 5-ml LB and grown overnight in a shaking water bath at 42°C. The next day, samples were diluted 105× and 100 μl was plated on LB agar plates and grown at 37°C. Colonies were then picked onto 100 μg/ml spectinomycin LB agar plates to verify loss of pTKRED.
Elimination of antibiotic resistance genes
After elimination of pTKRED, cells were transformed with pCP20 (6), which constitutively expresses FLP recombinase. Individual colonies were picked into 5-ml LB and grown overnight at 42°C. Samples were diluted 105×, and 100 μl of this dilution was plated on LB agar plates and grown overnight at 37°C. Colonies were screened against the retention of pCP20 and the chromosomally incorporated antibiotic marker. All colonies tested had lost both pCP20 and the chromosomal antibiotic marker.
RESULTS
Strategy
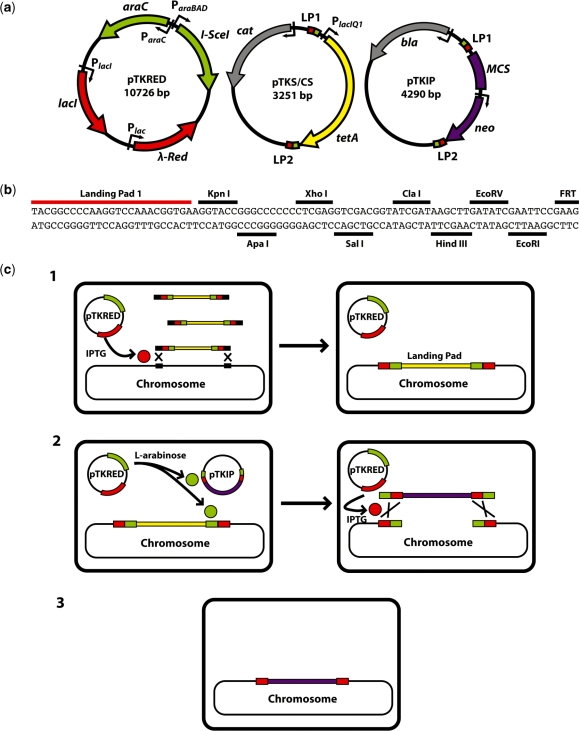
The three plasmids used in the protocol are diagrammed in Figure 2a, and the general strategy is outlined in Figure 2c. The first step is the transformation of the desired host strain with the helper plasmid pTKRED, containing the spectinomycin resistance marker aadA. pTKRED carries the genes and regulatory elements necessary for all downstream steps, including a constitutively expressed recA gene and the three λ-Red genes gam, bet and exo driven by a LacI regulated, IPTG inducible promoter. These genes are necessary for the integration of a landing pad at the desired integration site and the enhancement of fragment insertion via recombination. The constitutive expression of RecA allows for efficient recombineering of recA− host strains, such as the common laboratory strain DH5α (7). In addition, pTKRED harbors a ParaBAD-driven I-SceI gene inducible with l-arabinose. pTKRED bears a temperature-sensitive pSC101 replication origin, which maintains the plasmid at low copy number and allows for easy curing by growth at 42°C and screening against spectinomycin resistance.
Figure 2.
(a) Plasmids used in the integration protocol. The sequence size given is for pTKIP-neo; neo is exactly replaced with various antibiotic resistance genes for alternate versions of pTKIP. Small green boxes are I-SceI restriction sites; landing pad regions 1 and 2 are small red boxes labeled LP1 and LP2 respectively. (b) Annotated sequence of the pTKIP MCS showing LP1, available restriction sites, and the first four bases of the adjacent FRT site. (c) Strategy for large construct chromosomal integration. Step 1: the host strain is transformed with the helper plasmid pTKRED, bearing I-SceI endonuclease (green) and λ-Red (red). Linear landing pad fragments (yellow) are integrated into the chromosome at the desired location (black squares) when λ-Red expression is induced by IPTG. Step 2: the host strain is transformed with pTKIP bearing the fragment (purple) to be inserted into the landing pad. I-SceI expression is induced via the addition of l-arabinose, and the I-SceI recognition sites (green) in the donor plasmid and chromosome are cleaved. Integration of the fragment is facilitated by IPTG-induced λ-Red expression. Step 3: pTKRED is cured by growth at 42°C and screening against spectinomycin resistance.
