Abstract
Telomerase is a specialized reverse transcriptase that adds telomeric DNA repeats onto chromosome termini. Here, we characterize a new telomerase-specific motif, called motif 3, in the catalytic domain of telomerase reverse transcriptase, that is crucial for telomerase function and evolutionally conserved between vertebrates and ciliates. Comprehensive mutagenesis of motif 3 identified mutations that remarkably increase the rate or alter the processivity of telomere repeat addition. Notably, the rate and processivity of repeat addition are affected independently by separate motif 3 mutations. The processive telomerase action relies upon a template translocation mechanism whereby the RNA template and the telomeric DNA strand separate and realign between each repeat synthesis. By analyzing the mutant telomerases reconstituted in vitro and in cells, we show that the hyperactive mutants exhibit higher repeat addition rates and faster enzyme turnovers, suggesting higher rates of strand-separation during template translocation. In addition, the strong correlation between the processivity of the motif 3 mutants and their ability to use an 8 nt DNA primer, suggests that motif 3 facilitates realignment between the telomeric DNA and the template RNA following strand-separation. These findings support motif 3 as a key determinant for telomerase activity and processivity.
INTRODUCTION
Telomerase is a specialized reverse transcriptase (RT) responsible for adding telomeric DNA repeats onto the 3′-ends of chromosomes. Telomere elongation counter-balances the natural shortening of linear chromosomes due to the end-replication problem, preventing senescence, apoptosis and genome instability (1). The deficiency in telomerase function leads to limited renewal capacity in highly proliferative cells, and is associated with human diseases including dyskeratosis congenita, aplastic anemia and idiopathic pulmonary fibrosis (2,3).
Reconstitution of catalytically active telomerase in vitro requires two core components: the telomerase RNA (TR) and the telomerase reverse transcriptase (TERT) protein. The TR contains a short template region for the synthesis of telomeric DNA repeats, and conserved structural domains essential to in vivo biogenesis and assembly with the TERT protein. The TERT subunit is a multi-domain protein comprised of an N-terminal extension (NTE), a central catalytic RT domain and a C-terminal extension (CTE) (4). In most eukaryotes, the NTE consists of a TR binding domain and a telomerase-essential N-terminal (TEN) domain that binds telomeric DNA (5). However, the TEN domain is dispensable in certain species, such as insects (6). The catalytic RT domain encompasses seven essential motifs (1, 2, A, B, C, D and E) that are universally conserved among RTs (7).
Telomerase has the unique ability to add multiple telomeric repeats to a given primer before complete dissociation from the DNA, called ‘repeat addition processivity’ (abbreviated to ‘processivity’). Unlike conventional RTs which can utilize a variety of single-stranded RNA templates, telomerase uses only a short sequence from its intrinsic RNA component as template. During telomere DNA synthesis, the 3′-end of telomeric DNA base pairs with the RNA template forming an RNA/DNA duplex which is positioned within the catalytic site of TERT protein for nucleotide addition. When telomerase completes the synthesis of one telomeric DNA repeat, a ‘template translocation’ must occur whereby the RNA template dissociates from the DNA strand, translocates and realigns relative to the 3′-end of the DNA, providing an unoccupied template for the next round of repeat synthesis. This translocation process is the rate-limiting step in a processive telomerase reaction, as indicated by the strong pause after each round of repeat synthesis, giving rise to the characteristic ladder banding pattern of telomere products (8,9). While the repeat addition rate is determined by the rate-limiting translocation step, the processivity of the reaction is determined by the probability, or efficiency, of RNA/DNA realignment over complete product release during translocation.
The extent of telomerase processivity varies dramatically among species. Telomerase from ciliates and most vertebrates are highly processive (8–10). In contrast, telomerase from certain rodents and yeasts have little to no processivity (11–14). Reaction conditions and accessory proteins appear to contribute to the disparity in processivity observed in vitro (15–17). However, the intrinsic properties of TERT and TR components are by and large the major determinants for the varying degrees of processivity observed among species.
Mutations in both TERT and TR components have been found to affect the rate and processivity of the telomerase reaction. Several elements within TERT were shown to be crucial for telomerase processivity. The TEN domain contains an ‘anchor site’ that binds single-stranded telomeric DNA, preventing complete product release from the enzyme during template translocation (4,18). A recent study has shown that a mutation at Leu14 in the Tetrahymena TEN domain abolished processivity while leaving activity and DNA binding affinity intact. The Leu14 residue was proposed to function as an intra-molecular switch for processivity (19). In Saccharomyces cerevisiae, a motif called insertion in finger domain (IFD), located in the RT domain between motifs A and B, contains four conserved residues shown to be important for second repeat synthesis (20). Also within the RT domain, a point mutation in motif C of Tetrahymena TERT increases processivity by increasing protein–DNA primer affinity (21). Beyond the RT domain, a mutation in the CTE, a putative homolog to the HIV RT thumb, was shown to reduce repeat addition processivity (22). Within the TR, the template length and conserved structural elements also contribute to telomerase processivity, through affecting telomeric DNA/template RNA base-pairing interactions during template translocation (23,24) or the RNA/TERT protein interactions (25,26). A previous study by Drosopoulos et al. (27) showed that varying the template sequence can alter the rate of telomere repeat addition, possibly through modulating interactions between the template RNA, DNA product and TERT protein. Although the TERT protein was shown to contribute to the processivity of telomerase activity, its involvement in the regulation of telomere repeat addition rate is unclear.
In this study, we carried out a comprehensive alanine-substitution screening in a novel motif of the TERT protein and discovered mutations that surprisingly increased the rate or altered the processivity of telomere repeat addition. Characterization of the in vitro reconstituted telomerase enzymes containing these unusual hyperactive mutations indicates a higher rate of enzyme turnover or product dissociation. In addition, mutations that alter processivity alter the ability of the enzymes to use the short 8 nt primer as substrate, the use of which resembles the realignment of the 3′-end of DNA with the template RNA—the second step of the template translocation. We conclude that this novel TERT motif is an important determinant for telomerase activity and processivity, regulating both strand-separation and realignment of telomeric DNA and template RNA during template translocation.
MATERIALS AND METHODS
Sequence alignment analysis
The sequence alignment of the RT domain for TERT and other RTs was performed within the program BioEdit using the ClustalW algorithm, and further refined manually using the highly conserved RT motifs as anchor points. The alignment was carried out initially for individual groups of closely related species, then expanded to include sequences from more divergent species. The complete sequence alignment is available at the telomerase database (http://telomerase.asu.edu) (28).
Plasmid construction and mutagenesis
Specific mutations in the human TERT (hTERT) genes were introduced into pNFLAG-hTERT (a generous gift from Dr Vinayaka Prasad) by site-directed mutagenesis using an overlapping PCR strategy (29). For in vivo expression of TERT, gene fragments that contain specific mutations were sub-cloned from pNFLAG-hTERT into a modified pcDNA-hTERT vector (generous gifts from Dr Joachim Lingner) via two SacII sites within the TERT gene. Intended mutations were confirmed by sequencing.
Reconstitution of telomerase in vitro and in cells
Human telomerase was reconstituted in vitro using the TNT Quick Coupled rabbit reticulocyte lysate system (Promega) as described previously with minor modifications (30). To assemble telomerase, 1 µM of human TR (hTR) pseudoknot (nt 32–195) and CR4–CR5 (nt 239–328) RNA fragments were added to the hTERT synthesis reactions, and incubated at 30°C for 30 min.
