Abstract
The mTOR inhibitor everolimus (RAD001, Afinitor) is an orally active anticancer agent. Everolimus demonstrates growth-inhibitory activity against a broad range of tumor cell histotypes in vitro and has the capacity to retard tumor growth in preclinical tumor models in vivo through mechanisms directed against both the tumor cell and the solid tumor stroma components. These properties have rendered it to be a clinically active drug, with subsequent registration in renal cell carcinoma (Motzer et al. [2008]. Lancet 372, 449–456) as well as showing strong potential as a combination partner (André F et al. [2008]. J Clin Oncol 26. Abstract 1003). Although everolimus has a high specificity for its molecular target, the ubiquitous nature of mTOR and the multifactorial influence that mTOR signaling has on cell physiology have made studies difficult on the identification and validation of a biomarker set to predict and monitor drug sensitivity for clinical use. In this review, a summary of the preclinical and clinical data relevant to biomarker development for everolimus is presented, and the advantages and problems of current biomarkers are reviewed. In addition, alternative approaches to biomarker development are proposed on the basis of examples of a combination of markers and functional noninvasive imaging. In particular, we show how basal levels of pAKT and pS6 together could, in principle, be used to stratify patients for likely response to an mTOR inhibitor.
Introduction
Afinitor (generic name everolimus, also known as RAD001) is an orally active derivative of rapamycin that inhibits the Ser/Thr kinase, mTOR. This mTOR inhibitor (mTORi) was recently approved by the Food and Drug Administration and European Medicines Agency for treatment of renal cell carcinoma (RCC) after failure of vascular endothelial growth factor receptor (VEGF-R)-targetted therapy including the multikinase inhibitors sorafenib and sunitinib. Everolimus is also currently in a number of phase 2/3 clinical trials in many different cancer indications for its utility alone and in combination with other chemotherapeutics. Recently published studies have evaluated treatment regimens of everolimus in the clinic and monitored the activity of everolimus using various markers related to mTOR inhibition and cellular proliferation rates. Preclinical evaluation of these biomarkers, and pharmacokinetic/pharmacodynamic (PK/PD) modeling of preclinical in vivo studies, provided the basis for their clinical application and successfully predicted clinically useful treatment regimens for everolimus after the principle of determining optimal biologic regimens. In this report, we briefly review the mTOR literature to date, with an emphasis on everolimus, and explore alternatives to the further optimization of translational biomarkers that could be applied.
Mechanism of Action
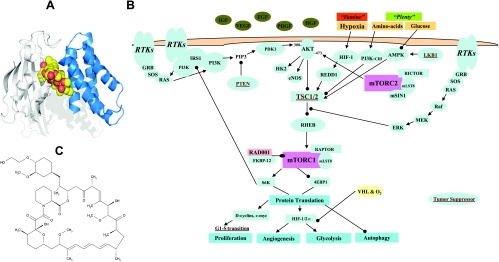
The target of the rapamycin class of agents is mTOR, a multifunctional signal transducing protein, which obtains signals from many upstream pathways and propagates the information through the regulation of multiple downstream pathways [1]. Rapamycins function to block mTOR activity by interacting with the immunophilin FKBP-12 (Figure 1A), which forms an inhibitory complex that binds with high affinity to mTOR. The signal transduction that occurs downstream from mTOR is rapamycin-sensitive and occurs through an mTOR-Raptor complex, known as TORC1. In addition, there is a TORC2 rapamycin-insensitive pathway when mTOR is complexed with Rictor (Figure 1B), which is also known as PDK2. However, besides serving a key role in normal cell physiology, mTOR has been implicated in cancer [2,3], and consequently, inhibition of this target has received considerable attention as an anticancer approach, leading to the clinical development of rapamycin analogs (rapalogs) with improved druglike properties such as everolimus [4,5].
Figure 1.
