Abstract
Alternative splicing can produce multiple protein products with variable domain composition from a single gene. The mouse Tcf7l2 gene is subject to alternative splicing. It encodes TCF4, a member of the T-cell factor (TCF) family of DNA-binding proteins and a nuclear interaction partner of β-catenin which performs essential functions in Wnt growth factor signalling. Multiple TCF4 isoforms, potentially exhibiting cell-type-specific distribution and differing in gene regulatory properties, could strongly influence tissue-specific Wnt responses. Therefore, we have examined mouse Tcf7l2 splice variants in neonatal tissues, embryonic stem cells and neural progenitors. By polymerase chain reaction amplification, cloning and sequencing, we identify a large number of alternatively spliced transcripts and report a highly flexible combinatorial repertoire of alternative exons. Many, but not all of the variants exhibit a broad tissue distribution. Moreover, two functionally equivalent versions of the C-clamp, thought to represent an auxiliary DNA-binding domain, were identified. Depending upon promoter context and precise domain composition, TCF4 isoforms exhibit strikingly different transactivation potentials at natural Wnt/β-catenin target promoters. However, differences in C-clamp-mediated DNA binding can only partially explain functional differences among TCF4 variants. Still, the cell-type-specific complement of TCF4 isoforms is likely to be a major determinant for the context-dependent transcriptional output of Wnt/β-catenin signalling.
INTRODUCTION
T-cell factors (TCF) constitute a large family of evolutionary conserved HMG-box-containing DNA-binding proteins and transcriptional regulators. Although the founding members of the family, TCF1 and lymphoid enhancer factor 1 (LEF1), were initially identified for their role in Wnt-independent control of gene expression in lymphocytes (1–3), at present, TCFs are most intensively studied as nuclear effectors of Wnt growth factor signalling. In this context, TCFs serve as assembly platforms for multifactorial transcription complexes, which, depending upon their composition, act either to repress Wnt target genes or to stimulate their expression [reviewed in refs (4–6)]. Wnt/β-catenin pathway activation induces compositional changes in these transcription factor complexes (7) by promoting intracellular accumulation and nuclear entry of β-catenin. This enables the formation of β-catenin::TCF complexes (8–10) through binding of β-catenin to a domain located at the N-termini of TCFs. Concomitantly, Grg/TLE transcriptional corepressors are displaced from an adjacent region in TCFs (11), and coactivators are recruited (4). Thereby, Wnt/β-catenin target genes are switched from inactive to active states.
The ability of Wnt signalling to elicit different responses at different time points and in different tissues critically depends upon its capacity to control target gene expression in a highly context-dependent manner. The mechanisms whereby this functional diversification is achieved are largely unknown. However, TCFs are likely to have a central part in the process of target gene selection and, thereby, in shaping cell-type-specific Wnt effects. In support of this, it has been shown that TCF3 and LEF1 have contrasting effects on formation of the embryonic body axis in Xenopus laevis (8,10,12–14). Furthermore, genome-wide and locus-specific chromatin immunoprecipitation (ChIP) studies uncovered distinct patterns of promoter occupancy by TCF family members in mouse and human cell lines (15–17). Moreover, TCF family members differ in their ability to support β-catenin-dependent transactivation at selected target gene promoters (16,18,19). Thus, there is increasing evidence for functional diversity and non-redundant activities among TCF family members (12–14,18–23).
Functional differences among TCFs most likely arise from variation in protein structure. In particular, regions outside of the β-catenin interaction site and the HMG-box DNA-binding domain show considerable amino acid sequence divergence. The structural diversity of TCFs is further increased due to dual promoter usage and extensive alternative splicing (5). Thus, each of the TCF genes can give rise to different protein isoforms which has clear functional implications. For example, the TCF1E splice variant harbours a so-called C-clamp domain C-terminally to the HMG-box. The C-clamp is a bipartite amino acid motif featuring four characteristic cysteines and two clusters of amino acids enriched in basic residues (24). It forms an auxiliary DNA-binding domain which enables TCF1E to recognize specific subsets of TCF-binding elements (TBEs). In contrast to TCF1E, the TCF1B splice form does not contain a C-clamp (24). As a result, TCF1B does not bind to the LEF1 promoter and, hence, is unable to transactivate it (5,24). Aside from the C-clamp other parts of TCFs, which are varied due to alternative splicing, have also been shown to influence their gene regulatory capacity (12,14). Thus, a growing body of information suggests that alternative splicing can generate a multitude of TCF protein isoforms with possibly unique capabilities to support differential gene expression by Wnt signalling.
In the human and mouse genomes, the TCF4 genes (official names TCF7L2 and Tcf7l2) consist of 17 exons. However, current information about variable exon composition of TCF4 transcripts is almost exclusively derived from studies using human cell lines and tumour samples (25–27). Three regions subject to alternative splicing were identified. The conditional exon 4 encodes a 23 amino acid protein domain without known function. Skipping of the splice donor at the 3′-end of exon 8 yields TCF4 transcripts, which give rise to TCF4N isoforms (28,29). The TCF4N isoforms consist of the N-terminal part of TCF4 including the β-catenin binding domain but lack the HMG-box and all other elements beyond it. The TCF4N variants likely function as dominant negative factors in Wnt signalling by diverting β-catenin from cognate target genes (28,29). In addition, variable use of alternative splice donor and acceptor sites at exons 7 and 9 leads to protein isoforms in which amino acid sequences derived from exon 8 are flanked by different combinations of LVPQ and SxxSS amino acid motifs. The presence or absence of these elements alters transcriptional properties of Xenopus TCFs (12,14). However, their role in mammalian TCFs is unknown. Finally, extensive alternative splicing of exons 13, 14, 15 and 16 in various combinations generates TCF4E (also known as TCF4L), TCF4M (also known as TCF4B) and TCF4S isoforms with highly diverging C-termini (25–27,30). Only TCF4E variants harbour a complete C-clamp and, in addition, contain two binding motifs for the transcriptional repressor Carboxy-terminal Binding Protein (CtBP) (31). All other TCF4 isoforms lack the CtBP-binding sites and possess either no C-clamp (TCF4M) or only an incomplete version of it (TCF4S) (25–27). To which extent the presence of variable C-termini alters the functional properties of TCF4 isoforms is not known.
Compared to their human counterparts, much less is known about the range of alternative splicing of mouse Tcf7l2 transcripts (30,32–34). Moreover, there is only very limited information about the tissue-specific distribution of Tcf7l2 splice variants, and about how altered domain architecture affects TCF4 protein function. Such knowledge, however, might provide invaluable insights into the tissue-specific activities of TCF4 as nuclear effector of Wnt/β-catenin signalling. Because of this, we have begun a systematic compilation of alternative splice forms of Tcf7l2 transcripts in different mouse tissues and cell types. Here we describe a multitude of Tcf7l2 splice variants, which are highly similar in design to the human gene transcripts, and which include hitherto unknown versions of the C-clamp. Most of the variants identified show a broad tissue distribution albeit relative proportions to each other can vary. Based on the presence or absence of C-terminal amino acid signature motifs, TCF4 splice variants fall into one of three main classes. Representatives of each of these were functionally tested with respect to promoter-specific transactivation and DNA binding. Our findings show that the extended C-terminal domains of ‘E’-type variants endow TCF4E isoforms with unique properties with respect to promoter recognition and activation.
MATERIALS AND METHODS
Cell culture, transient transfections and luciferase reporter gene assays
Human HEK293 and U2-OS cells (ATCC # CRL-1573, ATCC # HTB-96) were cultured in Dulbecco’s modified Eagle’s medium (PAN-Biotech) containing 10% fetal calf serum (Invitrogen), 10 mM Hepes buffer (PAN-Biotech), 1% MEM non-essential amino acids (Invitrogen) and 100 units/ml penicillin–streptomycin. The murine embryonic stem cell (ESC) line E14, murine C17.2 neural progenitor cells (NPCs) and C2C12 myoblasts (ATCC # CRL-1772) were cultured as previously described (35,36). For western blotting analysis, 5 × 105 HEK293 cells (per well of a 6-well plate) were transfected with 2.5 µg of expression vectors for TCF4 isoforms E2, E2ex4, E4, M1 and S2, respectively, using the FuGENE6 reagent (Roche Applied Science). For ChIP, 1.4 × 107 cells seeded in 15 cm dishes were transfected with 1.5 ml of a calcium phosphate–DNA coprecipitate containing 10 µg each of the TCF4 expression vectors and the various promoter constructs. For immunostaining, 2 × 104 U2-OS cells seeded per well of 24-well plates on gelatine-coated cover slips were transfected 4 h after plating using the FuGENE6 reagent (Roche Applied Science) and 100 ng of the TCF4E2, TCF4E2ex4, TCF4E4, TCF4M1 and TCF4S2 expression vectors, respectively. For luciferase assays, 1 × 105 HEK293 cells seeded per well of a 24-well plate were transfected with FuGENE6 reagent (Roche Applied Science) 4 h after plating. Cells received a mixture of 100 ng of firefly luciferase reporter plasmid with Wnt target gene promoters as indicated, 10 ng of the Renilla luciferase expression vector pRL-CMV (Promega), 100 ng of plasmid DNA for expression of a constitutively active form of β-catenin (18) and increasing amounts (1, 5, 25 and 125 ng) of expression vectors for TCF4 splice variants. Total amounts of transfected DNA were kept constant by adding appropriate amounts of the empty expression vector pCS2+. Where indicated, transfected cells received 200 ng/ml of recombinant Wnt3a (R&D Systems) 16 h prior to harvest. Reporter gene activities were determined as described (18), and Renilla luciferase activity was used for normalization. All results shown represent average values obtained from at least three independent experiments including standard deviations.
RNA isolation, reverse transcription polymerase chain reaction (RT–PCR) and cloning of RT–PCR products
RNA isolations from cell lines and tissue samples of three-days-old mice, cDNA synthesis and subsequent RT-PCRs were performed as described (16) using the NucleoSpin RNA II kit (Macherey and Nagel) and Superscript II reverse transcriptase (Invitrogen) following the manufacturer’s protocols. For tissues, 30 mg of starting material was used. PCR products were separated by electrophoresis on 6–10% polyacrylamide gels, or on 2% agarose gels. DNA fragments were isolated with the DNA and Gel Band Purification Kit (GE Healthcare). Subsequent cloning of PCR products into pCR4-TOPO vector was performed using the TOPO TA Cloning Kit for Sequencing (Invitrogen). Sequence analyses were carried out using the BigDye cycle sequencing Kit (Applied Biosystems) with the help of the sequencing core facility of the University Medical Center Freiburg, Germany.