To purify sufficient amount of mutant telomerase from cells for telomerase direct assay, we used the telomerase reconstitution system developed by Lingner’s group with minor modifications (31,32). Recombinant telomerase enzyme was reconstituted by over-expressing the hTERT and hTR genes in 293FT cells (Invitrogen) using pcDNA-hTERT and pBS-U1-hTR (generous gifts from Dr Joachim Lingner) as described previously. Cells were grown in DMEM media supplemented with 10% FBS at 37°C with 5% CO2. After reaching 80–90% confluency, cells were transfected with 1 µg of plasmid DNA (200 ng of pcDNA-hTERT and 800 ng of pBS-U1-hTR diluted in 50 µl of FBS-free DMEM media) and 4 µl of Fugene HD transfection reagent (Roche) in 12-well plates, following the manufacturer’s instruction. Two days post transfection, cells were harvested and lysed (31).
Telomerase activity assay
Telomerase reconstituted in vitro or in cells was assayed using the conventional direct primer-extension assay as previously described (30), with the exception of that 0.165 µM of [α-32P]dGTP (3000 Ci/mmol, Perkin Elmer) was used in a 10 µl reaction. Telomerase processivity was determined by measuring the intensity of each major band, normalized by the numbers of 32P-dGTP incorporated and plotted versus the repeat numbers as previously described (15). Processivity was calculated using the equation: Processivity = −ln 2/(2.303k), where k is the slope. The processivity of mutant telomerase was presented relative to the wild-type enzyme.
The pulse-chase time course experiments were carried out using in vitro reconstituted telomerase and the conventional direct assay as described above. The pulse-chase assay tracks the progressive extension of telomere products that were labeled (or pulsed) during the pulse reaction and extended by the processive telomerase enzymes during the chase reaction. During the pulse reaction, the enzymes add telomere repeats to the DNA primer with the incorporation of radioactive [α-32P]dGTP. In brief, telomerase was incubated with 4 µM (TTAGGG)3 primer in the presence of 1 × PE buffer (50 mM Tris–HCl, pH 8.3, 50 mM KCl, 2 mM DTT, 3 mM MgCl2 and 1 mM spermidine) at 30°C for 5 min to allow sufficient primer–enzyme complex formation prior to the pulse reaction. The pulse reaction was initiated by the addition of 1 mM dATP, 1 mM dTTP, 2 µM dGTP and 0.33 µM α-32P-dGTP (3000 Ci/mmol, Perkin Elmer) and incubated at 30°C for 5 min. The chase reaction was then carried out by adding non-radioactive dGTP to 100 µM in the reaction and incubated at 30°C for various amount of time as indicated. Upon addition of 50-fold excess non-radioactive dGTP, the initially labeled telomere products continue to be extended in the chase reaction by the same enzyme. During the chase reaction, the primers extended by the enzymes dissociated from the initially pulsed telomere products will not be seen. The chase reaction was terminated by ethanol precipitation and analyzed by gel-electrophoresis. For each chase reaction, 10 bands with highest intensity above initial pulse product bands were used to deduce a ‘modal band’ and calculate the extension rate as previously described (27). Because of the short extension time, the processivity cannot be accurately measured during the pulse reaction and the chase reactions at the early time points. Instead, telomerase processivity in the pulse-chase assay was determined from the first ten major bands (1–10 repeats added) in the chase reaction at the last time point (10 min).
For the short-primer telomerase assay, different telomere primers tel8, tel10, tel12, tel15, tel18 were used as indicated. For Km measurements, the primers were supplied at varying concentrations and the reaction mixture was incubated for 5–60 min, which falls in the linear range of product formation. A 32P-end-labeled oligonucleotide (50 nt) was used as the recovery and loading control. The product intensity of each reaction was quantitated, normalized with loading control and expressed as a relative activity compared to the reaction with highest primer concentration. The relative activities were plotted against primer concentration and the Michaelis–Menten equation, Y = Vmax × X/(Km + X), was used to fit the nonlinear curve to determine the Km (Prism 5, Graphpad Software).
To measure enzyme turnover rate, the in vitro reconstituted telomerase was pre-incubated with 10 µM tel7 primer (5′-AGGGTTA-3′) in the presence of 1× PE buffer at 30°C for 10 min. Telomerase reaction was initiated by addition of 2 µM dGTP and 0.33 µM [α-32P]dGTP (3000 Ci/mmol, Perkin Elmer), and aliquots were removed from the reaction mixture at different time points. The intensity of product was first adjusted by the TERT protein level, and normalized by the intensity of loading controls. The intensity of each band was normalized by that of the wild-type reaction at the last time point (10 min). The relative product intensities were then plotted against the amount of time. The slope of linear trend line represents the enzyme turnover rate. The enzyme turnover rates of mutant telomerases were indicated as relative values to the wild-type enzyme.
Western blot analysis
Ten micrograms of total protein of 293FT cell lysate was heated at 95°C for 5 min in 1 × Laemmli buffer (0.125 M Tris–HCl, pH 6.8, 2% SDS, 10% glycerol, 5% 2-mercaptoethanol and 0.0025% bromophenol blue), resolved on a 6 or 8% SDS–PAGE gel, and electro-transferred onto the PVDF membrane. Blocking (overnight at 4°C) and incubation with antibodies (1 h at room temperature) were carried out in 5% nonfat milk/1× TTBS (20 mM Tris–HCl, pH 7.5, 150 mM NaCl and 0.05% Tween 20). Anti-hTERT goat polyclonal antibody L-20 (Santa Cruz Biotechnology) and anti-GAPDH mouse monoclonal antibody 6C5 (Ambion) were used as the primary antibodies. After incubation with the HRP-conjugated secondary antibody (Santa Cruz Biotechnology), the blots were developed using the Immobilon Western Chemiluminescent HRP substrate (Millipore), and the blot images were acquired and analyzed using a Gel Logic440 system (Kodak).
Northern blot analysis
Total RNA was extracted from transfected cells using Tri-reagent (Molecular Research Center, Inc.) following manufacturer’s instruction. Three micrograms of total RNA was resolved on a 4% polyacrylamide/8 M urea denaturing gel and electro-transferred to the Hybond-XL membrane (GE Healthcare). Preparation of the riboprobes and hybridization of the blot were carried out as described previously (9).
Homology modeling
The RNA–DNA duplex was modeled into the Tribolium TERT structure (3DU6) by superimposition with the HIV1 p66 structure (1HYS) containing an RNA/DNA duplex. The two pdb files were superimposed in the CCP4 program using the following seven conserved resides within the RT domain as anchor points: three Asp residues in motifs A and C; Arg in motif 2; Glu and Gly in motif B; and Gly in motif E. These seven residues are highly conserved between TERTs and retroviral RTs.