Mechanism of action and structure of everolimus. (A) Everolimus (yellow-red) binds to the FK506-binding protein-12 immunophilin, FKBP-12 (white), and this complex interacts with the FRB domain (blue) of the large protein mTOR. (B) PI3K/AKT/mTOR pathway: the FKBP-12-RAD001 complex inhibits mTORC1 function. 4E-BP1 indicates eIF-4E (eukaryotic initiation factor 4E) binding protein 1; HIF-1, hypoxia-inducible factor 1 or 2; mTORC1/2, mammalian target of rapamycin complex 1 or 2; PI3K, phosphoinositide 3-kinase; PTEN, phosphatase and tensin homologue, dual-specificity lipid/protein phosphatase; S6K, 40S ribosomal S6 kinase; TSC1/2, tuberous sclerosis tumor suppressor genes; VHL, von Hippel-Lindau protein. Tumor suppressors are in brown and underlined. (C) Molecular structure of everolimus (RAD001).
The primary downstream targets of mTORC1 include eIF-4E-binding protein (4E-BP1) and S6 kinase 1 (p70S6K1), i.e., S6K. On mitogen-induced multisite phosphorylation, 4E-BP1 releases eIF-4E allowing cap-dependent translation of several mRNA encoding critical regulators of G1-phase progression. S6K functions in G1, through phosphorylation of the 40S ribosomal protein S6, to increase the translation of mRNA that largely encode ribosomal proteins and other elements of the translational machinery. Thus, through inhibition of mTOR function, rapalogs block these essential translational events, resulting in the inhibition of G1 progression (Figure 1B). Interestingly, inhibition of mTOR by rapalogs seems to predominantly signal through S6K, resulting in increased G1 block and decreased hypoxia-inducible factor (HIF) translation rather than through c-myc [6]. Furthermore, mTOR apparently resides in different subcellular locations and has several macromolecular structure(s) due to differential binding/association of other proteins. In congruence, some of the activities of mTOR seem to vary with different cell types [2], and consequently, the exact activities of rapalogs may also vary depending on the cell type. Rapalogs also possess antiangiogenic/antivascular activities [7–9]. The exact mechanism of action in endothelial cells, therefore, may be different than that occurring in tumor cells: for example, in addition to suppressing the effects of HIF-1 mediated endothelial cell proliferation, rapamycin can block CD40-mediated VEGF release and endothelial cell proliferation [10].
Chemical Structure, Properties, and Metabolism of Everolimus
Everolimus is a macrolide lactone, and the structural formula is shown in Figure 1C. Everolimus is a good substrate for the multidrug-resistant pump P-gp. After incubating with liver microsomes in vitro [11], everolimus is metabolized to more than 20 different metabolites, which is almost exclusively owing to the activity of the P450 enzyme CYP3A4 [12]. A similar metabolite pattern was seen in animals and humans after dosing with radioactive everolimus (unpublished observations). In humans, 24 hours after an oral dose of 3 mg, 40% of everolimus was unchanged; there were five major metabolites (each 4%-13%), and the other minor metabolites comprised 18%. The major metabolites showed little or no antiproliferative activity against activated lymphocytes or human tumor cells in vitro, with IC50 values more than 100-fold greater than everolimus (unpublished observations).
In experimental tumor models, everolimus is very well tolerated with no obvious clinical signs of toxicity and normally leads to slight body weight gains in comparison to vehicle-treated mice. The maximum tolerated dosage was not reached even when treating for up to 60 mg/kg per day by oral gavage.