Plasmids and generation of TCF4 expression constructs
The expression plasmids pCS2+ and pCS2+β-catS33A, as well as the luciferase reporters pGL3b-Axin2, pGL3b-Cdx1, pCycD1Δ-973XP2 and pS01234 were previously described (18,37–42). To generate eukaryotic expression vectors for TCF4 splice variants, the mouse TCF4M1 isoform in pFLAG-CMV-2 (32) was used as starting point. To produce TCF4 isoforms E2 and S2 from this vector, the entire plasmid including TCF4 sequences was amplified with divergent primers located in exons 12 and 17 using Pfu Ultra II HS DNA Polymerase (Stratagene; for primer sequences see Table 1). The linearized vector was then ligated with 5′ phosphorylated PCR products containing exons 13–14 and 13–14–15, respectively, which were derived from cloned RT-PCR fragments in pCR4-TOPO. Exon 4 sequences were introduced into TCF4E2 by a similar strategy. Primer pairs E4f/E4r and E3bluntr/E5bluntf (Table 1) were used for amplification of exon 4 sequences and the recipient vector backbone. TCF4E4 was derived from TCF4S2 by deleting exon 14 sequences. For this, plasmid sequences were PCR amplified with divergent primers in exons 13 and 15 (E13bluntr/E15bluntf; Table 1) followed by phosphorylation and religation. All expression cassettes for the TCF4 isoforms were sequence verified. For expression in vitro, the cDNAs for the corresponding TCF4 isoforms were cut out from pFLAG-CMV-2 expression vectors with EcoRI and ligated into the pZero-2 vector (Invitrogen).
Table 1.
Primer sequences
| Sequence | |
|---|---|
| Primer for RT–PCR | |
| E1f | 5′-AAACAGCTCCTCCGATTCC-3′ |
| E4f | 5′-CAATCCGGCAGCACTCATTAC-3′ |
| E5r | 5′-TCTGCTCTGGAGGCTTCCTG-3′ |
| E7f | 5′-TCCGCACCCTCCAGATATC-3′ |
| E9r | 5′-GAGTGAGCCGACGTCACTC-3′ |
| E12f | 5′-ACAAGCAGCCGGGGGAAAC-3′ |
| E16r | 5′-CATCATAATATTCCAAATTC-3′ |
| E17r1 | 5′-AGCCTAGCAGATGCGGTGAG-3′ |
| E17r2 | 5′-GTCTGTGACTTGGCGTCTTG-3′ |
| N3'r; | 5′-ATATCATATGTCTAGAGGGCCTCACAAGAAG-3′ |
| Primer for cloning | |
| E3bluntr | 5′-GGTTCGGGCGGTGGGCGAGAG-3′ |
| E5bluntf | 5′-TATCTCCAGATGAAATGGCCAC-3′ |
| E4bluntf | 5′-CTCCATTTTCAATCCGGCAG-3′ |
| E4bluntr | 5′-CTGAATTGCAATCTGGTGTTC-3′ |
| E17bluntf | 5′-GAGAAAAAAAAAGTGCGTTCG-3′ |
| E12bluntr | 5′-CGTTGGTTTCCCCCGGCTG-3′ |
| E13f | 5′-AACACAGCGAATGTTTCC-3′ |
| E13bluntr | 5′-CTGTGATCGGAGGAAGCG-3′ |
| E14bluntr | 5′-CTGCAGGGGCCGCACCAG-3′ |
| E15bluntf | 5′-ATGCAAATACGCCAAAGAAG-3′ |
| E15r | 5′-CTGCAGGGTTTGCACCATAAAG-3′ |
| Primer for ChIP | |
| Cdx1 −440 | 5′-CGTTTGAAGTCAGCCTTGCT-3′ |
| Cdx1 −307rev | 5′-CCTGGGTTACAGGTGGAGAA-3′ |
| SiaP for | 5′-GGTTCTGCTGCCAAGTTAGG-3′ |
| SiaP rev | 5′-ATGCACCCTGAAAGAATTGG-3′ |
| pGL3basic +7820 | 5′-CGCTTTCTTCCCTTCCTTTC-3′ |
| pGL3basic +8005rev | 5′-AAGGGCGAAAAACCGTCTAT-3′ |
| GAPDH | 5′-ACCACAGTCCATGCCATCACT-3′ |
| GAPDH rev | 5′-GTCCACCACCCTGTTGCTGTA-3′ |
Preparation of nuclear extracts
For the preparation of nuclear extracts, cells from two to three confluent 15 cm dishes were used. Cells were washed once with ice-cold PBS, scraped from the dishes and transferred into 15 ml centrifuge tubes. After centrifugation at 1200× g for 1 min, the cell pellet was resuspended in 1 ml of hypotonic swelling buffer [10 mM Hepes pH 7.9, 10 mM NaH2PO4, 1.5 mM MgCl2, 0.5 mM spermidine, 10 mM NaF, 0.1 mM Na3VO4 and Complete™ protease inhibitor (Roche Applied Science)]. Upon incubation on ice for 20 min, cells were disrupted by douncing in a glass-glass homogenizer with 50 strokes of a tight-fitting pestle. Thereafter, nuclei were pelleted at 800× g for 5 min. The cytoplasmic supernatant was removed and the nuclei were washed twice with 1 ml hypotonic swelling buffer prior to extraction with 250 µl high salt buffer [30 mM Hepes pH 7.9, 25% glycerol, 400 mM NaCl, 0.3 mM EDTA, 12 mM MgCl2 and Complete™ protease inhibitor (Roche Applied Science)] on ice for 45 min. The nuclear extract was cleared by centrifugation at 100 000× g and 4°C for 30 min and stored at −80°C until use.
Western blotting and indirect immunofluorescence
For western blotting analysis cells were lysed in IPN150 buffer [50 mM Tris/HCl pH 7.6, 150 mM NaCl, 5 mM MgCl2, 0.1% NP40, Complete™ protease inhibitor (Roche Applied Science), 1 mM DTT, 0.1 mM Na3VO4, 1 mM PMSF, 10 mM NaF] for 30 min on ice. Cell lysates were cleared by centrifugation at 20 000× g and 4°C for 10 min. Protein concentrations in cell lysates were determined using the DC protein assay kit (BioRad). Following SDS–PAGE, proteins were transferred onto nitrocellulose membranes using a semi dry blotting apparatus. As blocking reagent, 2% skim milk powder in TBS-T buffer was used. For detection of TCF4 isoforms, a mouse monoclonal antibody (6H5-3, Upstate) was used at a 1:1000 dilution in TBS-T buffer containing 0.025% BSA and 0.02% sodium azide. LEF1 and GSK3β were detected in nuclear extracts using mouse monoclonal antibodies (LEF1: clone 2D12, Upstate, 1:500; GSK3β: cat. no. 610201, BD Transduction Laboratories, 1:1000). For the detection of TCF1 a rabbit monoclonal antibody (clone C63D9, cell signaling: 1:2000) was utilized. A goat polyclonal antiserum was employed for the detection of TCF3 (M-20, Santa Cruz Biotechnology: 1:1000). Visualization of antibody::antigen complexes was done with appropriate species-specific horse radish peroxidase-coupled antibodies diluted 1:10 000 and a chemiluminescent substrate solution consisting of 0.1 M Tris/HCl, pH 8.6, 250 µg/ml luminol, 110 µg/ml p-hydroxycoumaric acid and 0.0105% H2O2. Substrate incubation time was 2 min prior to exposure. Alternatively, the LumiGLO substrate (Kirkegaard and Perry Laboratories) was used according to recommendations of the manufacturer. For immunolocalizations of the different TCF4 isoforms, U2-OS cells were immunostained 48 h after transfection as previously described (43). Mouse monoclonal TCF4 antibody 6H5-3 was used at a 1:250 dilution. The secondary antibody was an Alexa-555 conjugated goat anti-mouse IgG (molecular probes) at a 1:1000 dilution.
Transcription and translation in vitro and electrophoretic mobility shift assays
TCF4 splice variants were transcribed and translated in vitro using the TNT SP6 high-yield wheat germ protein expression system (Promega) with 6 µg of plasmid DNA in 50 µl reactions. Equal expression levels of the different isoforms were controlled by western blotting. Binding reactions for electrophoretic mobility shift assays (EMSAs) were composed as described (18), except that the total volume was 20 µl and 5′-biotinylated DNA probes were used at concentrations of 1.0 fmol/µl (Cdx1 probes) and 0.5 fmol/µl (Siamois probe), respectively. DNA probes consisted of complementary pairs of synthetic oligonucleotides covering Cdx1 promoter sequences, or were generated by PCR with biotinylated primers using Siamois promoter plasmid DNA as template. Sequences of the Cdx1 probes and the Siamois primers are listed in Table 2. For supershifts, binding reactions were supplemented with 2.0 µg of an anti-TCF4 antibody (N-20, Santa Cruz Biotechnology) and antibody::antigen complex formation was allowed for 10 min prior to adding the biotin-labelled probe. Binding reactions were resolved by electrophoresis on polyacrylamide gels in 0.5× TBE running buffer and transferred onto hybond N+ nylon membrane (GE Health Care) for 1 h at 340 mA in the same buffer. DNA was crosslinked to the membranes and detected according to the ‘Chemiluminescent Nucleic Acid Detection Module’ (Pierce). Images were recorded with the LumiImager and X-ray films.
Table 2.