Next, pTKS/CS is used as a PCR template to amplify a small 1.3-kb landing pad, consisting of a tetA tetracycline resistance gene flanked by I-SceI endonuclease recognition sites and small 25-bp landing pad regions. The small size of this construct allows the simple and reliable insertion of the landing pad into any location using previously established recombineering methods (6,7,10). tetA allows for both the easy selection of successful tetracycline-resistant integrants and later counterselection against landing pad retention using fusaric acid or nickel salts (25–27) after I-SceI stimulated replacement of the landing pad. However, in our hands the process has proven sufficiently effective to make counterselection unnecessary. The tight regulation of I-SceI by ParaBAD in the absence of l-arabinose (28) allows for the maintenance of pTKRED throughout the entire integration process, eliminating the need for tedious repeated transformations with pTKRED.
Finally, the host strain is transformed with the donor plasmid pTKIP. This plasmid contains the construct to be inserted flanked by I-SceI endonuclease recognition sites and the same 25-bp landing pad region contained within the landing pad. I-SceI expression is induced with l-arabinose, leading to cleavage of both the donor plasmid and the chromosome at the site of landing pad insertion. The incorporation of the insertion fragment into the landing pad is enhanced by the expression of the λ-Red enzymes and the introduction of double strand breaks in both the donor and chromosome. After overnight growth in l-arabinose and IPTG, the majority of surviving cells expressing the appropriate antibiotic marker have stably integrated the donor construct. Due to the small size of the landing pad regions, double-strand breaks caused by I-SceI cleavage are required for efficient integration, and the resulting destruction of the I-SceI sites allows for repeated insertions without the need for additional constructs containing novel landing pad regions.
Curing the donor plasmid
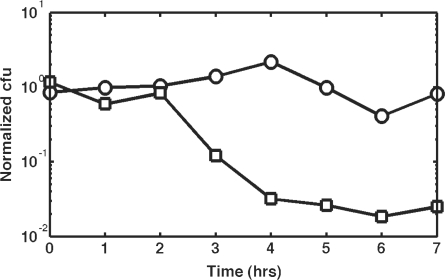
pTKRED includes a temperature sensitive pSC101 replication origin. This plasmid is thus easily cured by growth at 42°C and screening against spectinomycin resistance. Of more concern is the donor plasmid pTKIP, which is cured by I-SceI cleavage. To study the efficiency and kinetics of pTKIP curing by I-SceI expression, we transformed MG1655 with pTKRED and subsequently transformed the resulting cells with pTKIP-neo or pL451 without I-SceI recognition sites to serve as a control. These cells were inoculated into RDM medium containing 0.5% v/v glycerol, and I-SceI expression was induced by the addition of 0.2% w/v l-arabinose. Samples were taken every hour and plated on LB agar with and without plasmid antibiotic markers to measure the rate and completeness of plasmid curing. The results are shown in Figure 3; the curing of pTKIP is very efficient, with only ∼1% of cells retaining the donor plasmid.
Figure 3.
pTKIP is cured by in vivo I-SceI cleavage. Circles indicate the number of cells retaining pTKRED, squares indicate retention of pTKIP-neo. Normalized cfu is the ratio of surviving colonies on plates containing the appropriate antibiotic (100 μg/ml spectinomycin and 25 μg/ml kanamycin for pTKRED and pTKIP, respectively) to surviving colonies on LB plates without selection.