In Vitro Preclinical Pharmacology of Everolimus as a Single Agent
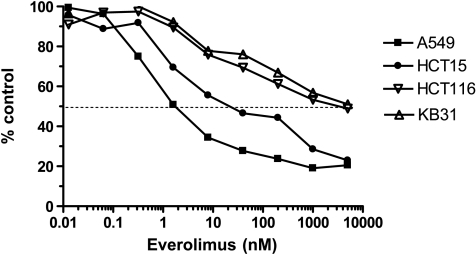
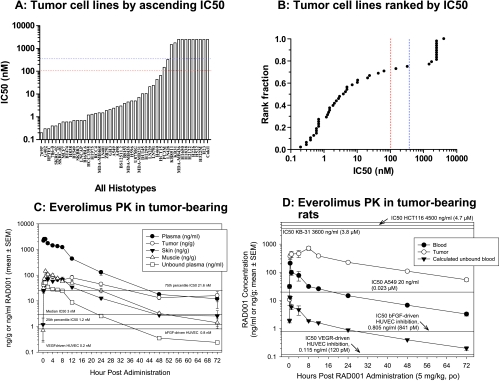
As a single agent, everolimus inhibits the in vitro growth of a wide variety of mammalian tumor cell lines. As for rapamycin, the dose-response curve for inhibition of proliferation is, for most cell lines, rather broad (Figure 2), although there are exceptions. In general, 100% cell kill is not seen, and the inflection point, which can be equivalent to the IC50, is often broad. Consequently, the IC50 represents the concentration that reduces cell proliferation (compared with control) by 50%, and if a 50% reduction is not reached, then no IC50 can be measured. Using this approach, everolimus produced a range of IC50 values that was independent of histotype in the 48 different cell lines tested (Figure 3A). A break point is apparent at ca. 100 nM, and indeed 71% (0.71 in Figure 3B) of the cell lines had an IC50 less than this (actually ≤65 nM) or 75% (IC50 actually ≤327 nM) using 350 nM as the cutoff (blue dotted line). This arbitrary approach allowed a classification of the cell lines into “sensitive” and “insensitive.” However, the PK of everolimus in mice supports this cutoff because the Cmax of the compound is 107 nM in subcutaneously grown human tumor xenografts after a single oral dose of 5 mg/kg (Figure 3C), and everolimus is eliminated from mouse plasma fairly rapidly with a half-life of ca. 5 hours. The PK of everolimus in rats is rather different (Figure 3D), and there is a much higher Cmax of 809 nM. Nevertheless, if the cell lines were grown in nude rats, 25% would still be well below the IC50.
Figure 2.
In vitro activity of everolimus: examples of IC50 determination. Results show the dose-dependent inhibition of cell growth by everolimus as a percentage of the control determined using methylene blue staining after 96 hours of incubation in four different human tumor cell lines, which can be regarded as sensitive (HCT-15, A549) and insensitive (KB-31 and HCT-116).
Figure 3.
In vitro activity of everolimus against mammalian cell lines and PK in mice and rats. Everolimus in vitro activity in 48 different cell lines assessed by IC50 values in ascending values (A) and ranked (B), showing cutoffs at 100 nM (red) or 350 nM (blue). Everolimus (RAD001) was administered once at 5 mg/kg per os (p.o.) to (C) female BALB/c athymic nude mice bearing s.c. KB-31 tumors or (D) Lewis rats bearing s.c. CA20498 tumors. Plasma/blood, tumors, and tissues were obtained at various time points after administration, and everolimus levels were determined using high-performance liquid chromatography/mass spectrometry. Data are expressed as mean ± SEM, n = 4. Horizontal lines represent the in vitro IC50 values shown in panels A and B, and the IC50 values for endothelial cells are from Lane et al. [9].
Consequently, on the basis of the PK of everolimus and individual IC50 values, tumor cells have been described in general as either sensitive or insensitive. However, isolated cells of both categories display inhibition of elements downstream from mTOR, notably reduction of pS6 and production of many bands of 4E-BP1 representing different stages of hypophosphorylation [9]. This suggests that decreases in pS6 and/or p4E-BP1 are markers of the target being hit by rapalogs but do not necessarily predict response. Interestingly, for everolimus, continuous exposure in vitro is not required for protracted activity against sensitive cell lines. For example, treatment of the sensitive cell line human lung A549 with a brief exposure of everolimus (20 nM for 30 minutes), followed by washout into compound-free cell culture medium, resulted in prolonged inactivation of the S6K [13]. Only 48 hours after removal of everolimus from the A549 cell culture did S6K activity start to recover. Furthermore, 96 to 120 hours after release, S6K1 activity did not reach vehicle control-treated levels (0.14–0.5 of vehicle controls).