Sequences of 5′-biotinylated oligonucleotides used to generate EMSA probes
| Sequencea | |
|---|---|
| TBE4 | 5′-CGAGGCTTCCCCCCGCTTTGAAATGCAAAGCCGCCC-3′ |
| TBE4 mTBE | 5′-CGAGGCTTCCCCCCGCTTTGgcATGCAAAGCCGCCC-3′ |
| TBE4 mCCG | 5′CGAGGCTTCCCCCCGCTTTGAAATGCAAAGCtGCCC-3′ |
| TBE3/4 | 5′-GGGCTTCCCCCTTTGATTCGCGGCCCCGAGGCT-TCCCCCCGCTTTGAAATGCAAAGCCGCCC-3′ |
| TBE3/4 mTBE | 5′-GGGCTTCCCCCTTTGgcTCGCGGCCCCGAGGCT-TCCCCCCGCTTTGgcATGCAAAGCCGCCC-3′ |
| TBE3/4 mCCG | 5′-GGGCTTCCCCCTTTGATTCGCGGCCCtGAGGCT-TCCCCCCGCTTTGAAATGCAAAGCtGCCC-3′ |
| TBE3′/3/4 | 5′-ATTTGTCTCCTTTTGAACCCCCTCGCCCGACGGGCTTCCCCCTTTGAT-TCGCGGCCCCGAGGCTTCCCCCCGCTTTGAAATGCAAAGCCGCCC-3′ |
| TBE3′/3/4 mTBE | 5′-ATTTGTCTCCTTTTGgcCCCCCTCGCCCGACGGGCTTCCCCCTTTGgc-TCGCGGCCCCGAGGCTTCCCCCCGCTTTGgcATGCAAAGCCGCCC-3′ |
| TBE3′/3/4 mCCG | 5′-ATTTGTCTCCTTTTGAACCCCCTCGCCtGACGGGCTTCCCCCTTTGAT-TCGCGGCCCtGAGGCTTCCCCCCGCTTTGAAATGCAAAGCtGCCC-3′ |
| SiaP(−233) sense | 5′-TGTCATCAGAATCATCAAAGGACC-3′ |
| SiaP(+5) antisense | 5′-CCAAAATGTTGGTCCACTCTGTCC-3′ |
aUnderlined nucleotides mark the TBE and 5′-RCCG-3′ motifs; lower case denotes mutated nucleotides.
ChIP
Solubilized, fragmented chromatin from transfected HEK293 cells was prepared following exactly the instructions of the ChIP-ITTM kit with the enzymatic chromatin shearing module (Active Motif). Digestion of chromatin was carried out for 30 min at 37°C with occasional mixing. For immunoprecipitation, 200 µg sheared chromatin and 2 µg of a goat polyclonal anti-TCF4 antibody (N-20; Santa Cruz Biotechnology) or 2 µg of goat IgG control antibodies, respectively, were combined, and samples were further processed as described (16). To monitor protein recovery of the TCF4 splice variants in immunoprecipitates, one twentieth of each sample was analysed alongside with one twentieth of the input chromatin by SDS–PAGE and western blotting using a mouse monoclonal anti-TCF4 antibody (6H5-3; Upstate). Images were recorded using the LumiImager and the LumiAnalyst software (Roche Applied Science) and relative quantities of the TCF4 isoforms were determined. This information was used to normalize the amounts of DNA which had been co-precipitated with each of the TCF4 isoforms, and which were used as templates in real-time PCR analyses. Primer pairs used to amplify the investigated promoters, a portion of the pGL3 basic control vector, and GAPDH as non-specific control, respectively, are listed in Table 1. Levels of promoter DNA and GAPDH control sequences which were precipitated by specific and non-specific antibodies, were calculated as fractions of the input DNA. GAPDH values were used for normalization. Results shown are average values and the corresponding standard deviations from at least three independent experiments.
RESULTS
Identification of Tcf7l2 splice variants in mouse tissues and cell lines
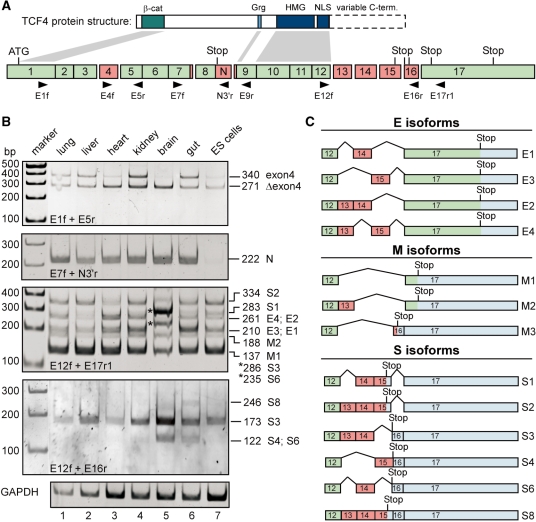
To examine the occurrence and tissue-specific distribution of splice variants derived from the mouse Tcf7l2 gene we reverse transcribed RNA from mouse neonatal tissues, ESCs, and NPCs. Sequence variability was examined between exons 1–9 and 12–17, where alternative splicing and variable use of splice signals has been extensively documented for the human and Xenopus genes (Figure 1A) (14,25–27). Tcf7l2-specific sequences were PCR-amplified with various combinations of exon-specific primer pairs (Figure 1A, Table 1). First, we analysed Tcf7l2 transcripts with or without exon 4. PCR products with the corresponding size differences were detected in all tissues analysed, as well as in ESCs and NPCs (Figure 1B, top panel, Supplementary Figure S1). Their identity was confirmed by subcloning and sequencing of PCR products from ESCs, NPCs, gut and brain samples. Interestingly, the relative proportions of transcripts containing or lacking exon 4 varied. Exon 4-containing transcripts are comparably less abundant in heart, brain and ESCs (Figure 1B). Furthermore, nested PCR amplification with primers located in exons 4, 12 and 17 yielded complex mixtures of products suggesting that exon 4 occurs in various combinations with the alternatively spliced exons 13–16 (Supplementary Figure S2). Thus, incorporation of exon 4 does not appear to be restricted to a particular subset of splice forms.
Figure 1.
Comparison of expression patterns of TCF4 splice variants in mouse tissues and ES cells. (A) Simplified structural schemes for TCF4 proteins (top) and the mouse Tcf7l2 gene (bottom). The Tcf7l2 gene consists of 17 exons, some of which are subject to alternative splicing (exons 4, 13–16; labelled in red). Constitutive exons are labelled in green. Additional variation of Tcf7l2 mRNA sequences is generated due to the use of alternative splice acceptor and donor sites, respectively, in exons 7, 8 and 9. Affected regions are also indicated by red boxes. The position of the start codon in exon 1 is marked. Which of the potential stop codons following exon 8 or within exons 15, 16 and 17 is used, depends on the particular exon combination present in the final mRNA. Retention of sequences following exon 8 (labelled N) in the final mRNA generates the TCF4N variant. Exon lengths are drawn approximately to scale. Horizontal arrowheads indicate the positions and the orientation of primers used in RT-PCR reactions. Some of the structural features of TCF4 proteins are indicated and linked with connectors to the exons by which they are encoded. Dashed lines skirt the highly variable C-terminal parts of TCF4 derived from exons 13–17. β-cat: β-catenin binding domain; Grg: binding motif for groucho-related gene (Grg) products (73); HMG: HMG box; NLS: nuclear localization signal. (B) Expression analyses of TCF4 splice variants in different mouse tissues and ES cells. Upon isolation of RNA from mouse ES cells and tissue samples from 3-days-old mice, cDNA was synthesized, and TCF4 splice variants were amplified by PCR using exon-specific primer combinations as indicated. Amplification of GAPDH sequences served to control RNA and cDNA integrity. PCR products were separated by gel electrophoresis, and visualized by ethidium bromide staining. To determine the exon combination represented by the various DNA fragments, PCR products were subcloned and sequenced. The deduced size of the PCR products and their assignment to TCF4 E, S and M groups of splice variants is indicated on the right side of the panels. PCR products from brain tissue labelled by asterisks represent isoforms S3 and S6. (C) Summary of the C-terminal TCF4 splice variants identified. Red exons are alternatively spliced whereas green exons are constitutively used. Blue areas indicate untranslated regions. Positions of stop codons used are shown. Splice variants were grouped depending upon whether the resulting translation products contain no C-clamp or complete and incomplete versions thereof, respectively (Figure 2). Accordingly, the isoform formerly denoted S5 (26) was reclassified as M3.
Next, we investigated sequence variations around exon 8. These can arise from the use of alternative splice donor and acceptor sites, or from retention of intronic sequences downstream of exon 8 (Figure 1A). In the first case, exon 8 can be flanked by additional sequences giving rise to the LVPQ and SFLSS amino acid motifs (14,25,44). Corresponding variants containing either both motifs or just the LVPQ motif were also identified for mouse Tcf7l2 (Supplementary Figure S1). Transcripts which lack both elements simultaneously or which incorporate just the SFLSS motif were not found. However, their existence, perhaps at a very low abundance, cannot be ruled out. Tcf7l2 transcripts in which the splice donor at the 3′-end of exon 8 is suppressed give rise to TCF4N proteins (28). Such transcripts were found in all tissue samples at similar levels. However, they were hardly detectable in ESCs (Figure 1B).
Finally, we turned to transcripts generated by alternative splicing of exons 12–17. PCR with primer pairs located in exons 12 and 17 yielded a complex pattern of PCR products (Figure 1B). To unequivocally determine their precise exon combinations, the PCR products obtained from brain and gut samples, as well as from ESCs and NPCs were excised from gels, subcloned and sequenced. Thereby, we identified splice variants E1–E4, M1–M3, and S1–S4, S6 and S8, and assigned them to specific PCR products (Figure 1B and C).
Within the limitations of the experimental approach, it appears that most of the Tcf7l2 exon 12–17 splice variants are expressed in every tissue or cell type investigated. As in case of exon 4, however, their relative amounts vary. The most prominent example for this is provided by transcripts S3 and S6 which appear particularly abundant in brain tissue (Figure 1B). S3 and S6 transcripts contain exon 16, which is absent from the majority of the other isoforms. This raised the possibility that exon 16 was incorporated in a brain-specific manner. Therefore, we more specifically examined the existence and tissue-specific distribution of exon 16-containing transcripts by PCR amplification with the primer combination E12f/E16r. Three PCR products with a length of 246, 173 and 122 bp, respectively, were detected. The 122 bp fragment, which based on its size is presumably derived from splice variants S4 or S6, was detected in cDNA from brain and to a lesser extent from gut. The 246 bp fragment, which corresponds in size to the splice variant S8, was observed at low levels only in RNA from gut. However, originally we identified the S8 isoform upon cloning and sequencing of a cDNA fragment derived from brain RNA. Therefore, TCF4S8 does not appear to be strictly gut-specific. In contrast to the tissue-restricted occurrence of the 122 and 246 bp bands, the 173 bp fragment could be amplified from RNA of all tissues analysed. Its size matches most closely to that of the S3 isoform. Alternatively, it might be derived from a variant with the C-terminal exon combination 12–13–15–16. Because the latter isoform was not detected by subcloning of PCR fragments and subsequent sequencing, we assume that the PCR fragment detected most likely is derived from TCF4S3. Irrespective of this, closer inspection of Tcf7l2 transcripts containing exon 16 reveals that some of these isoforms show a rather narrow range of tissue-specific expression, while others are widely distributed. Furthermore, while transcripts with exon 16 are indeed expressed at highest levels in brain, they are not exclusively present in this tissue.