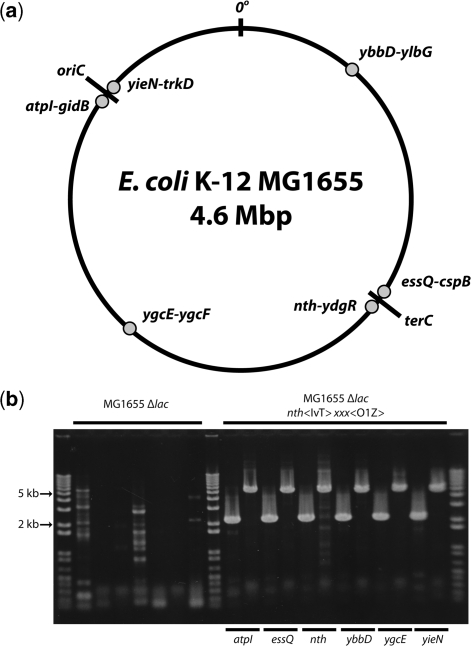
Large construct insertion in six unique chromosomal locations
We used the method outlined above to insert a 7-kb fragment into six unique locations distributed symmetrically about the E. coli replication origin (Figure 4a). The inserted fragment was a large transcriptional unit (the IvT-O1Z-neo cassette) constructed on pTKIP-neo composed of a lacI:venus:T1 translational fusion (22,29) and a lacZ reporter gene driven by the synthetic LacI regulated promoter PLlacO1 (21).
Figure 4.
(a) Chromosomal insertion positions. Fragments were inserted into six positions distributed symmetrically about the E. coli chromosomal origin of replication oriC. Insertion positions are between the marked genes at each position (dots). (b) Verification of chromosomal insertions. Colony PCR across insertion junctions for insertion of the PlacI-lacI:venus:T1-neo cassette (IvT-neo; 2.1-kb band) into the nth-ydgR position and the PLlacO1-lacZ-cat cassette (O1Z-cat; 5-kb band) at each site. The insertion of both the IvT-neo and O1Z-cat cassettes into the nth-ydgR position was accomplished by a single 7-kb insertion bearing the neo marker. Lanes 2–8 are MG1655 Δlac negative controls in the following order: lane 2: nth IvT-neo; lanes 3–8: O1Z in alphabetical order of insertion positions, corresponding to positive insertion lanes 10–20.
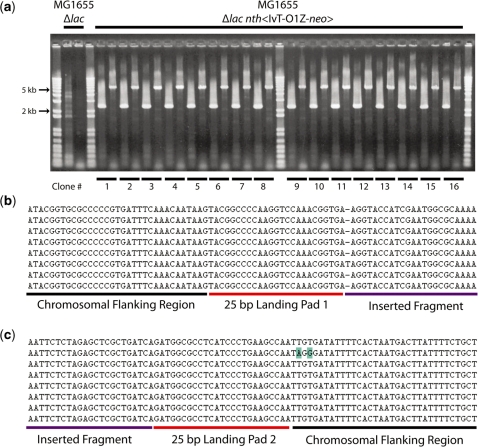
Cells with landing pad insertions at each position in Figure 4a were transformed with the donor plasmid pTKIP-IvT-O1Z and the insertion of the IvT-O1Z-neo cassette was performed as described in ‘Materials and Methods’ section. After insertion, cultures were diluted 105×, and 100 μl was plated on LB agar plates with 0.1% X-Gal, 2 mM IPTG and 25 μg/ml kanamycin and incubated overnight at 37°C. All of the resulting colonies were blue, and of the colonies picked for further analysis all showed the proper antibiotic resistances, indicating the insertion of the fragment and loss of the pTKIP plasmid and tetracycline landing pad. PCR across the integration junctions verified successful integration. Figure 5a shows the result of PCR verification of 16 clones after construct insertion at the atpI-gidB intergenic region.
Figure 5.