In Vivo Preclinical Pharmacology of Everolimus as a Single Agent
PK of Everolimus in Tumor-Bearing Mice and Rats
There are key differences in the PK profile between mice and rats [14]. In particular, there is greater partitioning of everolimus into erythrocytes in rats (60%) compared with just 2% in mice but greater plasma protein binding in mice (99.9%) compared with rats (92%). Furthermore, the plasma/blood half-life of everolimus is just 4 to 8 hours in mice but 17 to 20 hours in rats (Figure 3, C and D). These PK differences have a major impact on the levels of unbound everolimus that also is free in the plasma (not partitioned in red blood cells) of both rats and mice. Consequently, applying the data in Figure 3, C and D, shows that the free Cmax in both species is rather similar at ca. 1 nM with an AUC0–24 hours of 5 to 8 nM. However, there are also important differences in the volume of distribution and tissue/tumor penetration. Modeling of tumor and blood/plasma PK suggested that in mice, multiple daily administrations result in a ∼2-fold increase in tumor levels of everolimus at steady state, whereas in rats, a ∼7.9-fold increase would occur. In contrast, weekly high-dose regimens were predicted to not facilitate tumor accumulation in either species. Interestingly, total tumor levels of everolimus (Cmax and AUC) were four- to eight-fold greater in rats than in mice, which could be partially influenced by the more than two-fold greater plasma content and permeability of rat tumors compared with mouse tumors [13]. Overall, the PK of everolimus in rats was much more similar to that in humans than mouse to humans. Finally, in rodents, brain penetration of everolimus was poor but was dose-dependent and showed overproportional uptake in rats, with a longer half-life compared with the systemic circulation. These data imply that high intermittent doses may be more beneficial for brain tumors than lower daily doses.
In Vivo Activity of Everolimus in Tumor-Bearing Mice
In Vivo Target-Directed PD in Mice
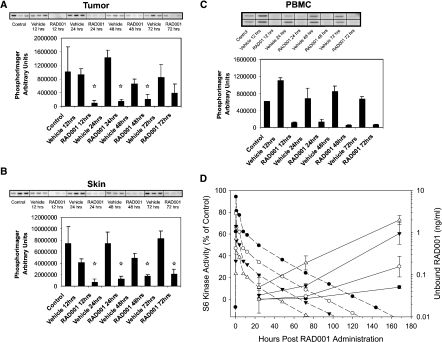
As already described, everolimus has been demonstrated to inhibit the phosphorylation and activity of the downstream mTOR-regulated S6K in tumor cells, and this also has been shown in tumors and surrogate tissues in both mice and rats [13]. Single administrations of everolimus reduced the levels of S6 in both tumors and skin in a dose-dependent manner, giving similar ED50 (effective dose reducing tumor volume by 50%) values (0.8–1.0 mg/kg) but were slightly higher than the ED50 of everolimus for antitumor effect against KB-31 tumors (0.32 ± 0.04).Overall, the data suggested that antitumor effects seemed to correlate with inactivation of the mTOR target S6K1 in treated tumors at the higher doses but not necessarily at lower doses, suggesting that a further antitumor mechanism may also exist such as antivascular activity (see below). Strikingly, everolimus-induced effects on skin-derived S6K activity were comparable with those obtained in tumor material derived from the same mice, suggesting that skin sampling in this context could provide a useful surrogate to tumor sampling. Furthermore, assuming a linear response between the measured data, there seemed to be an effect of concentration and time on the recovery rate of S6K activity in pericytes peripheral blood mononucleocytes (PBMCs).