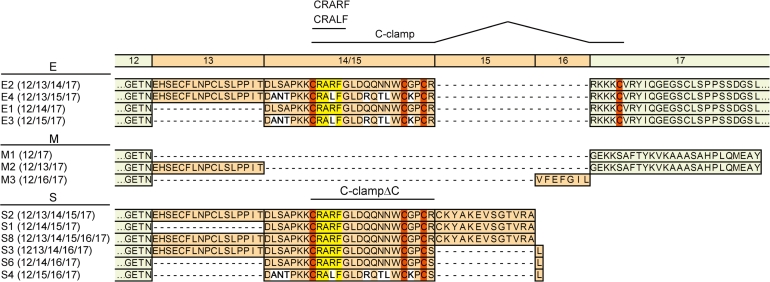
Depending upon the preceding exon combination, different open reading frames are used for translation of exons 15, 16 and 17. This distinction can be used to classify Tcf7l2 transcripts into three groups giving rise to TCF4E, TCF4M and TCF4S protein isoforms (Figures 1 and 2). The hallmark of the E isoforms (also known as L) (25–27,44) is the presence of a complete C-clamp. Interestingly, our analyses revealed that there are two variants of the C-clamp in TCF4. TCF4E1 and TCF4E2 harbour a CRARF motif together with the conserved cysteines, whereas TCF4E3 and TCF4E4 display a CRALF motif in conjunction with the cysteines (Figure 2). TCF4S variants contain only three of the four conserved cysteines. Again, these can be combined either with the CRARF or the CRALF motif. TCF4M variants (also denoted B) (25–27,44) lack both the cysteines and the CRARF/CRALF element.
Figure 2.
Amino acid sequence diversity at the C-terminus of TCF4 generated by alternative splicing. A schematic comparison of the C-terminal amino acid sequences derived from TCF4 splice variants identified in Figure 1 is shown. TCF4 splice variants are denoted, and their particular combination of exons is indicated by the numbers in brackets. Amino acid sequences encoded by alternative and constitutive exons are shaded green and red as before, and exon numbers are shown above the amino acid sequences. Exon 15 is differentially translated when it is preceded by exons 13 or 14, respectively. To facilitate the sequence comparison, amino acid sequences derived from exons 14 and 15, respectively, are either shown as pile up in the same column (variants E1–E4 and S3, 4, 6) or consecutively (variants S1, 2, 8). The four conserved cysteines of the C-clamp are highlighted in dark orange. Variable CRALF and CRARF motives are shaded yellow. White shading indicates amino acid sequences diverging among the splice variants.
To determine whether multiple variants of TCF4 exist at the protein level, we performed western blotting experiments with protein extracts from ESCs, NPCs and C2C12 myoblasts. Both, by use of a mouse monoclonal antibody and with a goat polyclonal antiserum against TCF4, two prominent protein bands were detected (Supplementary Figure S3, Figure 5C). The slower migrating band of ∼70 kDa corresponds in size to the expected molecular weights of TCF4E isoforms whereas the faster migrating band of ∼50 kDa matches the molecular weight of TCF4M/S variants. The same two protein bands were also revealed in reciprocal immunoprecipitation/immunodetection experiments with the two different anti-TCF4 antibodies (Supplementary Figure S3) further corroborating that they represent TCF4 protein forms. Some of the variants within the TCF4E and TCF4M/S groups, respectively, are nearly identical in molecular weight. Thus, it is possible that the 70 and 50 kDa bands each contain a collection of various TCF4E and TCF4M/S species. Other variants, however, should be resolvable by SDS–PAGE. The detection of only two dominant TCF4 bands therefore suggests that some isoforms are translated not at all or at much lower levels than others. Regardless of this, our results show that at least one representative of the TCF4E and TCF4M/S variants each is expressed in tissue culture cells.
Figure 5.
Differential activation of the Cdx1 promoter in HEK293 cells (A) and mouse embryonic stem (ES) cells (B) by recombinant Wnt3a. Cells were cotransfected with combinations of firefly and Renilla luciferase reporter genes, control vector, expression vector for a constitutively active form of β-catenin, and constructs coding for TCF4E2, TCF1B, TCF1E, LEF1 and TCF3 as indicated. A cDNA construct for the human TCF4E2 variant was used because it is present in the same vector backbone as the other TCF family members. Firefly luciferase expression was driven by the Cdx1 promoter. To induce reporter gene expression by endogenous pathway components transfected cells received 200 ng/ml recombinant Wnt3a (rWnt3a) 16 h prior to harvest. Control cells remained untreated. Reporter gene activities were determined 40 h post transfection. Bars represent relative luciferase activities (RLA) compared to controls transfected with luciferase reporter plasmids and empty expression vectors only. Average values and standard deviations from at least three independent experiments are given. hu: human; m: mouse. (C) Expression of endogenous LEF/TCF proteins in different cell lines. Nuclear extracts from HEK293 cells, ES cells, C17.2 neural progenitors (NPC) and C2C12 myoblasts were used for western blotting and immunodetection of TCF4, TCF1, LEF1 and TCF3. Antibodies against GSK3β were used to monitor equal loading. As a control for antibody specificity, epitope-tagged TCF1E, LEF1, TCF3 and TCF4E were used (controls). For this, HEK293 cells were transfected with expression vectors for HA-tagged LEF/TCF proteins, whole cell lysates were prepared 40 h after transfection, and aliquots containing roughly equal amounts of the reference proteins were analysed in parallel with the nuclear extracts. The presence of the HA-tag accounts for size differences between controls and cellular TCFs. Mw: molecular-weight standard; n.a.: not analysed.
In summary, the analyses performed so far reveal that the mouse Tcf7l2 gene exhibits the same extensive range of alternative splicing and splice site usage as the human gene. Although there is some tissue-specific variability in their relative abundance, most transcript isoforms appear to be present in a wide range of tissues. The near absent levels of TCF4N encoding transcripts in embryonic stem cells and the tissue-specific expression of some exon 16-containing isoforms may be notable exceptions. Thus, the Tcf7l2 gene has the potential to give rise to a large variety of TCF4 protein species including several forms containing variants of the C-clamp.
Wnt/β-catenin target gene activation by different TCF4 isoforms
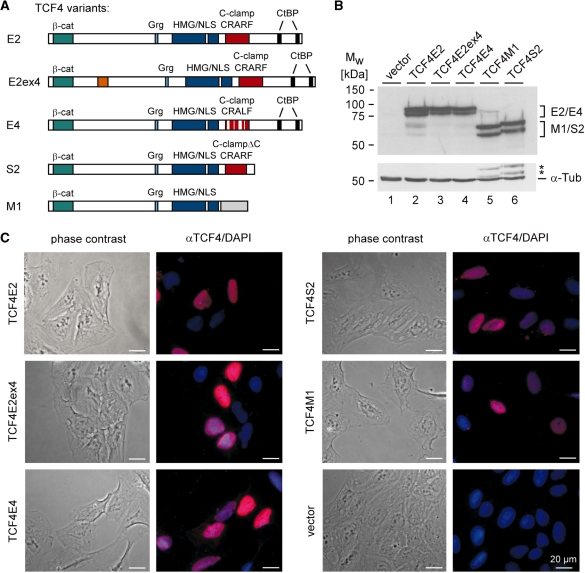
The structural divergence and different domain compositions strongly suggest that TCF4 isoforms also differ in function. However, an extended functional comparison of TCF4 splice variants particularly in the context of natural Wnt/β-catenin target gene promoters has not yet been undertaken. Therefore, we generated expression constructs for mouse TCF4E2, TCF4M1 and TCF4S2, representative for the three main groups of splice variants. In addition, to obtain information about the functional properties of amino acid sequences encoded by exon 4 and the CRALF variant of the C-clamp, expression vectors for TCF4E2ex4 and TCF4E4 were made (Figure 3A). TCF4N was not included in these studies because its functional properties have been extensively characterized before (29). Western blotting was used to demonstrate that all TCF4 variants were expressed at similar levels after transfection of HEK293 cells (Figure 3B). Furthermore, all TCF4 protein variants were located in the nuclei of transfected U2-OS cells consistent with the fact that each of them has an intact HMG box and a nuclear localization signal (Figure 3C).
Figure 3.
Structure and expression of TCF4 protein isoforms chosen for functional analyses. (A) Schematic representation of TCF4 protein isoforms E2, E2ex4, E4, S2, and M1. The location of binding domains for β-catenin (β-cat), Grg and CtBP corepressors, the HMG box, and the nuclear localization signal (NLS) are indicated. The presence of a complete or incomplete (ΔC) C-clamp is denoted. The TCF4E2, TCF4E2ex4 and TCF4S2 variants contain a CRARF motif as shown whereas in TCF4E4 a CRALF motif is present. The variant TCF4E2ex4 incorporates additional amino acid sequences derived from exon 4 (orange box). TCF4M1 does not share amino acid sequences with TCF4E and TCF4S variants C-terminally to the NLS as shown by the grey box. (B) Expression analyses of TCF4 isoforms. HEK293 cells were transfected with expression vectors for TCF4E2, TCF4E2ex4, TCF4E4, TCF4M1 and TCF4S2. Control samples received the empty expression vector (lane 1). TCF4 proteins were detected in total cell lysates 40 h after transfection by western blotting. The same blot was subsequently probed with antibodies against α-Tubulin (α-Tub) as loading control. Asterisks label residual TCF4S2 and TCF4M1 signals from the previous detection round. (C) U2-OS cells were transfected with expression vectors for TCF4E2, TCF4E2ex4, TCF4E4, TCF4S2 and TCF4M1, respectively. Control samples received the empty expression vector. TCF4 isoforms were detected by indirect immunofluorescence using a monoclonal antibody to TCF4 and an Alexa-555-labelled secondary antibody. Nuclei were counterstained with DAPI. Phase contrast images (left) and an overlay (right) of Alexa-555 (αTCF4) and DAPI stainings are shown. Bars: 20 µm.