(a) Verification of large chromosomal insertions. Colony PCR of 16 randomly picked colonies obtained from insertion of IvT-O1Z-neo between atpI and gidB. Lane 2: MG1655 Δlac pTKRED negative control atpI proximal junction. Lane 3: MG1655 Δlac pTKRED negative control gidB proximal junction. Lanes 5–20 and 22–37 are IvT-O1Z-neo cassette integrants. Lanes are alternating pairs of atpI proximal junctions (2.1-kb bands; lacI:venus:T1 amplified) and gidB proximal junctions (5-kb bands; lacZ-neo amplified) for each clone. (b, c) Alignment of insertion junction sequences obtained from first eight clones shown in (a). (b) Junction proximal to atpI. (c) Junction proximal to gidB. Sequences of the flanking chromosomal regions, 25-bp landing pad regions, and the inserted fragment are labeled and indicated by a black, red or purple underscore, respectively. Mismatches to the expected sequence are highlighted in blue.
The products obtained from PCR verification of the atpI-gidB integration junctions were subsequently sequenced, and the alignment of eight representative sequences is shown in Figure 5b and c. The correct chromosomal sequence is obtained in the flanking regions, with the sequence of the integrated fragment flanked by the 25-bp landing pad regions. The sequence of the inserted fragment was identically correct for all eight clones and is not shown in Figure 5b and c for brevity. The I-SceI sites of both the landing pad and the donor fragment were eliminated, although in two instances 1 or 2 bp flanking the 25-bp landing pad region were incorrect. Due to the high sequence fidelity of the inserted construct, we speculate that these isolated mismatches flanking the 25-bp landing pad region are due to imprecise elimination of the I-SceI sites.
Repeated insertion of constructs at multiple locations
To demonstrate the repeatability of the protocol, we first inserted the IvT-neo cassette into the nth-ydgR intergenic region. This insertion was verified via PCR and the resulting clones were kanamycin resistant. We then cloned PLlacO1-lacZ into pTKIP-cat and attempted to insert this O1Z-cat cassette into each of the remaining positions indicated in Figure 4a. After insertion, the cultures were diluted 105× and plated on LB plates including X-Gal, 2 mM IPTG, 25 μg/ml kanamycin and 34 μg/ml chloramphenicol. All resulting colonies were blue, indicating insertion of the O1Z-cat cassette. Colonies were picked onto LB plates containing 25 μg/ml kanamycin and 34 μg/ml chloramphenicol, 100 μg/ml ampicillin or 10 μg/ml tetracycline. All colonies displayed the correct antibiotic resistances, and representative PCR verifications across the insertion junctions are shown in Figure 4b.
Insertion without antibiotic selection
Selection of successful chromosomal integrants of a large construct is a straightforward process because the inserted fragment includes a unique antibiotic marker. However, other studies in which double-strand breaks have been induced in the E. coli chromosome have shown such damage to be lethal (30). We, therefore, reasoned that after induction of chromosomal breaks by I-SceI, successful integration of fragments resulting in chromosomal repair would provide sufficient selective pressure to eliminate all but successful integrants from the culture, eliminating the need for antibiotic selection.