RESULTS
Sequence conservation of motif 3
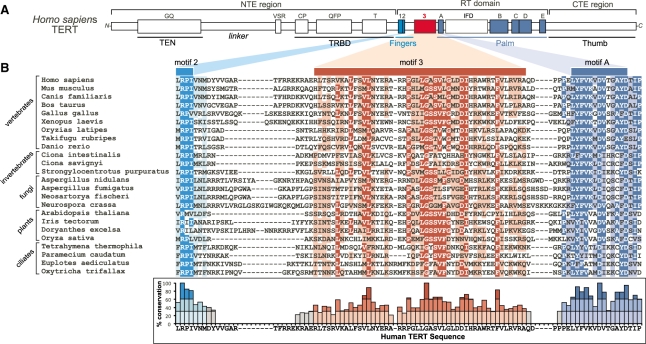
The novel motif 3, located in the catalytic RT domain of TERT between motifs 2 and A (Figure 1A), was previously found conserved in vertebrate and invertebrate chordates (33). To determine the extent of conservation of this motif in species beyond chordates, we extended the sequence alignment analysis to include additional groups of eukaryotes (detailed in ‘Materials and Methods’ section). Remarkably, the sequence of motif 3 is conserved also in non-yeast fungi, plants and ciliates (Figure 1B). The conservation of motif 3 sequences over a large evolutionary distance, from vertebrates to ciliates, implies necessity for telomerase function. Secondary-structure prediction of the motif 3 sequences from a large number of species suggested a putative helix–coil–helix fold (Supplementary Figure S1A). Interestingly, the recent crystal structure of TERT from an insect, Tribolium castaneum, contains a helix–coil–helix structure between motif 2 and A, supporting the secondary-structure prediction (34). Despite the apparent similarity in predicted secondary-structure, TERTs from insects, nematodes and yeasts did not show the same degree of sequence conservation, particularly in the central region of motif 3 (Supplementary Figure S2). Previously in the RTs closely related to the TERT, including the Penelope-like retrotransposons, non-LTR retrotransposons and group II introns, a conserved motif between motif 2 and A was identified as motif 2a (35–37), which however shares no common sequence to the TERT motif 3 (Supplementary Figure S2). Since motif 3 appears conserved specifically in vertebrates and ciliates whose telomerase is highly processive, but not in yeasts whose telomerase is not processive, we speculated that motif 3 might be important for telomerase processivity.
Figure 1.
Multiple sequence alignment of TERT motif 3. (A) Schematic of domain and motif organization of human TERT protein. Motif 3 (red) and the conserve RT motifs 1, 2 and A–E (cyan and blue) are colored. (B) Sequence alignment of TERT motif 3 from vertebrates, invertebrates, fungi, plants and ciliates. Shading indicates a minimum of 55% identity (dark cyan, red and blue) and 55% similarity (light cyan, red and blue) conservation. The degree of identity conservation with the human sequence at each residue is shown below the sequence alignment. No conservation determined where more than two sequences had gaps present. Darker shading indicates greater identity conservation with the human sequence (<30% light, 30–60% medium and >60% dark).
Mutations in motif 3 affect telomerase activity and processivity independently
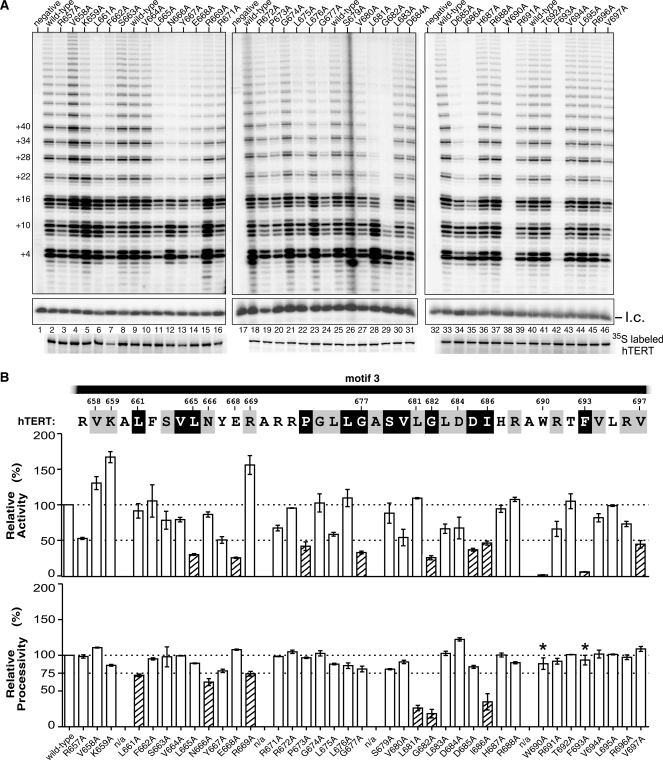
To experimentally determine the function of motif 3, we conducted comprehensive alanine substitution mutagenesis on human TERT and analyzed the telomerase mutants reconstituted in rabbit reticulocyte lysate (RRL) for activity and processivity (see ‘Materials and Methods’ section). Certain residues in motif 3 appear to be critical for telomerase catalysis, as the alanine substitutions W690A and F693A nearly abolished telomerase activity (Figure 2A, lanes 38 and 42). This is not unexpected for the F693 residue as it is one of the three most conserved residues in motif 3 (Figure 1B). The W690 residue, although not as highly conserved as F693, is naturally substituted with hydrophobic leucine or valine in most species (Figure 1B).
Figure 2.
Alanine substitution screening of motif 3. (A) Activity assays of the motif 3 mutants. Human telomerases with alanine substitutions in motif 3 were reconstituted in vitro and assayed for activity. Numbers on the left (+4, +10, +16, etc.) of the gel indicate the number of nucleotides added to the primer in each major band. l.c.: loading control, a 32P-end-labeled 15-nt DNA oligonucleotide, shown with the contrast adjusted. Below the gel, the [35S] methionine labeled TERTs analyzed by SDS–PAGE for quantitation are shown. (B) Quantitation of activity and processivity of motif 3 mutants. The residues in the human motif 3 sequence are shaded according to their identity and similarity as shown in Figure 1B. Below the sequence, the bar graph shows the activity and processivity of each mutant relative to wild-type. The dash line across the graph indicates the wild-type level of activity and processivity. The shaded bars indicate a relative activity lower than 50% or a processivity lower than 75% of the wild-type level. Alanine residues in the wild-type motif 3 sequence were omitted from the analysis and labeled as n/a (not available). Error bars indicate the standard deviation derived from 2–4 independent experiments. Asterisk: for the mutants with an extremely low activity, a special assay (3-fold more enzyme and reagents, and longer exposure time of the gel) was performed to determine processivity.
An intriguing finding from analyzing these motif 3 mutants was that telomerase activity and processivity can be independently altered (Figure 2). Three mutations (V658A, K659A and R669A) in the N-terminal region of motif 3 increased telomerase activity up to 1.7-fold, but had different effects on processivity (Figure 2B). For example, mutant V658A is hyperactive and hyper-processive, compared to wild-type enzyme (Figure 2A, lane 4). In contrast, hyperactive mutants K659A and R669A had reduced processivity (Figure 2A, lanes 5 and 15). Unlike V658A, mutants E668A, D684A and V697A showed greater processivity, but lower activity (Figure 2B). Mutations L661A, N666A, R669A, L681A, G682A and I686A, while all reduced processivity, altered activity differently (Figure 2B). In summary, the analysis of motif 3 mutants showed no correlation between changes in activity and changes in processivity, suggesting that telomerase activity and processivity are independent and possibly regulated through separate mechanisms.