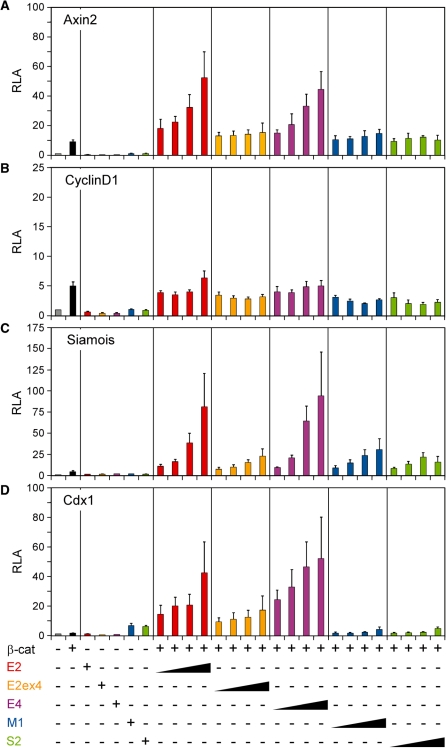
To compare TCF4 variants with respect to their transactivation capacities we performed reporter gene assays. HEK293 cells were cotransfected with luciferase reporters, a construct encoding a β-catenin mutant with alanine substitutions at its GSK3 and CK1 phospho-acceptor sites to mimic an active Wnt/β-catenin pathway, and expression vectors for the different TCF4 variants. Promoter regions of the reporter constructs were derived from the well-characterized Wnt/β-catenin target genes Axin2, Cdx1, Cyclin D1 and Siamois. The Axin2 gene is considered to be a ubiquitous Wnt/β-catenin target without tissue-specificity (45). The Cyclin D1 promoter, too, exhibits no cell-type specificity and has been reported to be activated by Wnt/β-catenin signalling in different contexts (46–48). In contrast, the Siamois and Cdx1 promoters are derived from highly tissue- and stage-specific Wnt target genes (39,40,49).
Expression of constitutively active β-catenin was sufficient to increase luciferase expression from the Axin2, Cyclin D1 and Siamois promoters by 5–10-fold presumably by making use of endogenous TCF family members in HEK293 cells (Figure 4A–C). In contrast, the Cdx1 promoter did not respond to the β-catenin mutant alone (Figure 4D). Co-expression of β-catenin and the TCF4 splice variants E2 and E4 strongly increased activity of the Axin2, Siamois and Cdx1 promoters in a dosage-dependent manner, irrespective of their C-clamp version (Figure 4A, C and D). Presence of amino acid sequences encoded by exon 4 dampened the transactivation capacity of TCF4E2. The TCF4M1 and TCF4S2 variants differed from the TCF4E isoforms in several aspects. They could not cooperate with β-catenin at the Axin2 and Cdx1 promoters and stimulated reporter activity beyond levels obtained by β-catenin expression alone only with the Siamois construct (Figure 4). Importantly, the functional differences among the TCF4 splice variants are not related to differences in their interactions with β-catenin. In coimmunoprecipitation experiments, all TCF4 isoforms interacted with β-catenin with similar efficiencies (Supplementary Figure S4). TCF4M1 and TCF4S2 are also unique in that they significantly elevated Cdx1 promoter activity in the absence of exogenous β-catenin (Figure 4D). Finally, none of the TCF4 variants was able to synergize with β-catenin at the Cyclin D1 promoter, nor did they influence its basal activity. However, the β-catenin-induced Cyclin D1 activation does involve a TCF factor as it could be blocked by dominant negative LEF1 and TCF4E2 lacking their β-catenin-binding domains (Supplementary Figure S5). Additionally, this also implies that TCF4E2 is able to bind to the Cyclin D1 promoter, albeit non-productively. Taken together, these analyses showed that TCF4 splice variants differ considerably in their gene regulatory properties depending upon their precise domain composition. Furthermore, promoter context appears to be important in determining whether TCF4 isoforms can effectuate their transactivation potential.
Figure 4.
TCF4 splice variants differentially activate Wnt/β-catenin target gene promoters. HEK293 cells were cotransfected with combinations of firefly and Renilla luciferase reporter genes, control vector, expression vector for a constitutively active form of β-catenin, and increasing amounts of expression vectors for TCF4 splice variants as indicated. Firefly luciferase expression was driven by promoters from the Wnt/β-catenin target genes Axin2 (A), Cyclin D1 (B), Siamois (C) and Cdx1 (D). Reporter gene activities were determined 40 h post transfection. Bars represent relative luciferase activities (RLA) compared to controls transfected with luciferase reporter plasmids and empty expression vectors only. Average values and standard deviations from at least three independent experiments are given.
The Cdx1 reporter construct was the only one whose activity could not be increased by constitutively active β-catenin and an endogenous TCF family member in HEK293 cells although it did respond to exogenous TCF4E variants. Transcription of the endogenous Cdx1 gene can be stimulated by Wnt3a in mouse ESCs. To gain further insight into the TCF requirements of the Cdx1 promoter, we performed additional transfection experiments in HEK293 and ESCs. Unlike in HEK293 cells, both treatment of transfected cells with recombinant Wnt3a and expression of β-catenin robustly stimulated the reporter in ESCs (compare Figures 5A and B). To correlate this Cdx1 response with TCF expression and activity, we examined the expression of LEF1, TCF1, TCF3 and TCF4 in HEK293 and ESCs by western blotting and asked which of the TCF family members could synergize with β-catenin in Cdx1 reporter activation. Only TCF4E2 and TCF1E enhanced β-catenin-dependent Cdx1 promoter activity (Figure 5A and B). Interestingly, compared to HEK293 cells, ESCs possess a greater abundance of TCF1 protein variants which, based on their molecular weights, likely represent ‘E’-type isoforms (Figure 5C). Overall, these results suggest that the failure of β-catenin and Wnt3a to activate the Cdx1 promoter in HEK293 cells is due to insufficient levels of endogenous transactivation-competent TCF4 and TCF1 isoforms. On the other hand, they confirm the particular transactivation capacities of TCF proteins containing an extended ‘E’-tail.
DNA-binding properties of TCF4 protein isoforms at different Wnt/β-catenin target genes
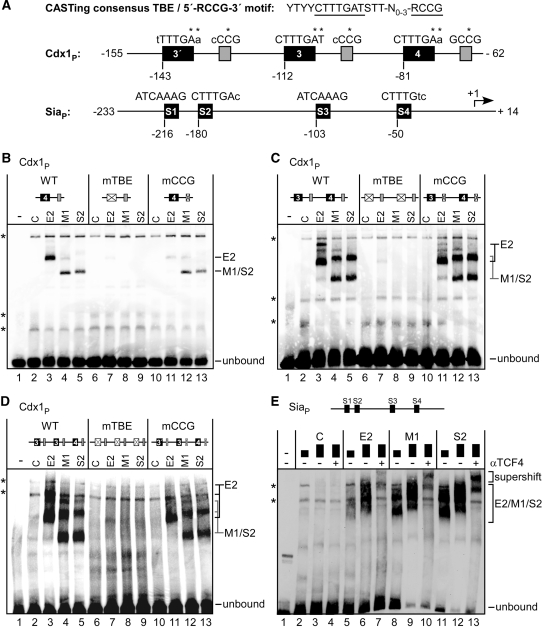
The C-clamp is thought to facilitate transactivation by TCFs through mediating extra DNA contacts with a 5′-RCCG-3′ motif situated adjacent to canonical TBEs (24,50). The observed functional differences between the TCF4 isoforms thus could be related to differences in promoter recognition according to the presence or absence of the C-clamp at their C-termini. To test this possibility, we first analysed the DNA-binding capacities of the different TCF4 isoforms in vitro by EMSAs. TCF4E2, TCF4M1, and TCF4S2 were expressed in vitro using a wheat germ based transcription and translation system. Immunoblotting with a mouse monoclonal antibody which recognizes all three TCF4 variants was used to verify their expression at similar levels (Supplementary Figure S6). EMSA probes used were a Siamois promoter fragment with four potential TBEs, and synthetic oligonucleotides covering mouse Cdx1 promoter sequences which contained the core TCF-binding motif TBE4, a combination of TBEs 3 and 4, or the three TBEs 3′, 3 and 4 together (18). Whereas there are no 5′-RCCG-3′ motifs discernable next to the core TBEs at the Siamois promoter, all three Cdx1 core TBEs are flanked by these elements (Figure 6A, Table 2).
Figure 6.
TCF4 isoforms differentially bind to Wnt/β-catenin target gene promoters. (A) DNA probes used for EMSAs. Top: The extended consensus TBE sequence derived from CASTing experiments and the 5′-RCCG-3′ motif recognized by the TCF1 C-clamp (24). Nucleotide sequences representing the heptameric TBE core (74,75) and the 5′-RCCG-3′ element are underlined. Promoter fragments from the Cdx1 and Siamois genes are shown schematically. TBEs and their relative positions are indicated. Coordinates of the DNA fragments are given relative to the transcription start site (+1; marked by an arrow at the Siamois promoter). Grey boxes indicate 5′-RCCG-3′ elements in the vicinity of the Cdx1 TBEs. Capital letters: matches between core consensus and the actual TBEs, lower case: mismatches. Asterisks indicate the positions which were mutated in core TBE or 5′-RCCG-3′ motifs. (B–E) In vitro transcribed and translated TCF4 isoforms E2, M1 and S2, or a mock programmed wheat germ extract (C) were used in EMSAs as indicated. A PCR fragment with Siamois promoter sequences (SiaP), or synthetic oligonucleotides harbouring Cdx1 promoter sequences with single, double or triple combinations of TBEs 3′, 3 and 4, were used as probes as schematically indicated on top of each panel. The Cdx1 probes contained all wild-type sequences (WT) or base pair exchanges in every TBE (mTBE) or 5′-RCCG-3′ motif (mCCG) present as indicated. At the Siamois promoter, two different protein amounts were used (black boxes). For supershifts in panel E, a goat polyclonal TCF4 antibody (N-20) was added to the binding reactions (αTCF4). Specific protein::DNA complexes and positions of the unbound DNA probes are marked. Asterisks indicate non-specific bands.