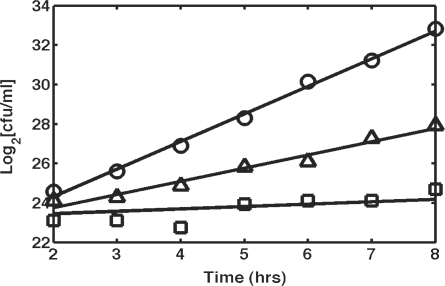
To test the lethality of I-SceI-mediated chromosomal breaks, landing-pad integrants were inoculated into RDM medium with 0.5% v/v glycerol, 2 mM IPTG and 0.2% w/v l-arabinose. Another tube was prepared in parallel with identical conditions using MG1655 Δlac pTKRED as a control. Samples were taken every hour and plated on LB plates to measure the viability of the cells. The results for landing pad integrants in the atpI-gidB intergenic region (squares) and the essQ-cspB intergenic region (triangles) are shown in Figure 6. The growth curves display site-specific sensitivity to chromosomal cleavage, since the growth of atpI-gidB landing pad integrants is arrested by expression of I-SceI (doubling time 8.3 h), whereas essQ-cspB landing-pad integrants grow at a significantly reduced rate (doubling time 1.5 h) compared to cells with no landing-pad insertion (doubling time 43 min). Colonies grew after replica printing onto low nutrient fusaric acid plates (25), verifying excision of the landing pad. We postulate that the ability of cells to survive in spite of chromosomal cleavage is due to repair via recombination of the chromosomal break mediated by overexpression of λ-Red enzymes. This hypothesis is supported by the observation that repeating the above experiment in the absence of IPTG and plating on fusaric acid LB agar plates results in a complete lack of growth for all landing-pad integration strains studied thus far. It is, however, unclear what determines the site-specificity of the ability to repair the chromosome.
Figure 6.
I-SceI-induced chromosomal breaks are not lethal in the presence of λ-Red. Solid lines are linear regression fits used to calculate doubling time. Circles: MG1655 Δlac pTKRED without landing pad, doubling time 43 min; triangles: essQ-cspB landing pad insertion, doubling time 1.5 h; squares: atpI-gidB landing pad insertion, doubling time 8.3 h.
We next attempted the insertion of the same 7-kb IvT-O1Z-neo cassette used above without antibiotic selection. The insertion was performed as above in medium supplemented with 0.2% l-arabinose with and without the addition of 2 mM IPTG to determine the degree of enhancement of recombination stimulated by the expression of λ-Red. After overnight growth, cultures were diluted 105× and plated on LB agar plates with 0.1% X-Gal and 2 mM IPTG. The resulting blue/white colony counts are given in Table 1. In instances where chromosomal breaks are lethal (e.g. atpI-gidB and nth-ydgR) markerless insertion is extremely effective, with ∼100% successful integration with or without the addition of 2 mM IPTG. However, when the chromosomal break occurs in non-lethal sites, the insertion statistics are less favorable. Induction of λ-Red increases the efficiency of insertion considerably, with the least effective insertion position (essQ-cspB) yielding 19.2% successful integrants. Without the induction of λ-Red, the efficiency of fragment insertion drops to 5.9–14.5%.
Table 1.
Selectionless insertion statistics for insertion of the 7-kb IvT-O1Z-neo cassette at the indicated positions
| Insertion location | IPTG (λ-Red induction) | Total (blue/white) | % Integrants |
|---|---|---|---|
| atpI-gidB | + | 705/4 | 99.4 |
| − | 1010/44 | 95.8 | |
| yieN-trkB | + | 642/15 | 97.7 |
| − | 461/163 | 73.9 | |
| ygcE-ygcF | + | 39/100 | 28.6 |
| − | 184/1084 | 14.5 | |
| ybbd-ylbG | + | 178/133 | 57.2 |
| − | 354/2295 | 13.4 | |
| nth-ydgR | + | 191/0 | 100 |
| − | 362/16 | 95.8 | |
| essQ-cspB | + | 377/1583 | 19.2 |
| − | 43/685 | 5.9 |
Samples were plated on LB agar plates +0.1% X-Gal, 2 mM IPTG without antibiotic selection; these plates were replica printed onto LB agar plates with 25 μg/ml kanamycin, 100 μg/ml ampicillin or 10 μg/ml tetracycline.
% integrants: the number of blue colonies displaying appropriate antibiotic sensitivities divided by total number.
DISCUSSION
Our protocol allows large fragments to be inserted into any desired position and orientation in the chromosome. Our method takes ∼1.5–2 weeks to accomplish from start to finish, which does not include the time required for engineering the donor plasmid. It should be noted that due to the requirement to clone the insertion fragment into a donor plasmid, our method suffers from the general weaknesses inherent in molecular cloning, such as MCS/insert compatibility.