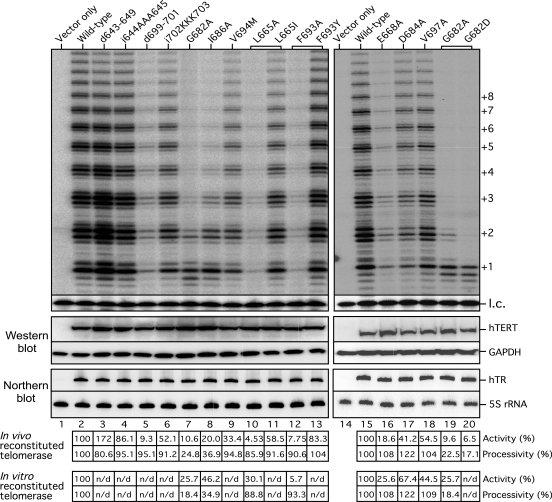
To determine if the telomerase mutants assembled in vitro and in cells behave similarly, we assayed two hypo-processive mutants (G682A and I686A) and three hyper-processive mutants (E668A, D684A and V697A). The mutant telomerases were reconstituted by over-expressing the full-length hTR and mutant hTERT genes in the 293FT cells to generate high telomerase activity sufficient for direct telomerase assay, a system developed by Cristofari and Lingner (31,32). The endogenous telomerase activity from 293FT cells transfected with the empty vector was undetectable by the direct primer-extension assay (Figure 3, lane 1). The telomerase mutants reconstituted in RRL and in human cells exhibited similar levels of activity and processivity (Figure 3, lanes 7, 8, 16, 17 and 18), confirming the results from the in vitro reconstituted enzymes. Additionally, we rescued two nearly inactive alanine-substituted mutants, L665A and F693A, with substitutions of conservative amino acids L665I and F693Y (Figure 3, compare lanes 10 to 11 and 12 to 13), suggesting the bulky hydrophobic side-chains of these two residues are required for telomerase function.
Figure 3.
Activity assay of telomerase mutants reconstituted in cells. Top panel: Mutant telomerases were reconstituted in 293FT cells and analyzed for activity. The TERT mutants d643-649 and d699-701 contain deletions of 7 and 3 residues, respectively. The TERT mutants i644AAA645 and i702KKK703 contain insertions of three alanine residues and three lysine residues between 644–645 and 702–703, respectively. The TERT mutants that contain different amino acid substitutions at the same residue are indicated by brackets. Numbers on the right (+1, +2, +3 etc.) indicate the number of repeats added to the telomeric primer. A 32P-end-labeled 15-mer DNA oligonucleotide is used as a loading control (l.c.). Middle panel: Expression level of hTERT protein in the transfected cells was analyzed by western blots of hTERT and GAPDH using anti-hTERT L-20 and anti-GAPDH antibodies. The level of GAPDH was used as a loading control. Bottom panel: Expression level of hTR in the transfected cells was analyzed by northern blots of hTR and 5S ribosomal RNA (rRNA) using riboprobes against hTR or 5S rRNA. The level of 5S rRNA was used as a loading control. The endogenous hTR is not visible in the vector-only sample (lane 1) due to the short exposure time. Quantitation of telomerase activity and processivity of telomerase reconstituted in cells in relation to the wild-type TERT are shown below the gel (in vivo reconstituted telomerase). Activity and processivity of telomerase mutants reconstituted in vitro analyzed in Figure 2 is shown at bottom for comparison (in vitro reconstituted telomerase). n/d: not determined.
We also tested two disease-associating motif 3 mutations, G682D and V694M, previously identified in aplastic anemia patients (38,39). While both mutations significantly reduced telomerase activity (Figure 3, lanes 9 and 20), the G682D mutation also caused a significant reduction in processivity (Figure 3, lane 20). The reduced telomerase activity and processivtiy of the disease-associating motif 3 mutants, together with the shortened telomere length in the patients harboring the mutations, support the importance of motif 3 for telomerase function and telomere maintenance in vivo.
The length of N-terminal linker of motif 3 affects telomerase activity
Based on the sequence alignment, the upstream linker connecting motif 3 to 2 is more variable in length than the downstream linker connecting motif 3 to A (Figure 1B). To assess the functional importance of the motif 3 flanking linkers, we generated TERT mutants with insertions (i644AAA645 and i702KKK703) or deletions (d643–649 and d699–701) in the linker regions and assayed the in vivo reconstituted enzymes for telomerase activity.
Both linker regions are more sensitive to deletions than insertions. Insertions i644AAA645 (N-terminal linker) and i702KKK703 (C-terminal linker) did not significantly decrease telomerase activity or processivity (Figure 3, lanes 4 and 6). In contrast, deletions in the linker regions caused dramatic alterations in telomerase activity. The 3-residue deletion d699–701 in the C-terminal linker 3/A nearly abolished activity (Figure 3, lane 5). Surprisingly, a 7-residue deletion d643–649 in the N-terminal linker increased activity by nearly two-fold without significant changes in processivity (Figure 3, lane 3). The N-terminal linker appeared more flexible than of the C-terminus, as a 12-residue deletion in the N-terminal linker did not affect telomerase activity (data not shown). Notably, the hyperactive alanine-substitution mutations, V658A, K659A, R669A, are located within the N-terminal portion of motif 3 near the upstream linker, implicating a similar role for the N-terminal linker and the N-terminal portion of motif 3 in regulating telomerase activity.
Hyperactive motif 3 mutants exhibit faster rates of repeat addition
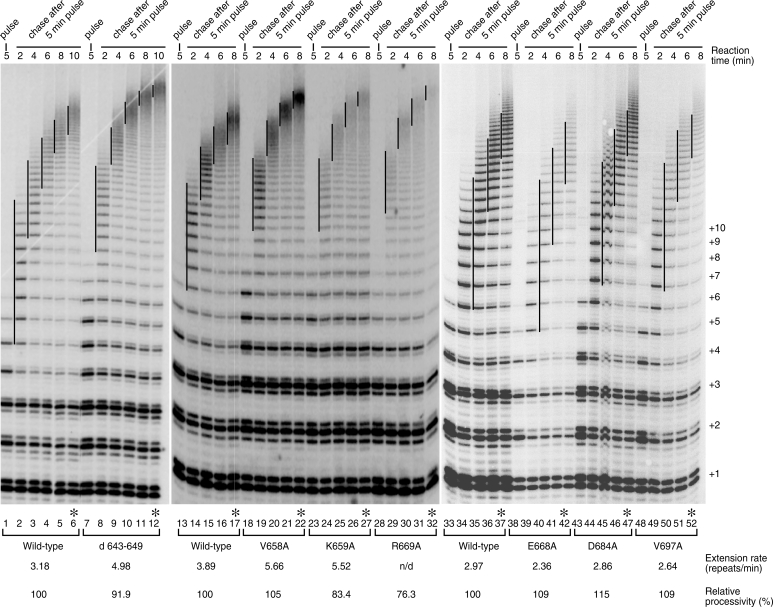
To determine if the greater activity observed within the hyperactive mutants was due to faster repeat addition, we carried out a pulse-chase time-course assay to measure repeat addition rate. Our results demonstrate that, regardless of their differences in processivity, the hyperactive d643–649, V658A, K659A and R669A mutants add telomere repeats at a faster rate of 5–6 repeats/min, higher than the wild-type enzyme, 3–4 repeats/min (Figure 4, lanes 7–32). Conversely, the hypoactive mutants E668A, D684A and V697A present slower repeat addition rates than the wild-type enzyme (Figure 4, lanes 38–52). These results suggest a critical role for motif 3 in regulating repeat addition rate of telomerase enzyme.
Figure 4.