At the Siamois promoter, two different amounts of the TCF4 proteins and antibody supershifts were used to distinguish TCF4 isoform-dependent DNA binding from endogenous activity in the wheat germ lysate (Figure 6E). Indeed, dose-dependence of the signals and the appearance of additional retarded bands upon application of an anti-TCF4 antibody identified specific TCF4::DNA complexes. Importantly, no major differences in the respective binding capacities of TCF4E2, TCF4M1 and TCF4S2 were evident. Similarly, all three TCF4 protein variants showed robust interactions with the Cdx1 probes regardless of the presence or absence of a C-clamp (Figure 6B–D, lanes 3–5). To determine to which extent core TBEs and 5′-RCCG-3′ motifs contributed to these interactions, additional EMSAs with mono-, di- or trimeric Cdx1 probes were performed in which either all core TBEs or all 5′-RCCG-3′ motifs, respectively, had been mutated simultaneously. As expected, the core TBE mutations completely abolished DNA binding by TCF4E2, TCF4M1 and TCF4S2 (Figure 6B–D, lanes 7–9). DNA binding by TCF4M1 and TCF4S2 variants lacking a C-clamp was not affected by mutation of the 5′-RCCG-3′ motifs (Figure 6B–D, lanes 12 and 13). In contrast, the ability of TCF4E2 to interact with the monomeric TBE4 probe was severely reduced in the absence of the 5′-RCCG-3′ motif (Figure 6B, compare lanes 3 and 11). However, at the di- and trimeric probes the requirements of the 5′-RCCG-3′ motifs for efficient TCF4E2::DNA complexation was considerably less pronounced (Figure 6C and D, compare lanes 3 and 11). Apparently, the presence of multiple core TBEs within the physiologically more relevant multimeric DNA probes can at least partially compensate for mutation of the 5′-RCCG-3′ motifs.
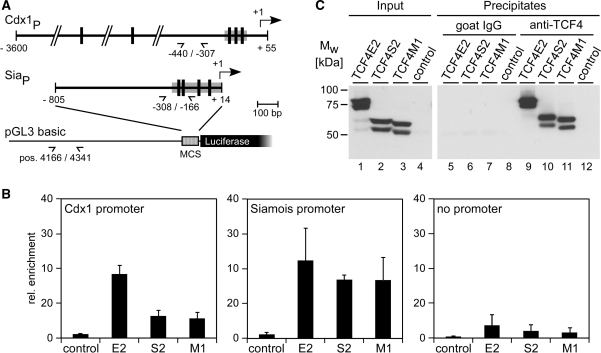
To analyse the promoter-binding capacities of TCF4 variants also in living cells, we performed ChIPs. Expression vectors for TCF4 isoforms were introduced into HEK293 cells together with luciferase reporter gene constructs containing either the Siamois promoter or the Cdx1 promoter, respectively. As a control the promoter-less parental plasmid pGL3-basic was used (Figure 7A). After formaldehyde crosslinking and preparation of fragmented chromatin, parallel immunoprecipitations with an anti-TCF4 antibody and non-specific goat IgGs were carried out. Plasmid DNA present in the immunoprecipitates was quantified by real-time PCR after normalization based on TCF4 protein content of the immunoprecipitates as determined by western blotting (Figure 7C). Sequences of the Cdx1 and Siamois promoters were strongly enriched upon ChIP of all three TCF4 variants (Figure 7B). In the absence of specific Cdx1 or Siamois promoter sequences, recovery of plasmid DNA by TCF4-ChIP was much reduced, but not eliminated. Presumably, cryptic TCF binding sites in the vector backbone mediate the residual enrichment. Importantly, however, precipitation of promoter DNA strictly depended on expression of exogenous TCF4 demonstrating the specificity of the ChIP. Interestingly, whereas all three TCF4 variants bound to the Siamois promoter in a similar manner, Cdx1 sequences were more efficiently precipitated by TCF4E2. Thus, superior transactivation by TCF4E2 is reflected to some extent by enhanced promoter binding in case of the Cdx1 construct but not with the Siamois promoter. Taken together, the results of the DNA-binding experiments in vitro and in vivo suggest that the extended C-terminus of TCF4E splice variants does have some influence on TCF4E2::DNA interactions but, in addition, it appears to harbour further functions critical for promoter activation.
Figure 7.
Promoter-binding capacity of TCF4 isoforms in transfected cells. (A) Schemes of the Cdx1 and Siamois (Sia) promoter regions present in the pGL3 basic luciferase vector. Vertical bars mark the location of TBEs. Positions of primers used for quantitative PCR are shown (demi-arrows). Coordinates of primers and DNA fragment end points are given relative to the transcription start sites (+1; arrows) of the Cdx1 and Siamois promoters. DNA sequences used as EMSA probes are marked by gray boxes. At the pGL3 basic backbone positions of primers refer to the parental vector. (B) HEK293 cells were cotransfected with combinations of expression vectors for TCF4 isoforms and reporter gene constructs harbouring Siamois or Cdx1 promoter sequences as indicated. Control samples received the empty expression vector and a promoter-less construct, respectively. To determine promoter occupancy, cells were crosslinked 40 h post transfection and solubilized fragmented chromatin was prepared. ChIP was done with a goat polyclonal antibody against TCF4 (N-20) or non-specific goat control IgGs. Promoter DNA recovered by ChIP was quantified by real-time PCR. Relative enrichments represent the ratio of DNA amounts obtained by specific versus non-specific precipitations. Results from at least three independent experiments and the corresponding standard deviations are shown. (C) Abundance of TCF4 splice variants in solubilized fragmented chromatin from transfected HEK293 cells and their recovery by ChIP. Levels of TCF4 splice variants were monitored by western blotting using fractions of the input chromatin and of the material immunoprecipitated by either goat control IgGs or the goat polyclonal anti-TCF4 antibody N-20. Detection was performed with the mouse monoclonal anti-TCF4 antibody 6H5-3 and appropriate peroxidase-conjugated secondary antibodies. Results from one representative experiment are shown.
DISCUSSION
The Wnt/β-catenin pathway is capable of eliciting a large number of different tissue-specific responses (45). The predominant Wnt effect in target cells consists of changes in gene expression at the transcriptional level and many of the Wnt/β-catenin target genes are subject to cell-type-specific control (45,51,52). One mechanism whereby the Wnt/β-catenin pathway can address different sets of target genes, involves the use of multiple nuclear effector proteins with different functional properties. In vertebrate genomes, several TCF genes exist and in some cases the complement of TCF proteins can be additionally expanded by alternative splicing (5,6,25–27,53). To further explore the extent of TCF structural variability and its potential importance in transcriptional control by Wnt/β-catenin signalling, we have undertaken a comprehensive characterization of splice variants derived from the mouse Tcf7l2 gene.
Sequence diversity of Tcf7l2 splice variants
Previous studies concentrated on the analysis of alternative splicing of the human TCF7L2 gene (25–27). Recently, a more extensive study of transcript variants in the mouse was reported (30). We further complement and extend this work. Overall, the mouse and human genes turn out to be highly similar both with respect to the regions, which exhibit sequence variability, and in view of the combinatorial patterns of alternative exons. However, some differences appear to exist. At present, there is no evidence that alternative splice acceptors exist in exons 16 and 17 of Tcf7l2 as previously reported for the human gene (this study, 30). It is not clear whether this is a species-specific difference or whether this reflects aberrant splice events in human tumour cells. In support of the latter possibility, it has also been described that in brain tumours a novel exon 16′ is incorporated into TCF7L2 transcripts which is not found in other cell types or healthy tissues (26). A further difference concerns transcript variants which encode TCF4N. So far, these have only been reported in mouse but not in human cells. The strong emphasis on splice events affecting exons 13–16 may have precluded their detection in the human system up to now. Given the unique functional properties of TCF4N (28,29,54) and its intriguing tissue distribution might make it worthwhile to determine whether TCF4N variants also occur in human cell types.
Even by combining the results of all currently available studies it is not clear whether the full range of all theoretically possible alternatively spliced Tcf7l2 mRNA species is actually realized. For example, variable use of alternative splice donors and acceptors in exons 7 and 9 could bring about four different transcript variants. To date, three of these four variants have been found (this study, 30). Similarly, out of 16 possible arrangements of exons 12–17, only 13 have been experimentally confirmed by sequence analyses. As a novel aspect, we show here that exon 4 can be found in multiple TCF4E, TCF4M and TCF4S isoforms. Although this does not exclude the possibility that some combinations among exons 4 and 13–17 are mutually exclusive, it seems that exon 4 is not specifically excluded from one of the three main groups of TCF4 isoforms.
An unexpected observation was that there are two different amino acid versions of the first part of the C-clamp derived from exon 14 or 15, respectively. Exons 14 and 15 have the same lengths and they are highly related in sequence. Still, their translation products differ at seven out of 25 amino acid positions. The majority of these alterations are non-conservative exchanges. Most notably, the CRARF element is converted into a CRALF motif. Nonetheless, our functional analyses did not detect any striking difference between CRARF and CRALF versions of the C-clamp with respect to their transactivation properties. Perhaps significantly, the variant positions spare amino acids, which were found to be highly conserved in evolution and which appear crucial for C-clamp function (19,24). Thus, the CRALF version of the C-clamp may result from an exon duplication event (25) followed by mutagenesis and selection of neutral exchanges. This naturally occurring sequence variation could help to further distinguish functionally important residues within the C-clamp.