Evaluation of Everolimus in Mouse Models of Cancer
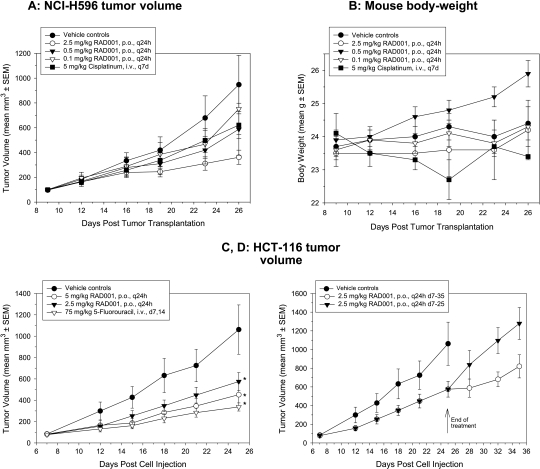
Everolimus is orally active in both mice and rats, producing an antitumor effect that is characterized by dramatic reduction in tumor growth rates as opposed to producing tumor regressions, although regressions can occur, for example, in subcutaneous A549 lung tumor xenografts, a cell line classed as sensitive in vitro. On the basis of data generated in Novartis Laboratories, Table 1 provides a detailed summary of the activity of everolimus against subcutaneous tumor xenografts in immunodeficient nude mice. Daily treatment (2.5–10 mg/kg) is well tolerated and can produce antitumor effects similar to conventional cytotoxic agents such as paclitaxel, 5-fluorouracil (5-FU), and doxorubicin but without the associated body weight losses (Figure 4). Strikingly, activity can also be observed in xenografts created from insensitive cell lines (i.e., HCT-116, KB-31, and KB-8511; Table 1). Indeed, when the IC50 values of 17 cell lines were compared with the maximal activity achievable in vivo, there was no significant correlation (r = 0.25, P = .34 [9]), and there was no correlation between in vitro IC50 and in vivo ED50 in seven cell line/tumors [14]. These data, in addition to several different models of angiogenesis both in vitro and in vivo, demonstrated a likely antiangiogenic/antivascular activity of everolimus, which would contribute to its activity in vivo [9]. Note that the IC50 for everolimus was essentially the same for both the low and high P-gp-expressing cell lines KB-31 and KB-8511 (Table 1), suggesting that P-gp may play little role in the sensitivity of cells to everolimus.
Table 1.
Anticancer Activity of Everolimus as a Single Agent In Vitro and in Athymic Nude Mice In Vivo.
| In Vitro | In Vivo | |||||
| Tumor Origin | Cell Line | IC50 (nM) | Optimal Treatment Regimen | Tumor Response | Host Response | |
| T/C | % Body Weight Change (Controls) | |||||
| Sensitive in vitro | Lung | A549 | 2.4* | Everolimus: 2.5 mg/kg per day | -0.41† | 1 ± 1 (6 ± 2†) |
| Doxorubicin: 9 mg/kg i.v., once every 7 days | 0.49 | -18 ± 1† | ||||
| NCI-H520 | 42.6* | Everolimus: 5 mg/kg per day | 0.14† | 4 ± 1 (6 ± 2) | ||
| Cisplatin: 5 mg/kg i.v., once every 7 days | 0.34† | -6 ± 2† | ||||
| NCI-H596 | 5 ± 2 | Everolimus: 5 mg/kg per day | 0.18† | 10 ± 1† (9 ± 1†) | ||
| Paclitaxel: 15 mg/kg i.v., three times per week | -0.33† | -32 | ||||
| Melanoma | B16/BL6* | 0.7 ± 0.2 | Everolimus: 5 mg/kg per day | 0.24† | 9 ± 1† (-0.3 ± 3) | |
| Insensitive in vitro | Colon | HCT-116 | 4125 ± 1853 | Everolimus: 5 mg/kg per day | 0.50 | -6 ± 3 (7 ± 5) |
| 5-FU: 75 mg/kg i.v., once every 7 days | 0.30† | -13 ± 4 | ||||
| Epidermoid | KB-31 | 1778 ± 800 | Everolimus: 2.5 mg/kg per day | 0.26† | 6 ± 1† (5 ± 2†) | |
| Doxorubicin: 9 mg/kg i.v., once every 7 days | 0.30† | -8 ± 2† | ||||
| KB-8511 | 1489 ± 806 | Everolimus: 5 mg/kg per day | 0.17† | 2 ± 1 (6 ± 3†) | ||
| Paclitaxel: 15 mg/kg i.v., three times per week | 0.71 | -2 ± 5 | ||||