Here, we have demonstrated a substantial increase in capacity over previously reported recombineering attempts (13–16). We have not yet explored the very upper limits of insertion size. However, our method bears some resemblance to the ALFIRE method for BAC subcloning (19), which was shown to allow the transfer between BACS of large fragments up to 55 kb in size. We think it likely that our method will allow for the insertion of similarly large constructs into the chromosome using a donor BAC. In addition, even considering the introduction of the two 25-bp landing pad regions, our method is ‘cleaner’ than phage-derived insertion strategies (12,17), which generally result in integration of the entire donor plasmid into the phage attachment site. Consequently, many extraneous features, such as the plasmid replication origin, are inserted along with the desired fragment. Our method, in contrast, results in only the insertion of the homology region-flanked fragment, the sequence of which can be controlled precisely. In instances where the landing pad sequences cannot be tolerated, or exact replacement is required, the landing pad and donor plasmid can be modified to use the appropriate chromosomal sequences as the necessary landing pad regions. For applications, such as the one described here, where disruption of the genome is a tangential concern, the introduction of these sequences is likely to be insignificant, and the ease of use afforded by the standardized sequences in our view outweighs the repeated modification of the donor and landing pad that would be necessary to eliminate these scars.
A potential problem with the sequential use of the standardized landing pad regions is that repeated insertions into the same strain could lead to replacement of previously inserted fragments. The 25-bp size of the landing pad regions was chosen specifically to reduce the efficiency of λ-Red mediated homologous recombination 1000–10 000× below that obtained with larger (>40 bp) homology regions (7). This reduction in recombination efficiency reduces the efficiency of replacement of previously inserted fragments via homologous recombination at the standardized landing pad regions. The use of I-SceI to introduce double-strand breaks in the landing pad and donor increases the efficiency of recombination at these sites by ∼5000× (18). In addition, the maintenance of selective pressure by incubating the cells in the presence of the appropriate antibiotics ensures retention of all inserted fragments.
We have also shown that in many instances it is possible to perform markerless insertion. If an identifiable reporter gene is being inserted (such as lacZ in our case), successful integrants can be easily recognized without antibiotic selection. In all of the cases studied here, when recombination of the insertion fragment is enhanced by the induction of λ-Red, the percentage of successful integrants is ∼20% or greater. It therefore seems that even without an easily identifiable reporter, successful integrants can be found via hybridization or with a limited number of PCR screens. In such cases, and if the landing pad regions are altered to coincide with the desired integration site, it should be possible to insert very large fragments into the chromosome without any additional extraneous sequence. The internal boundaries of landing pad regions 1 and 2 are marked by KpnI and BclI restriction sites; in such cases where markerless insertion is desired, the construct can be cloned between these two sites, leaving little extraneous sequence to serve as potential competing recombination targets upon subsequent application of the method.
For routine chromosomal insertion, we have constructed a set of donor plasmids expressing a variety of easily selectable antibiotic markers. These markers are flanked by FRT recombination sites and, after insertion, they can be easily removed via the expression of FLP recombinase. Since an intact 82–85-bp FRT site is left as a scar after excision of the marker (6), care must be taken in performing multiple repeated insertions in close proximity, as expression of FLP can result in the excision of the entire intervening region.
The in vivo cleavage of the donor plasmids by I-SceI expression has many advantages over introduction of linear DNA fragments by direct transformation. The replication of the donor plasmid is subject to the repair and editing mechanisms employed by the cell during chromosomal and plasmid replication, allowing for higher fidelity replication of the desired fragment than can be achieved by PCR (20). In addition, the transformation of supercoiled donor plasmid is highly efficient, and maintenance of relatively high plasmid copy numbers (∼50 copies for the pMB1 origin used in pTKIP) allows for the maintenance of a much higher intracellular fragment concentration than can be achieved by transformation with linear PCR fragments.