In the past, different nomenclatures were used to denominate splice variants with different combinations of exons 13–16. In some cases the sorting criterion was the presumed size of the protein products which were classified as ‘L’ (long), ‘M’ (medium) and ‘S’ (short) (25–27). Others distinguished between ‘E’ and ‘B’ forms in analogy to transcripts derived from the TCF7 gene (5,6,18,28,32–34). We propose a unified classification of transcripts according to the C-terminal amino acid composition of their translation products. Transcripts which code for TCF4 isoforms containing a complete C-clamp and the two CtBP-binding motifs, represent the E-type in reminiscence of the structural and functional similarities to the TCF1E variant (5,6). Transcripts which give rise to TCF4 variants with a partial C-clamp (C-clampΔC) form the type ‘S’ group. Splice products coding for TCF4 isoforms lacking all of the C-clamp constitute the M-type (hitherto ‘B’). Overall, the proposed classification equalizes the nomenclatures used for mouse and human transcripts by using structurally and functionally relevant criteria while largely maintaining the labelling previously assigned to the human splice variants. One notable exception is the splice variant with exon combination 12–16–17 to which we now refer as TCF4M3.
Tissue-specific expression of TCF4 splice variants
Cell-type-specific expression of protein isoforms with variable domain composition could be one way whereby the transcriptional output of Wnt/β-catenin signalling is diversified. While our comparative analyses show that many Tcf7l2 transcript isoforms exhibit a broad tissue distribution, they also reveal a few notable exceptions with more restricted expression patterns. Transcripts containing exon 4 are considerably less abundant in brain and ES cells. Amino acids encoded by exon 4 appear to have a dampening effect on TCF4 transactivation capacity. The underrepresentation of exon 4-containing transcripts in brain and ES cells may indicate a need to achieve comparably higher expression levels of TCF4-regulated genes in these cells. A second example for TCF4 transcripts with a pronounced tissue-specific expression is provided by the variants encoding TCF4N which lack a DNA-binding domain and a nuclear import sequence. In functional tests, TCF4N diminishes transcriptional activity of β-catenin/TCF-regulated promoters and redirects β-catenin to other regulatory elements where it potentiates transcriptional activity of non-TCF transcription factors (28,29). In view of this, the reduced abundance of TCF4N variants in embryonic stem cells may indicate the requirement of an unrestrained availability of β-catenin for TCF-controlled Wnt target genes (55–57). A third example for transcripts with a more conspicuous tissue distribution are variants with exon 16 (25–27,30). Some of these (TCF4S3, TCF4S6) appear to be particularly abundant in brain. The functional significance of this is unclear, though. Inclusion of exon 16 sequences is incompatible with the production of TCF4E isoforms. Instead, predominantly TCF4S isoforms are generated. Their tissue-specific representation and their relative amounts compared to other splice forms, thus, might be of importance in shaping a cell-type-specific transcriptional response of the Wnt/β-catenin pathway in the brain.
Promoter-specific transactivation by TCF4 splice variants
By comparing the transactivation potential of LEF1, TCF1B, TCF1E and TCF3, it was found earlier that only TCF1E was able to activate the P1 promoter of the human LEF1 gene (19,24). Similarly, functional analyses of LEF1 and TCF4E revealed the particular ability of TCF4E to synergize with β-catenin and p300 in Cdx1 promoter activation (18). Here, we have extended the functional characterization of TCF4 to additional target gene promoters and included representatives of all three major categories of TCF4 splice isoforms. We found that, in principle, all splice variants tested can function as transcriptional activators in conjunction with β-catenin. However, promoter context strongly influences the ability of TCF4 proteins to synergize with β-catenin as shown by the example of the Cyclin D1 promoter which can be bound by TCF4 but which is not stimulated by it. Also, TCF4M1 and S2 which were ineffective at the Axin2 and Cdx1 promoters, possessed limited transactivation capacity at the Siamois promoter. However, at promoter elements where TCF4 does function as a transactivator, TCF4E variants generally induced much higher activity than TCF4M1 and TCF4S.
What could be the molecular basis for promoter specificity of TCF4 transcriptional activity and the functional differences between TCF4E and TCF4M/S variants? It was suggested that TCF1E and Drosophila dTCF are capable of dual DNA-recognition which is mediated by both the HMG box which interacts with the TBE core motif, and the C-clamp which contacts 5′-RCCG-3′ sequences or so-called helper elements present at somewhat flexible distances and orientations relative to some core TBEs (19,24). Due to this mode of bipartite DNA-recognition, TCF1E is thought to be able to function at promoter elements which cannot be bound by other TCF family members or by TCF1 isoforms lacking a C-clamp (19). However, TCF4M1 and TCF4S2 variants interacted with the Cdx1 promoter in vitro and in vivo despite the absence of a C-clamp, although they could not activate this promoter. Moreover, TCF4E2, TCF4M1 and TCF4S2 bound in an indistinguishable manner to the Siamois promoter whose TBEs lack discernable 5′-RCCG-3′ sequences. Yet again, transactivation by TCF4E variants vastly exceeded that by TCF4M1 and TCF4S2. From this we conclude that differences in promoter recognition are not responsible for differences in promoter activation by TCF4 isoforms. Consistent with this view, it was also previously shown that LEF1 which does not have a C-clamp and which does not transactivate the Cdx1 and LEF1 P1 promoters, nonetheless binds to these DNA elements (18,19).
Interestingly, it appears from our analyses that 5′-RCCG-3′ sequences next to a core TBE promote DNA binding only in case of the C-clamp-containing TCF4E2 variant. The requirement for the 5′-RCCG-3′ motif was at least partially alleviated with Cdx1 and Siamois promoter probes containing multiple core TBEs. TCF4M1 and TCF4S2 without C-clamp and other ‘E’-type-specific amino acid sequences were seemingly indifferent towards the 5′-RCCG-3′ motif. One interpretation of this could be that C-terminal amino acid sequences of TCF4E isoforms function as intramolecular inhibitors of DNA binding. Interaction between the C-clamp and 5′-RCCG-3′ sequences or intermolecular interactions with TCF4E proteins nearby appear to alleviate this inhibition. Obviously, this mechanism would be obsolete in case of TCF4 variants lacking the extended C-termini of ‘E’-type variants. From our observations it also follows that promoter recognition and transcriptional activation in a manner depending upon β-catenin and promoter context are separable functions associated with the extended ‘E’-type sequences. This could occur by protein::protein interactions between TCF4 and other transcription factors such as p300 (18). In summary, the ability of individual TCF proteins to interact with regulatory DNA elements and, beyond this, to participate in promoter activation appears to depend on TCF4 protein domain composition, architectural differences among Wnt-inducible promoters with respect to the arrangement of TBEs and adjacent DNA elements, and possibly additional protein cofactors (18,58–60).
Role of Tcf7l2 splice variants in control of proliferation and differentiation
In mice, TCF4 deficiency results in perinatal lethality due to depletion of a proliferative cell compartment in the intestinal epithelium (61). Expression of dominant negative dnTCF1 or dnTCF4 induces cell-cycle arrest in human colorectal cancer cells (24,48,62). In ChIP experiments, TCF4 was found to occupy cis-regulatory elements of genes which promote cell proliferation (15,63,64). These observations are in agreement with the view that TCF4 proteins support the oncogenic activities of the Wnt/β-catenin signalling cascade. Recent studies, however, ascribe tumour suppressor properties to TCF4 (65,66). By mutational analyses the tumour inhibitory function of TCF4 was mapped to a C-terminal domain which encompasses the C-clamp (66). This fits with the observation that a homopolymeric stretch of nine consecutive adenosines at the 5′-end of exon 17 is frequently shortened or extended in tumour cells (26,67–69). The frame shifts which ensue, interfere with the production of TCF4E variants with a complete C-clamp and CtBP-binding motifs. Seemingly, the tumour inhibitory functions of TCF4 are specifically linked to full-length TCF4E variants. Consistent with this idea, TCF4E isoforms are distinguished by their unique transactivation properties at the Axin2 and Cdx1 genes, which contribute to the suppression of tumourigenesis (70–72). The assumption that different TCF4 isoforms are required for the control of distinct groups of genes would allow to reconcile the apparently conflicting activities of TCF4 in maintaining cell proliferation and in suppressing tumourigenesis (61,62,65,66). These opposing functions may simply reflect promoter-specific activities of different TCF4 splice variants. To gain further insight into this and to precisely define the functions of TCF4 splice variants under physiological and pathological conditions, the full range of TCF4-regulated genes in different cellular settings and the dependency of their expression on the availability of specific TCF4 isoforms will have to be analysed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (He 2004/6-1 and He 2004/6-2 to A.H.). Funding for open access charge: Institutional resources and extramural funding (DFG He 2004/6-2).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to M. Schrempp for excellent technical assistance and to R. Shivdasani for gift of the mTCF4M1 expression construct.