In vitro IC50 determinations. Each cell line was added to 96-well plates (1000–3000 cells/well in 100 µl of medium) and incubated for 24 hours. Subsequently, everolimus was added in a two-fold dilution series, and the cells were reincubated for 3 days. Methylene blue staining was performed on day 4, and the amount of bound dye (proportional to the number of surviving cells that bind the dye) was measured. IC50 values were determined using the SoftmaxPro program. In vivo evaluations used s.c. xenografts of human tumor cells in nude mice and also the murine B16/BL6 cell line growing intradermally in the ear as a syngeneic orthotopic model.
T/C: mean increase of tumor volumes of treated animals divided by the mean increase of tumor volumes of control animals.
Percent body weight change is (g body weight at end of experiment - g body weight at beginning / g body weight at beginning) expressed as a percentage.
i.v. indicates intravenous.
Mean of duplicate determinations; all others mean ± SEM of n = 4–7 determinations.
P < .05 versus controls, but for tumor volume changes, the statistical significance used . tumor volumes (tumor volume at end of experiment - tumor volume at beginning).
Figure 4.
Efficacy and tolerability of everolimus in mice bearing s.c. human tumor xenografts. (A and B) When tumors reached 100 mm3, everolimus was administered daily at 0.1, 0.5, or 2.5 mg/kg p.o. to mice bearing NCI-H596 human lung tumor xenografts. Alternatively, mice received 5 mg/kg cisplatin intravenously once perweek. Results show the mean ± SEM (n = 8) for tumor volume(A) and bodyweight (B). Only the 2.5-mg/kg per day regimen of everolimus produced a statistically significant reduction in tumor volume (Dunnett's test vs vehicle controls). (C and D) When HCT-116 tumors reached 100 mm3, everolimus was administered daily at 2.5 or 5 mg/kg from day 7 to day 25 (C) or (D) until day 35 in comparison to only day 7 to day 25. Alternatively, mice received 75 mg/kg 5-FU once per week. Results show the mean ± SEM (n = 7) for tumor volume, where *P < .05 versus controls by Dunnett's one-way ANOVA.
Consistent with the fact that the target needs to be consistently hit, intermittent treatment with everolimus of mice bearing human colon HCT-116 tumors reduced effectiveness. Furthermore, on cessation of treatment, tumors quickly regained growth rates similar to those of the vehicle control (Figure 4, C and D).
In general, similar observations have been described in the literature for everolimus. Thus, although regression from monotherapy is seldom seen, strong growth inhibition was observed in models of ovarian cancer [7,15], colon cancer [8,16], prostate cancer with bone metastases [17], leiomyosarcoma [18], gastrointestinal stromal tumors [19], endometrial cancer [20], breast cancer [21], lymphoma [22], leukemia [23], and thyroid cancer [24]. Activity normally matched that of conventional agents and was increased in combination with cytotoxics such as cisplatin [7,15] or paclitaxel [17] but also with other targeted agents such as gefitinib [16], imatinib [19], and zoledronate [17] and also ionizing radiation [8]. These preclinical observations are now being verified in the clinic, for example, in the combination of everolimus with trastuzumab and paclitaxel in breast cancer [25].