The stable integration of synthetic constructs into any desired chromosomal site should prove useful, for example, in cases where the multi-copy dosage resulting from plasmids is problematic, or where measurements with precision greater than plasmid copy number noise will allow are required. We believe that extension of this method should prove straightforward; we are currently attempting to adapt the system to other microorganisms and to apply the method to study the impact of prokaryotic chromosomal organization on gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (GM078591, GM071508); and the Howard Hughes Medical Institute (52005884). Funding for open access charges: National Institutes of Health and Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Frederick Blattner, Dr Michele Calos, Dr Steven Elledge, Dr Atsushi Miyawaki and Dr Sunney Xie for kindly providing requested strains and constructs and the required permissions for their use. We also thank Dr Justin Kinney and the lab of Dr Frederick Blattner for useful discussions and suggestions.
REFERENCES
- 1.Lin-Chao S, Bremer H. Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli. Mol. Gen. Genet. 1986;203:143–149. doi: 10.1007/BF00330395. [DOI] [PubMed] [Google Scholar]
- 2.Paulsson J, Ehrenberg M. Noise in a minimal regulatory network: plasmid copy number control. Q. Rev. Biophys. 2001;34:1–59. doi: 10.1017/s0033583501003663. [DOI] [PubMed] [Google Scholar]
- 3.Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 4.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 5.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl Acad. Sci. USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu D, Sawitzke JA, Ellis H, Court DL. Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate. Proc. Natl Acad. Sci. USA. 2003;100:7207–7212. doi: 10.1073/pnas.1232375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu BJ, Kang KH, Lee JH, Sung BH, Kim MS, Kim SC. Rapid and efficient construction of markerless deletions in the Escherichia coli genome. Nucleic Acids Res. 2008;36:e84. doi: 10.1093/nar/gkn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pohl T, Uhlmann M, Kaufenstein M, Friedrich T. Lambda Red-mediated mutagenesis and efficient large scale affinity purification of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 2007;46:10694–10702. doi: 10.1021/bi701057t. [DOI] [PubMed] [Google Scholar]
- 15.Orford M, Nefedov M, Vadolas J, Zaibak F, Williamson R, Ioannou PA. Engineering EGFP reporter constructs into a 200 kb human beta-globin BAC clone using GET recombination. Nucleic Acids Res. 2000;28:E84. doi: 10.1093/nar/28.18.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan K, Warburton PE. DNA modification and functional delivery into human cells using Escherichia coli DH10B. Nucleic Acids Res. 2003;31:e51. doi: 10.1093/nar/gng051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl Acad. Sci. USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li MZ, Elledge SJ. MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nat. Genet. 2005;37:311–319. doi: 10.1038/ng1505. [DOI] [PubMed] [Google Scholar]
- 19.Rivero-Muller A, Lajic S, Huhtaniemi I. Assisted large fragment insertion by Red/ET-recombination (ALFIRE)–an alternative and enhanced method for large fragment recombineering. Nucleic Acids Res. 2007;35:e78. doi: 10.1093/nar/gkm250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herring CD, Glasner JD, Blattner FR. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene. 2003;311:153–163. doi: 10.1016/s0378-1119(03)00585-7. [DOI] [PubMed] [Google Scholar]
- 21.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calos MP, Miller JH. The DNA sequence change resulting from the IQ1 mutation, which greatly increases promoter strength. Mol. Gen. Genet. 1981;183:559–560. doi: 10.1007/BF00268783. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. p. 1.147. [Google Scholar]
- 25.Maloy SR, Nunn WD. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 1981;145:1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bochner BR, Huang HC, Shieven GL, Ames BN. Positive selection for loss of tetracycline resistance. J. Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podolsky T, Fong ST, Lee BT. Direct selection of tetracycline-sensitive Escherichia coli cells using nickel salts. Plasmid. 1996;36:112–115. doi: 10.1006/plas.1996.0038. [DOI] [PubMed] [Google Scholar]
- 28.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 30.Posfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.