Pulse-chase time course analysis to measure repeat addition rates of the motif 3 mutants. In vitro reconstituted wild-type, hyperactive (d643-649, V658A, K659A and R669A) or hypoactive (E668A, D684A and V697A) telomerase mutants were incubated with (TTAGGG)3 primer in the pulse reaction for 5 min in which the [α-32P]dGTP is incorporated to the newly synthesized telomere repeats. After 5 min of pulse reaction, non-radioactive dGTP was added to 100 µM to initiate the chase reactions and the reactions were terminated at different time points (2–10 min). The vertical lines on the gel denote the major bands of telomere products synthesized and labeled in the initial 5-min pulse reaction, and extended in the following chase reactions. Numbers on the right (+1, +2, +3, etc.) indicate the number of repeats added to the telomeric primer. Repeat-extension rate, expressed as repeats per minute, of each enzyme were calculated (see ‘Materials and Methods’ section) and indicated below the gel. Asterisk denotes the 10 min chase reaction from which the processivity of each mutant was measured based on the first 10 major bands (repeats 1–10) (see ‘Materials and Methods’ section).
The increase or decrease in repeat addition rate is independent of the processivity level of the mutants. In the pulse-chase assay, the extent of telomerase processivity was measured at the last time point where the processive enzyme–product complexes have already moved up to the top of the gel and separated from the products dissociated from the enzyme during the time course. While the hyperactive V658A mutant was more processive, the other two hyperactive mutants, K659A and R669A, were less processive (Figure 4, compare lanes 17, 22, 27 and 32). Moreover, by combining two motif 3 mutations (d643–649 and V658A) and an hTR-57C template mutation that increases processivity (23), we generated a telomerase mutant that is super-active and super-processive, demonstrating an additive effect for these mutations in telomerase activity and processivity (Supplementary Figure S3, lane 6).
Low-processivity motif 3 mutants are defective in utilizing short DNA primers
The motif 3 mutations that altered processivity presumably affected either the template realignment or product release step of translocation, as telomerase processivity correlates to the probability of template realignment over product release. To determine if the low-processivity mutations affect the template realignment step, we designed a short primer assay to analyze the ability of telomerase to use short primers that, when base-pair with the RNA template, leave no single-stranded overhang for the TEN domain anchor site to bind (Figure 5B). This assay thus discounts the effect of the TEN domain on substrate binding, as it binds to the upstream single-stranded region of a longer telomeric DNA primer (40). The base-pairing between the short tel8 DNA primer and the template RNA resembles the realignment of the 3′-end of telomeric DNA with the RNA template during translocation. By using a short primer, i.e. the 8-nt tel8 primer, in the telomerase assay, we can then determine solely the contribution of motif 3 in facilitating formation, or recognition, of the RNA/DNA duplex inside the active site. Thus, a low-processivity mutant with an inability to complete the realignment step in a translocation cycle would be predicted unable to use a short primer as substrate.
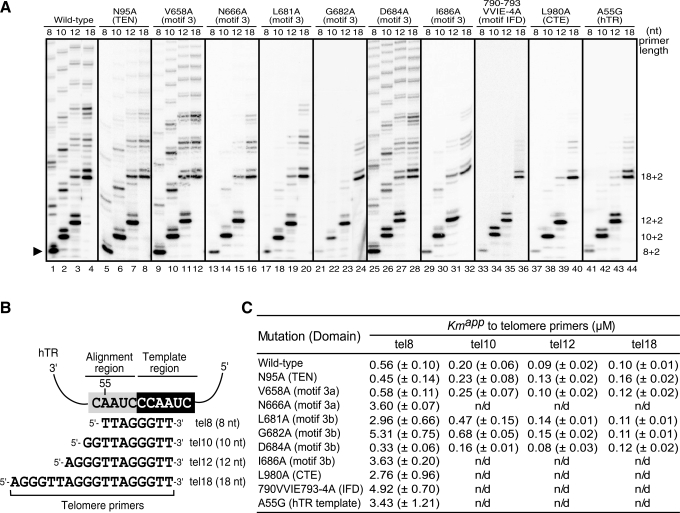
Figure 5.
Activity assay of telomerase mutants using primers with various lengths. (A) Telomerases with specific mutations in TERT or TR indicated were assayed for activity using telomere primers, tel8, tel10, tel12 or tel18, with length ranging from 8 to 18 nt. Due to the difference in overall activity between mutants, the gel image of each mutant was adjusted to have similar intensity for better comparison of the products of different primers. The numbers (8 + 2, 10 + 2, 12 + 2, etc.) labeled on the right of the gel indicate the length of the primer plus the number of nucleotides added. The black triangle on the left of the gel indicates the first repeat product extended from the tel8 primer. (B) The four different primers are aligned with hTR template sequence. The alignment (nt 52–56) and template (nt 46–51) regions are shaded in grey and black, respectively. (C) The 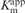 values for telomere primers of various lengths. The apparent Km values are determined from experiments using tel8, tel10, tel12 or tel18 primers at various concentrations and by fitting the data to the Michaelis–Menten equation (see ‘Materials and Methods’ section). Standard deviations (n = 3–4) are given in parentheses. n/d: not determined.
values for telomere primers of various lengths. The apparent Km values are determined from experiments using tel8, tel10, tel12 or tel18 primers at various concentrations and by fitting the data to the Michaelis–Menten equation (see ‘Materials and Methods’ section). Standard deviations (n = 3–4) are given in parentheses. n/d: not determined.
Using the short-primer assay, we tested six motif 3 mutants (V658A, N666A, L681A, G682A, D684A and I686A) with either increased or decreased processivity. In addition to motif 3, we also analyzed low-processivity mutations that are located in other parts of TERT, or in the TR component. Mutations N95A (TEN), L980A (CTE), 790-VVIE-793-4A (IFD) and hTR-A55G (RNA template) have been previously shown to reduce telomerase processivity (20,22,23,41). The wild-type telomerase can utilize all primers (8, 10, 12 or 18 nt) tested with similar activity (Figure 5A, lanes 1–4). Remarkably, the low-processivity mutants, N666A, L681A, G682A, I686A, 790-VVIE-793-4A, L980A and hTR-A55G, that can extend longer primers normally, had little to no activity when using the short tel8 primer (Figure 5A, lanes 13, 17, 21, 29, 33, 37 and 41). This suggests these mutations compromised the ability of TERT in promoting RNA/DNA duplex formation or positioning the duplex into the active site for the first repeat synthesis. As would be expected, the hyper-processive mutants V658A and D684A utilized the short tel8 and the longer primers with equal efficiency (Figure 5A, lanes 9 and 25). The TEN domain N95A mutant, while having low processivity, can however extend the short tel8 primers efficiently (Figure 5A, lane 5). The TEN domain thus does not appear to play a role in facilitating primer/template realignment, instead preventing product release during template translocation through DNA binding. When using tel10 and tel12 primers, all enzymes gave rise to stronger first bands (10 + 2 and 12 + 2) than the subsequent bands, indicating a lower efficiency for the first translocation event (Figure 5A). This phenomenon is also consistent with the notion that TEN domain binding to the longer DNA primer facilitates template translocation.