REFERENCES
- 1.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterman ML, Fischer WH, Jones KA. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 3.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 4.Stadeli R, Hoffmans R, Basler K. Transcription under the control of nuclear Arm/beta-catenin. Curr. Biol. 2006;16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 5.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 6.Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J. Cell. Sci. 2007;120:385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 7.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 9.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech. Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 10.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 11.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 12.Gradl D, Konig A, Wedlich D. Functional diversity of Xenopus lymphoid enhancer factor/T-cell factor transcription factors relies on combinations of activating and repressing elements. J. Biol. Chem. 2002;277:14159–14171. doi: 10.1074/jbc.M107055200. [DOI] [PubMed] [Google Scholar]
- 13.Liu F, van den Broek O, Destree O, Hoppler S. Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/beta-catenin signalling in mesoderm development. Development. 2005;132:5375–5385. doi: 10.1242/dev.02152. [DOI] [PubMed] [Google Scholar]
- 14.Pukrop T, Gradl D, Henningfeld KA, Knochel W, Wedlich D, Kuhl M. Identification of two regulatory elements within the high mobility group box transcription factor XTCF-4. J. Biol. Chem. 2001;276:8968–8978. doi: 10.1074/jbc.M007533200. [DOI] [PubMed] [Google Scholar]
- 15.Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wohrle S, Wallmen B, Hecht A. Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors. Mol. Cell. Biol. 2007;27:8164–8177. doi: 10.1128/MCB.00555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht A, Stemmler MP. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 2003;278:3776–3785. doi: 10.1074/jbc.M210081200. [DOI] [PubMed] [Google Scholar]
- 19.Atcha FA, Munguia JE, Li TW, Hovanes K, Waterman ML. A new beta-catenin-dependent activation domain in T cell factor. J. Biol. Chem. 2003;278:16169–16175. doi: 10.1074/jbc.M213218200. [DOI] [PubMed] [Google Scholar]
- 20.Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes & Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 22.Roel G, Hamilton FS, Gent Y, Bain AA, Destree O, Hoppler S. Lef-1 and Tcf-3 transcription factors mediate tissue-specific Wnt signaling during Xenopus development. Curr. Biol. 2002;12:1941–1945. doi: 10.1016/s0960-9822(02)01280-0. [DOI] [PubMed] [Google Scholar]
- 23.Standley HJ, Destree O, Kofron M, Wylie C, Heasman J. Maternal XTcf1 and XTcf4 have distinct roles in regulating Wnt target genes. Dev. Biol. 2006;289:318–328. doi: 10.1016/j.ydbio.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol. Cell. Biol. 2007;27:8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duval A, Rolland S, Tubacher E, Bui H, Thomas G, Hamelin R. The human T-cell transcription factor-4 gene: structure, extensive characterization of alternative splicings, and mutational analysis in colorectal cancer cell lines. Cancer Res. 2000;60:3872–3879. [PubMed] [Google Scholar]
- 26.Howng SL, Huang FH, Hwang SL, Lieu AS, Sy WD, Wang C, Hong YR. Differential expression and splicing isoform analysis of human Tcf-4 transcription factor in brain tumors. Int. J. Oncol. 2004;25:1685–1692. [PubMed] [Google Scholar]
- 27.Shiina H, Igawa M, Breault J, Ribeiro-Filho L, Pookot D, Urakami S, Terashima M, Deguchi M, Yamanaka M, Shirai M, et al. The human T-cell factor-4 gene splicing isoforms, Wnt signal pathway, and apoptosis in renal cell carcinoma. Clin. Cancer Res. 2003;9:2121–2132. [PubMed] [Google Scholar]
- 28.Douglas KR, Brinkmeier ML, Kennell JA, Eswara P, Harrison TA, Patrianakos AI, Sprecher BS, Potok MA, Lyons R.H., Jr, MacDougald OA, et al. Identification of members of the Wnt signaling pathway in the embryonic pituitary gland. Mamm. Genome. 2001;12:843–851. doi: 10.1007/s00335-001-2076-0. [DOI] [PubMed] [Google Scholar]
- 29.Kennell JA, O’Leary EE, Gummow BM, Hammer GD, MacDougald OA. T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors. Mol. Cell. Biol. 2003;23:5366–5375. doi: 10.1128/MCB.23.15.5366-5375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nazwar TA, Glassmann A, Schilling K. Expression and molecular diversity of Tcf7l2 in the developing murine cerebellum and brain. J. Neurosci. Res. 2009;15:1532–1546. doi: 10.1002/jnr.21989. [DOI] [PubMed] [Google Scholar]
- 31.Valenta T, Lukas J, Korinek V. HMG box transcription factor TCF-4’s interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res. 2003;31:2369–2380. doi: 10.1093/nar/gkg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho EA, Dressler GR. TCF-4 binds beta-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech. Dev. 1998;77:9–18. doi: 10.1016/s0925-4773(98)00131-2. [DOI] [PubMed] [Google Scholar]
- 33.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YJ, Swencki B, Shoichet S, Shivdasani RA. A possible role for the high mobility group box transcription factor Tcf-4 in vertebrate gut epithelial cell differentiation. J. Biol. Chem. 1999;274:1566–1572. doi: 10.1074/jbc.274.3.1566. [DOI] [PubMed] [Google Scholar]
- 35.Snyder EY, Deitcher DL, Walsh C, Arnold-Aldea S, Hartwieg EA, Cepko CL. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell. 1992;68:33–51. doi: 10.1016/0092-8674(92)90204-p. [DOI] [PubMed] [Google Scholar]
- 36.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 37.Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 38.Aulehla A, Wehrle C, Brand-Saberi B, Kemler R, Gossler A, Kanzler B, Herrmann BG. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell. 2003;4:395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 39.Lickert H, Domon C, Huls G, Wehrle C, Duluc I, Clevers H, Meyer BI, Freund JN, Kemler R. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development. 2000;127:3805–3813. doi: 10.1242/dev.127.17.3805. [DOI] [PubMed] [Google Scholar]
- 40.Brannon M, Gomperts M, Sumoy L, Moon RT, Kimelman D. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herber B, Truss M, Beato M, Muller R. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:1295–1304. [PubMed] [Google Scholar]
- 42.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch C, Campano LM, Wohrle S, Hecht A. Canonical Wnt signaling transiently stimulates proliferation and enhances neurogenesis in neonatal neural progenitor cultures. Exp. Cell Res. 2007;313:572–587. doi: 10.1016/j.yexcr.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Young RM, Reyes AE, Allende ML. Expression and splice variant analysis of the zebrafish tcf4 transcription factor. Mech. Dev. 2002;117:269–273. doi: 10.1016/s0925-4773(02)00180-6. [DOI] [PubMed] [Google Scholar]
- 45.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 46.Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- 47.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze'e;v A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 49.Pilon N, Oh K, Sylvestre JR, Savory JG, Lohnes D. Wnt signaling is a key mediator of Cdx1 expression in vivo. Development. 2007;134:2315–2323. doi: 10.1242/dev.001206. [DOI] [PubMed] [Google Scholar]
- 50.Chang MV, Chang JL, Gangopadhyay A, Shearer A, Cadigan KM. Activation of wingless targets requires bipartite recognition of DNA by TCF. Curr. Biol. 2008;18:1877–1881. doi: 10.1016/j.cub.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aoki M, Hecht A, Kruse U, Kemler R, Vogt PK. Nuclear endpoint of Wnt signaling: neoplastic transformation induced by transactivating lymphoid-enhancing factor 1. Proc. Natl Acad. Sci. USA. 1999;96:139–144. doi: 10.1073/pnas.96.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vleminckx K, Kemler R, Hecht A. The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis. Mech. Dev. 1999;81:65–74. doi: 10.1016/s0925-4773(98)00225-1. [DOI] [PubMed] [Google Scholar]
- 53.Van de Wetering M, Castrop J, Korinek V, Clevers H. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell. Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brinkmeier ML, Potok MA, Cha KB, Gridley T, Stifani S, Meeldijk J, Clevers H, Camper SA. TCF and Groucho-related genes influence pituitary growth and development. Mol. Endocrinology. 2003;17:2152–2161. doi: 10.1210/me.2003-0225. [DOI] [PubMed] [Google Scholar]
- 55.Galceran J, Farinas I, Depew MJ, Clevers H, Grosschedl R. Wnt3a-/–like phenotype and limb deficiency in Lef1(-/-)Tcf1(-/-) mice. Genes Dev. 1999;13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV, Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 57.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nature Genetics. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 58.Tutter AV, Fryer CJ, Jones KA. Chromatin-specific regulation of LEF-1-beta-catenin transcription activation and inhibition in vitro. Genes Dev. 2001;15:3342–3354. doi: 10.1101/gad.946501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beland M, Pilon N, Houle M, Oh K, Sylvestre JR, Prinos P, Lohnes D. Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol. Cell. Biol. 2004;24:5028–5038. doi: 10.1128/MCB.24.11.5028-5038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. PITX2, beta-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J. Cell. Sci. 2005;118:1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- 61.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nature Genetics. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 62.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 63.Kirmizis A, Bartley SM, Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol. Cancer Therapeutics. 2003;2:113–121. [PubMed] [Google Scholar]
- 64.Yochum GS, Cleland R, Goodman RH. A genome-wide screen for beta-catenin binding sites identifies a downstream enhancer element that controls c-Myc gene expression. Mol. Cell. Biol. 2008;28:7368–7379. doi: 10.1128/MCB.00744-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shulewitz M, Soloviev I, Wu T, Koeppen H, Polakis P, Sakanaka C. Repressor roles for TCF-4 and Sfrp1 in Wnt signaling in breast cancer. Oncogene. 2006;25:4361–4369. doi: 10.1038/sj.onc.1209470. [DOI] [PubMed] [Google Scholar]
- 66.Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc. Natl Acad. Sci. USA. 2008;105:9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cuilliere-Dartigues P, El-Bchiri J, Krimi A, Buhard O, Fontanges P, Flejou JF, Hamelin R, Duval A. TCF-4 isoforms absent in TCF-4 mutated MSI-H colorectal cancer cells colocalize with nuclear CtBP and repress TCF-4-mediated transcription. Oncogene. 2006;25:4441–4448. doi: 10.1038/sj.onc.1209471. [DOI] [PubMed] [Google Scholar]
- 68.Duval A, Gayet J, Zhou XP, Iacopetta B, Thomas G, Hamelin R. Frequent frameshift mutations of the TCF-4 gene in colorectal cancers with microsatellite instability. Cancer Res. 1999;59:4213–4215. [PubMed] [Google Scholar]
- 69.Ruckert S, Hiendlmeyer E, Brueckl WM, Oswald U, Beyser K, Dietmaier W, Haynl A, Koch C, Ruschoff J, Brabletz T, et al. T-cell factor-4 frameshift mutations occur frequently in human microsatellite instability-high colorectal carcinomas but do not contribute to carcinogenesis. Cancer Res. 2002;62:3009–3013. [PubMed] [Google Scholar]
- 70.Crissey MA, Guo RJ, Fogt F, Li H, Katz JP, Silberg DG, Suh ER, Lynch JP. The homeodomain transcription factor Cdx1 does not behave as an oncogene in normal mouse intestine. Neoplasia. 2008;10:8–19. doi: 10.1593/neo.07703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo RJ, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Can. Biol. Ther. 2004;3:593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- 72.Bonhomme C, Calon A, Martin E, Robine S, Neuville A, Kedinger M, Domon-Dell C, Duluc I, Freund JN. Cdx1, a dispensable homeobox gene for gut development with limited effect in intestinal cancer. Oncogene. 2008;27:4497–4502. doi: 10.1038/onc.2008.78. [DOI] [PubMed] [Google Scholar]
- 73.Arce L, Pate KT, Waterman ML. Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer. 2009;9:159. doi: 10.1186/1471-2407-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giese K, Amsterdam A, Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991;5:2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- 75.van de Wetering M, Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992;11:3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.