In Vivo Activity of Everolimus in Tumor-Bearing Rats
Everolimus has been extensively characterized with respect to dose and scheduling using a syngeneic pancreatic tumor, CA20498, grown subcutaneously in Lewis rats. The everolimus-dependent reduction in S6 levels in rat CA20498 tumors, skin, and peripheral circulating leukocytes was extensively studied [13] and modeled [26], based in part on a previously developed PK model to predict and interpret clinical trial data.
In the rat CA20498 model, daily treatment with everolimus (0.5 or 2.5 mg/kg) dose-dependently inhibited growth, and intermittent dosing using a higher dose of 5 mg/kg (once or twice per week) also showed similar antitumor efficacy [13]. Inhibition by everolimus was characterized by sustained suppression rather than regression and was not associated with any body weight loss.
A single 5-mg/kg dose of everolimus (optimal for antitumor efficacy) caused a dramatic reduction in S6 phosphorylation after 24 hours. In contrast, a similar reduction was not observed in skin and PBMC extracts because these tissues exhibited no detectable S6 phosphorylation at baseline, which may reflect the relatively low proliferative index of these tissues compared with the highly proliferative CA20498 tumors.
In the tumors, significant inhibition of S6K1 activity was maintained for up to 48 hours after everolimus treatment. At this time, S6K1 activity started to recover, but levels were still reduced compared with vehicle controls at 72 hours (Figure 5). In comparison, S6K1 derived from skin samples taken from the same rats remained significantly inhibited for up to 72 hours. These data indicate that everolimus results in prolonged inhibition of S6K1 in tumors and skin derived from tumor-bearing rats, providing mechanistic support for the observed antitumor potency of intermittent dosing schedules used in this rat model and suggesting the potential of skin sampling as a source of material for biomarker evaluation in the clinic.
Figure 5.
Duration (A, B, C) and recovery (D) of the effects of everolimus on S6K1 activity in CA20498 tumors, rat skin, and PBMCs after a single dose (5 mg/kg). Lewis rats bearing CA20498 tumors were treated with a single dose of 5 mg/kg everolimus or vehicle. At the times indicated, tumor (A) and skin (B) extracts from three rats were prepared separately, and PBMC (C) extracts were prepared from pooled blood samples. p70S6K1 activity was measured using 40S ribosomal subunits as an in vitro substrate. In each case, autoradiographs of [32P]phosphate incorporation into S6 protein are shown. Graphs represent PhosphorImager quantification of the autoradiographs, where *P < .05 versus untreated controls (Dunnett's test); from Boulay et al. [13]. (D) S6 kinase activity was determined in circulating PMBCs from rats treated with 0.5, 1, 2, and 5 mg/kg everolimus once per day. The PK profiles were based on a single administration of everolimus at 5 mg/kg, and the everolimus concentrations obtained with doses were estimated assuming a linear PK profile. Inspection of the data suggests that 0.03 ng/ml blood seems to be a minimal value below which recovery of S6K activity begins. Back extrapolation of the S6 phosphorylation curves suggests that, by the first measured time point, the recovery process of S6K has already begun with the 0.5mg/kg dose, with the estimated rate of recovery being 0.425, and the estimated blood everolimus (RAD001) level was approximately 0.05 ng/ml. However, considering the other dose groups, the recovery of S6K activity seems to occur when the drug level is approximately 0.03 ng/ml. The recovery after 1, 2, or 5 mg/kg everolimus began at 24, 72, and approximately 125 hours after everolimus administration, respectively. The S6K recovery rate was 0.408, 0.284, and 0.244 for 1, 2, and 5 mg/kg everolimus. The time where blood levels drop below 0.03 ng/kg everolimus was correlated to the S6K recovery rate (setting the initiation of recovery at 12 hours for the 0.5-mg/kg dose) (r = -0.968, P = .0316). The total amount of S6K inhibition (AUC) was correlated to the estimated everolimus AUC at each dose (r = 0.96, P = .038) and the everolimus AUC was also correlated to the rate of S6K recovery (r = 0.95, P = .046).