To quantitatively determine the ability of these processivity mutants to utilize short primers as substrate, we measured the Km of the wild-type and mutant enzymes to the tel8 DNA primer. All mutants that failed to extend the tel8 primer had higher Km values ranging from 2.76 to 5.31 µM, 5–10-fold higher than the 0.56 µM of the wild-type enzyme (Figure 5C). In contrast, mutants that retained the ability to extend the tel8 primer had Km values similar to or lower than the wild-type enzyme. For example, the most processive mutant D684A has a Km of 0.33 µM, significantly lower than that of the wild-type (Figure 5C). Compared to tel8, primers tel10 and tel12 can still be used efficiently by the wild-type and mutant enzymes, with Km ranging from 80 to 680 nM (Figure 5A and 5C). When the longer tel18 primer (18 nt) was used, the wild-type enzyme and all mutants, with the exception of the TEN N95A mutant, had similar Km values ∼100 nM (Figure 5C).
The hTR-A55G template mutation resulted in a mismatch between the RNA template and the DNA substrate (Figure 5B), leading to a high Km for the tel8 primer (Figure 5C) and lower processivity (Figure 5A, lanes 41–44). The IFD motif, found first in yeast, also contributes to the repeat addition processivity (20). Based on the sequence alignment, we divided the IFD into three regions, termed IFD-a, -b and -c (see Supplementary Figures S1B and S4). Our human IFD-b mutant (790-VVIE-793-4A) causes a phenotype similar to motif 3, CTE and the hTR template mutants, confirming that IFD-b is indeed required for processivity (Figure 5A, lanes 33–36).
The TEN N95A mutant had a higher Km value of 0.16 µM to the 18 nt tel18 primer, presumably due to a reduced binding affinity to the 5′-end single-strand region of the longer DNA primer (Figure 5C). This supports that the TEN domain is a major contributor for the overall substrate affinity as proposed previously. The strong correlation between the processivity and the Km to the tel8 primer of the motif 3, IFD and CTE mutants ascertain that these elements contribute to the formation or positioning of an extendable RNA/DNA duplex substrate in the active site.
Hyperactive telomerase mutants have higher enzyme turnover rates
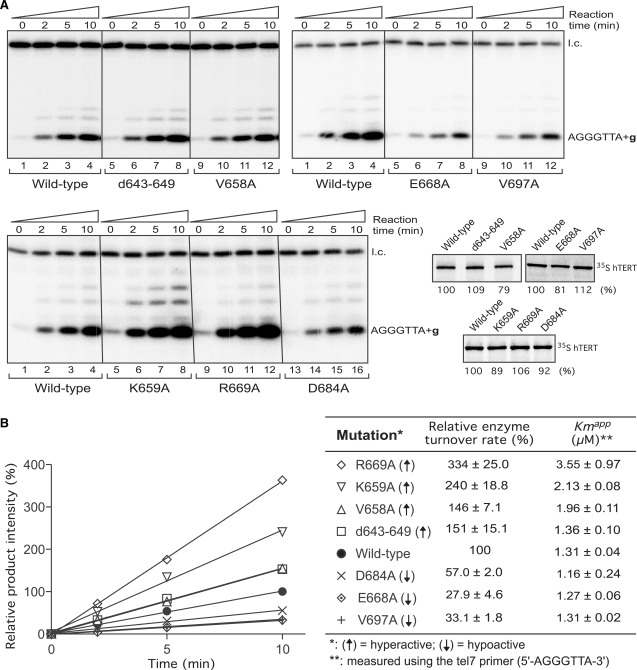
Since template translocation is the rate-limiting step in the processive telomerase reactions, the increased repeat addition rate observed with the hyperactive mutants should result from a greater translocation rate. Strand-separation between the telomeric DNA and the template RNA is a crucial step of template translocation. Taking advantage of the short primer assay, we asked if the hyperactive mutants have a faster dissociation rate for extended telomere product from the template, which resembles the strand-separation step of template translocation. Since the tel8 primer still gave rise to multiple repeats products, indicating successful translocation events, we thus used an even shorter tel7 DNA primer, 5′-AGGGTTA-3′ (7-nt) and only dGTP in the reaction to prevent any possible realignment of the telomeric DNA product with the RNA template after one round of repeat synthesis. Without the interference from TEN domain or other DNA binding sites, this time-course assay focused primarily on the rate of product dissociation from the active site.
Our results from this time-course analysis indicated that all hyperactive mutants d643-649, V658A, K659A and R669A have higher enzyme turnover rates than the wild-type enzyme (Figure 6). The higher turnover rates of these mutants were not due to higher substrate binding affinity, as these hyperactive mutants had a Km higher or similar to that of the wild-type enzyme (Figure 6B) and the reactions were performed at a saturated substrate concentration of 10 µM (see ‘Materials and Methods’ section). The high Km values of the hyperactive mutants K659A and R669A were consistent with their low processivity as shown above (Figures 4 and 6B). Conversely, the hypoactive mutants E668A, D684A and V697A that showed lower repeat addition and template translocation rates (Figure 4) had lower enzyme turnover rates (Figure 6B). Together, these results suggest that the increased template translocation rates of the hyperactive motif 3 mutants are likely due to the higher dissociation rates of products from the active site after each round of repeat synthesis.
Figure 6.
Enzyme turnover rates of the hyperactive and hypoactive motif 3 mutants. (A) Telomerase activity time course analysis of the hyperactive (d643–649, V658A, K659A and R669A) and hypoactive (E668A, D684A and V697A) mutant telomerases. The reactions were performed using a 7-nt telomere primer 5′-AGGGTTA-3′ and incubated for various amounts of time (0, 2, 5 and 10 min) as indicated. The reactions were carried out in the presence of only 32P-dGTP nucleotide to prevent processive reactions. A 32P-end-labeled 15-nt DNA oligonucleotide is used as the loading control (l.c.). The in vitro synthesized TERT proteins (wild-type, d643–649, V658A, K659A, R669A, D684A, E668A and V697A) were labeled with [35S]-methionine and quantitated after SDS–PAGE. (B) Quantitation of enzyme turnover rates of the hyperactive and hypoactive telomerase mutants. For each telomerase enzyme, the intensity of products was adjusted with protein amount and normalized by the intensity of loading control. For each set of reactions, the product intensities are further normalized to that of the wild-type reaction at the 10 min time point. The relative product intensities were then plotted against the amount of time. Wild-type (filled circle); d643–649 mutant (square); V658A (triangle); K659A (reverse triangle); R669A (diamond); D684A (cross); E668A (dotted diamond); V697A (plus). The relative enzyme turnover rates were determined from slopes of the linear trend lines in relation to that of the wild-type enzyme. The apparent Km values of different telomerase mutants toward the tel7 primer were determined by fitting the data to Michaelis–Menten equation. The standard deviation was derived from three independent experiments.
DISCUSSION
The repeat addition processivity of telomerase relies on a unique template translocation mechanism that presumably requires novel structural elements within the TERT protein. The telomerase-specific motif 3 we characterized in this study has been overlooked in the past, in part due to the low degree of sequence conservation among eukaryotic lineages, the presence of variable linkers and the inefficiency of alignment algorithms (Figure 1 and Supplementary Figure S2). In this study, through a comprehensive mutagenesis analysis within motif 3 (Figure 2), we identified mutations that unusually increased the rate or altered the processivity of telomere repeat addition (Figures 3 and 4). By using a novel short-primer assay to determine the binding affinity of the mutants to short DNA primers (Figure 5) and the time-course analysis to measure enzyme turnover rates (Figure 6), we showed that motif 3 mutations affect repeat addition rate and processivity, suggesting a crucial role for motif 3 in strand-separation and realignment during template translocation.
