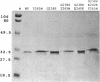
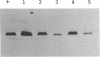
Abstract
TEM beta-lactamase variants with the amino acid substitutions R164S, E104K, G238S, and E240K (ABL numbering) display increased activity toward extended-spectrum cephalosporins. The T265M substitution is frequently found to be associated with the above substitutions in extended-spectrum beta-lactamases. However, the residue is located away from the active site in the three-dimensional structure and has been assumed to have no effect on catalysis. To examine the effect of the substitution on the structure and function of TEM beta-lactamase we constructed the following mutants: G238S, T265M, T265M:G238S, and T265M:G238S:E240K. Each enzyme was purified to homogeneity and the kinetic parameters kcat, Km and kcat/Km were determined for cefotaxime, ceftazidime, cephaloridine, and ampicillin. The results indicate that the T265M mutation has little effect on hydrolysis. In addition, we used immunoblotting to show that the substitution has little or no effect on the in vivo steady-state levels of beta-lactamase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Coulson A. F., Frère J. M., Ghuysen J. M., Joris B., Forsman M., Levesque R. C., Tiraby G., Waley S. G. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991 May 15;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 1, 2a, 2b, and 2b'. Antimicrob Agents Chemother. 1989 Mar;33(3):264–270. doi: 10.1128/aac.33.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Singer S. B. Biochemical characteristics of extended broad spectrum beta-lactamases. Infection. 1989 Nov-Dec;17(6):429–433. doi: 10.1007/BF01645566. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A., Medeiros A. A. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991 Sep;35(9):1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsch C., Mourey L., Masson J. M., Samama J. P. Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution. Proteins. 1993 Aug;16(4):364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mabilat C., Courvalin P. Development of "oligotyping" for characterization and molecular epidemiology of TEM beta-lactamases in members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1990 Nov;34(11):2210–2216. doi: 10.1128/aac.34.11.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros A. A. Beta-lactamases. Br Med Bull. 1984 Jan;40(1):18–27. doi: 10.1093/oxfordjournals.bmb.a071942. [DOI] [PubMed] [Google Scholar]
- Naumovski L., Quinn J. P., Miyashiro D., Patel M., Bush K., Singer S. B., Graves D., Palzkill T., Arvin A. M. Outbreak of ceftazidime resistance due to a novel extended-spectrum beta-lactamase in isolates from cancer patients. Antimicrob Agents Chemother. 1992 Sep;36(9):1991–1996. doi: 10.1128/aac.36.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965 Sep;240(9):3685–3692. [PubMed] [Google Scholar]
- Palzkill T., Botstein D. Probing beta-lactamase structure and function using random replacement mutagenesis. Proteins. 1992 Sep;14(1):29–44. doi: 10.1002/prot.340140106. [DOI] [PubMed] [Google Scholar]
- Palzkill T., Le Q. Q., Venkatachalam K. V., LaRocco M., Ocera H. Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of beta-lactamase. Mol Microbiol. 1994 Apr;12(2):217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]
- Sougakoff W., Petit A., Goussard S., Sirot D., Bure A., Courvalin P. Characterization of the plasmid genes blaT-4 and blaT-5 which encode the broad-spectrum beta-lactamases TEM-4 and TEM-5 in enterobacteriaceae. Gene. 1989 May 30;78(2):339–348. doi: 10.1016/0378-1119(89)90236-9. [DOI] [PubMed] [Google Scholar]
- Urban C., Meyer K. S., Mariano N., Rahal J. J., Flamm R., Rasmussen B. A., Bush K. Identification of TEM-26 beta-lactamase responsible for a major outbreak of ceftazidime-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother. 1994 Feb;38(2):392–395. doi: 10.1128/aac.38.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]