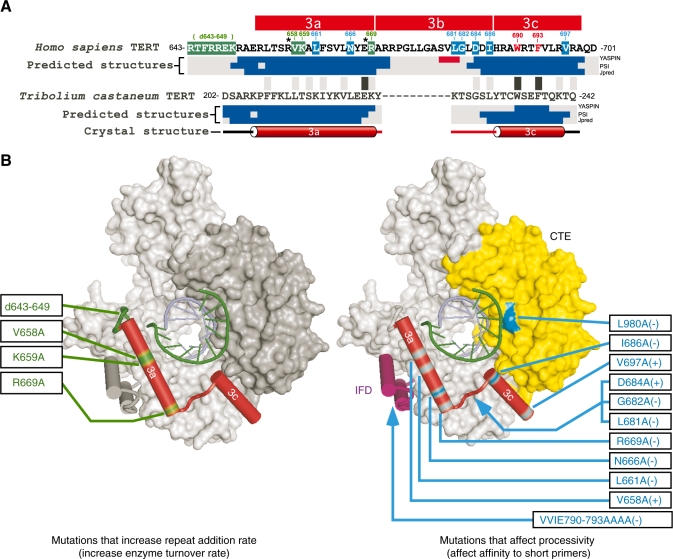
Our sequence alignment analysis and secondary-structure prediction on motif 3 provide useful insights into the function and evolution of this motif. The secondary structures of motif 3 predicted from different TERT homologs are surprisingly conserved (Supplementary Figure S1A) and consistent with the crystal structure of Tribolium castaneum TERT (34). The secondary-structure prediction of TERT sequences from all available species suggests that the motif 3 region consists of two α-helices separated by a conserved linker (Supplementary Figure S1A and data not shown), consistent with the Tribolium crystal structure. To display the physical location of motif 3 in relation to other TERT domains and the RNA/DNA duplex, we mapped the sequence of the two putative α-helices of human motif 3 onto the crystal structure Tribolium TERT based on the structural and sequence homology. To specifically denote the different structural features, we divided motif 3 into three sub-motifs, 3a, 3b and 3c, where 3a and 3c designate the two separate α-helices and 3b designates the spanning linker (Figure 7A). While the sequences of helices 3a and 3c are well conserved in most organisms, the sequence of linker 3b is conserved most specifically within vertebrates, non-yeast fungi, plants and ciliates (Figures 1B and 7A). This group-specific sequence conservation of linker 3b suggests a role important for telomerase function in most species, yet dispensable and lost in species including nematodes, insects and yeasts (Supplementary Figure S2).
Figure 7.
Homologous locations of human TERT mutations on the Tribolium TERT structure. (A) Sequence alignment and predicted secondary structures of human and Tribolium motif 3. Residues critical for repeat addition rate are shaded in green, while critical residues for processivity are shaded in blue; asterisk indicates both. The residues that abolish activity when mutated are colored red. The secondary structure (α-helix shown as a cylinder) based on the crystal structure of Tribolium TERT is shown below the predicted secondary structures. The predicted secondary structures (α-helix shown in blue and β-sheet shown in red) based on three algorithms: YASPIN, PSI and JPred (see Supplementary Figure S1A). Black/grey boxes located between human and Tribolium sequences indicate identity/similarity. The alignment is based on optimal positioning within the predicted helix sequence. (B) Mutations in human TERT that affect repeat addition rate and processivity are mapped onto the crystal structure of Tribolium TERT. Left panel: mutations that increase repeat addition rate are located in helix 3a and its N-terminal linker that connects motif 2 to helix 3a. Right panel: mutations that affect processivity and RNA/DNA duplex formation are dispersed in the IFD, motif 3 and the CTE. The blue arrowheads indicate putative locations for the human TERT sequences (IFD-b and motif 3b) absent from the Tribolium TERT. The (+) and (−) following the mutations denote an increase or decrease in processivity. Superimposed hetero-duplex of RNA strand (green) and DNA strand (blue).
Our comprehensive mutagenesis surveyed the functional effects of alanine-substitution at individual residues of motif 3. Although alanine-substitution at most of the highly conserved residues resulted in significant changes in telomerase activity or processivity, mutations at the residues V664A, L676A and S679A showed no dramatic changes (Figure 2B). It was however expected that alanine substitutions do not always give the same degree of effects for all conserved residues, due to unique structural and chemical properties within various amino acids. Substitutions to amino acids other than alanine will presumably produce different phenotypes.
The mutations (L681A, G682A and I686A) that severely impair processivity are located in linker 3b (Figures 2B and 7B, right), suggesting a primary role for this linker in regulating the template translocation efficiency and repeat addition processivity. In comparison, mutations (d643–649, V658A, K659A and R669A) that significantly increase telomerase activity are located in the helix 3a and its N-terminal linker (Figure 7B, left), suggesting that helix 3a is more important in regulating the rate of template translocation and repeat addition. However, helix 3a might play an additional role in telomerase processivity as several mutations (V658A, L661A, N666A, E668A and R669A) in motif 3a also substantially altered processivity.
The role of motif 3 on repeat addition rate
The three mutations in motif 3a, V658A, K659A and R669A, and the deletion d643–649 in N-terminal linker of motif 3a remarkably increased the repeat addition rate (Figure 4), presumably due to an increase in template translocation rate. This increase in repeat addition rate is independent of the processivity, as some hyperactive mutants have decreased processivity (Figure 4). The combination of a high addition-rate mutation with a low-processivity mutation did not reduce the repeat addition rate of the enzyme (data not shown), supporting that the rate and processivity of telomerase are regulated separately, as previously proposed (27).
The higher enzyme turnover rates measured in the non-processive short primer assay suggest faster product dissociation (or strand-separation) for the motif 3 hyperactive mutants (Figure 6), assuming that the product dissociation is the rate-limiting step. This is consistent with the observation of the hyperactive mutants showing faster template translocation in the processive pulse-chase assay, in which the translocation is the rate-limiting step (Figure 4). We propose that the putative helix 3a and its N-terminal linker regulate the strand-separation step of template translocation and thus modulate the rate of repeat addition. It remains unclear if the strand-separation of the RNA/DNA hybrid involves a conformational change to helix 3a or its N-terminal linker. We hypothesize that the long N-terminal linker could function in allowing helix 3a to swing away from the active site, permitting the RNA/DNA duplex to dissociate from the active site and the two strands to separate from each other (Figure 7B). A source of energy for such a conformational change could originate from the movement and distortion of DNA/RNA duplex during repeat synthesis as previously proposed (42).
The role of motif 3 on repeat addition processivity
Mutations at conserved residues within motif 3, the CTE and the IFD of TERT as well as the template region of TR affected telomerase processivity and the ability to use short telomere primers (Figures 5 and 6). The retained ability of the wild-type enzyme to extend the short tel7 and tel8 primers indicates that the catalytic core of TERT protein alone is capable of promoting the formation of, or recognizing, the RNA/DNA duplex substrate inside the active site independent of the TEN domain. The ability of telomerase to use the short primer correlates to the processivity of repeat addition as it resembles the second step of the translocation event, where the RNA and DNA realign to form the hetero-duplex inside the active site for the next round of repeat synthesis. The fact that the TEN N95A mutant can efficiently elongate the tel8 primer is consistent with the notion that this TEN mutation impairs the binding to the upstream single-stranded region of telomeric DNA primer, representing a different mechanism to affect telomerase processivity.
It was however unexpected that the wild-type enzyme would be capable of adding more than one telomere repeats to the 8-nt DNA primer in the absence of upstream single-stranded sequence for TEN binding (Figure 5A, lane 1). The synthesis of multiple repeats indicates the occurrence of template translocation after the synthesis of the first repeat. Although the tel8 primer does not initially leave a single-stranded overhang when base-paired with the RNA template, it would potentially have the 5′-end unpaired from the template during cycles of nucleotide addition, while maintaining a constant number of base-pairings between the telomeric DNA and the template RNA, as previously proposed (43). Since the TEN domain requires a longer single-stranded DNA overhang for binding, a more adjacent DNA binding site (the template-proximal anchor site) in the RT domain would thus seem responsible for binding the partially unpaired 5′-end of the short tel8 DNA primer (41,44).
Our results suggest that motif 3, CTE and IFD contribute to the realignment of telomeric DNA and the RNA template, i.e. the reformation of RNA/DNA duplex. In support of this notion, the crystal structure of Tribolium TERT shows that these three motifs are located adjacent to the RNA/DNA duplex, forming a horseshoe shaped structure encircling the duplex (Figure 7B). The majority of the motif 3 mutations that severely impaired the enzyme’s processivity and short primer usage are located in linker 3b (Figure 7). We envision the conserved motif 3b would act as a molecular hinge, positioning helix 3a and the CTE to facilitate the RNA/DNA duplex formation or positioning the duplex within the active site (Figure 7B). Since motif 3b is not conserved in insects, a crystal structure of a vertebrate or ciliate TERT would be necessary to elucidate the structural and functional purpose of motif 3b in template translocation.
Our phylogenetic and biochemical studies of motif 3 shed light upon the molecular mechanism of the translocation process for the processive telomerase reaction. The implication of our data provides testable hypotheses and elicits critical questions for future studies of telomerase action. Moreover, our hyperactive and hyper-processive motif 3 mutants demonstrate the feasibility of enhancing telomerase enzymatic activity through motif 3 targeting. Altering telomerase function can possibly affect the proliferative capacity of adult stem cells. Drugs that augment enzymatic activity and processivity of telomerase, similar to our mutants, might provide treatments for patients suffering from telomerase-insufficiency diseases. Additionally, the abated telomerase motif 3 mutants provide potential drug target locals for anti-cancer therapies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (CAREER award MCB0642857 to J.J.-L.C.). Funding for open access charges: National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Blake Atkinson and Drs Jim Allen, Carol Greider and Juan Alfonzo for critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Shawi M, Autexier C. Telomerase, senescence and ageing. Mech. Ageing Dev. 2008;129:3–10. doi: 10.1016/j.mad.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Armanios MY, Chen J.JL, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips J.A., 3rd, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 3.Walne AJ, Dokal I. Dyskeratosis Congenita: a historical perspective. Mech. Ageing Dev. 2008;129:48–59. doi: 10.1016/j.mad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 6.Osanai M, Kojima KK, Futahashi R, Yaguchi S, Fujiwara H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle) Gene. 2006;376:281–289. doi: 10.1016/j.gene.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 7.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greider CW. Telomerase is processive. Mol. Cell. Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 10.Xie M, Mosig A, Qi X, Li Y, Stadler PF, Chen J.JL. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J. Biol. Chem. 2008;283:2049–2059. doi: 10.1074/jbc.M708032200. [DOI] [PubMed] [Google Scholar]
- 11.Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 12.Prowse KR, Avilion AA, Greider CW. Identification of a nonprocessive telomerase activity from mouse cells. Proc. Natl Acad. Sci. USA. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lue NF, Peng Y. Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25:4331–4337. doi: 10.1093/nar/25.21.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosoy D, Lue NF. Yeast telomerase is capable of limited repeat addition processivity. Nucleic Acids Res. 2004;32:93–101. doi: 10.1093/nar/gkg943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 16.Aigner S, Cech TR. The Euplotes telomerase subunit p43 stimulates enzymatic activity and processivity in vitro. RNA. 2004;10:1108–1118. doi: 10.1261/rna.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Lopez-Guajardo CC, Quada J, Hurley LH, Von Hoff DD. Regulation of catalytic activity and processivity of human telomerase. Biochemistry. 1999;38:4037–4044. doi: 10.1021/bi982249n. [DOI] [PubMed] [Google Scholar]
- 18.Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 19.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat. Struct. Mol. Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lue NF, Lin YC, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol. Cell. Biol. 2003;23:8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryan TM, Goodrich KJ, Cech TR. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 2000;275:24199–24207. doi: 10.1074/jbc.M003246200. [DOI] [PubMed] [Google Scholar]
- 22.Huard S, Moriarty TJ, Autexier C. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 2003;31:4059–4070. doi: 10.1093/nar/gkg437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavory G, Farrow M, Balasubramanian S. Minimum length requirement of the alignment domain of human telomerase RNA to sustain catalytic activity in vitro. Nucleic Acids Res. 2002;30:4470–4480. doi: 10.1093/nar/gkf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriarty TJ, Marie-Egyptienne DT, Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drosopoulos WC, Direnzo R, Prasad VR. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J. Biol. Chem. 2005;280:32801–32810. doi: 10.1074/jbc.M506319200. [DOI] [PubMed] [Google Scholar]
- 28.Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen J.JL. The telomerase database. Nucleic Acids Res. 2008;36:D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge L, Rudolph P. Simultaneous introduction of multiple mutations using overlap extension PCR. Biotechniques. 1997;22:28–30. doi: 10.2144/97221bm03. [DOI] [PubMed] [Google Scholar]
- 30.Alder JK, Chen J.JL, Lancaster L, Danoff S, Su SC, Cogan JD, Vulto I, Xie M, Qi X, Tuder RM, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc. Natl Acad. Sci. USA. 2008;105:13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 32.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Yates JA, Chen J.JL. Identification and characterization of sea squirt telomerase reverse transcriptase. Gene. 2007;400:16–24. doi: 10.1016/j.gene.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 35.Malik HS, Eickbush TH. The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol. Biol. Evol. 1998;15:1123–1134. doi: 10.1093/oxfordjournals.molbev.a026020. [DOI] [PubMed] [Google Scholar]
- 36.Zimmerly S, Hausner G, Xc W. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–1250. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arkhipova IR. Distribution and phylogeny of Penelope-like elements in eukaryotes. Syst. Biol. 2006;55:875–885. doi: 10.1080/10635150601077683. [DOI] [PubMed] [Google Scholar]
- 38.Liang J, Yagasaki H, Kamachi Y, Hama A, Matsumoto K, Kato K, Kudo K, Kojima S. Mutations in telomerase catalytic protein in Japanese children with aplastic anemia. Haematologica. 2006;91:656–658. [PubMed] [Google Scholar]
- 39.Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 40.Moriarty TJ, Ward RJ, Taboski MA, Autexier C. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell. 2005;16:3152–3161. doi: 10.1091/mbc.E05-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyatt HD, Lobb DA, Beattie TL. Characterization of physical and functional anchor site interactions in human telomerase. Mol. Cell. Biol. 2007;27:3226–3240. doi: 10.1128/MCB.02368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greider CW. In: Telomeres. Blackburn EH, Greider CW, editors. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1995. pp. 35–68. [Google Scholar]
- 43.Forstemann K, Lingner J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 2005;6:361–366. doi: 10.1038/sj.embor.7400374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finger SN, Bryan TM. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 2008;36:1260–1272. doi: 10.1093/nar/gkm866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.