A similar but stronger reduction in S6K1 was seen in PBMCs isolated 2 and 12 hours after administration, which was maintained for up to 72 hours (Figure 5). Furthermore, an experiment investigating the duration of effect of a single suboptimal (0.5 mg/kg) versus an optimal (5 mg/kg) antitumor dose of everolimus indicated that the time required for the recovery of S6K1 was dose-dependent. Thus, only efficacious doses of everolimus result in sustained, significant S6K1 inactivation for up to 7 days after administration (Figure 5D). These data suggested a correlation between prolonged S6K1 inactivation in PBMCs and optimal everolimus doses, eliciting significant antitumor responses with intermittent (weekly) treatment schedules; a hypothesis confirmed by a more detailed analysis presented by Boulay et al. [13]. Hence, blood sampling could provide a source of material for biomarker evaluation in the clinic.
Rapamycin is known to promote 4E-BP1 dephosphorylation, resulting in the repressive association of 4E-BP1 with the eIF-4E translational activator [27]. Everolimus in vitro also promotes 4E-BP1 dephosphorylation in a number of cell lines, and consequently, a similar analysis to that described above for S6K1 was performed. In all tissues, reductions in threonine-70 4E-BP1 phosphorylation were observed after everolimus treatment, and this correlated with changes in the mobility of the 4E-BP1 protein (particularly striking in PBMCs) and a trend for increased association with eIF-4E. These data demonstrated activation of 4E-BP1 as a translational repressor in tissue derived from everolimus treated tumor-bearing animals (unpublished observations).
Development of a PK/PD Model Using S6K Activity in PBMC as a Biomarker for Everolimus
Detection of S6K1 Activity in Human PBMCs
To assess the potential of using mTOR effectors as biomarkers to evaluate everolimus dosing schedules, basal S6K1 activity was measured in human PBMC extracts obtained from healthy volunteers [13]. Using EDTA as an anticoagulant, a coefficient of variation (for PBMC preparation and kinase assay) of just 10% was obtained, indicating good reproducibility of preparation. Treatment of healthy volunteer blood with 2 nM everolimus for 30 minutes diminished S6K1 activity compared with vehicle controls (44% and 63% inhibition in donors 1 and 2, respectively). Furthermore, increasing everolimus concentrations led to almost complete inactivation of S6K1 (≥95% inhibition with ≥20 nM everolimus). In contrast, no 4E-BP1 phosphorylation could be detected in human PBMCs, and because 4E-BP1 is not an enzyme, a quantitative assay is not readily available. Consequently, unlike S6K1, 4E-BP1 protein may not be applicable as a biomarker for monitoring everolimus-specific effects in human PBMCs.
Everolimus is a targeted, highly specific agent with an IC50 for binding to isolated FKBP-12, or FKBP-12 complexed to mTOR of 5 to 6 nM, and no significant activity against other protein kinases (D. Fabbro, PhD, T. Meyer, PhD, and H.A. Lane, PhD, unpublished observations). Thus, this affords the opportunity to administer a dosage regimen that provides a sufficient drug level to inhibit a biologic end point, providing an optimal biologic dose (OBD) and, consequently, avoiding the adverse events likely to be associated with dosing dictated by the maximum tolerated dose. With the information described above, a PK/PD model, which could be used to guide clinical administration regimens to reduce the time for initial clinical trials by aiming for an OBD and regimen, was sought [26]. PK/PD models associate dose-concentration relationships (PK) with concentration-effect (PD) to provide a predictive model for the time course of drug effects, which can be used to optimize clinical regimens. The key assumption of this work is that a clinically effective everolimus administration regimen should provide a degree and duration of S6K1 inhibition in PBMC and tumor tissues that has been associated with an antitumor effect, as was observed in rats. Rat data were also used because everolimus PK in this species resembles that of humans much more than that of mice [14].
Modeling of the rat data (blood and tumor PK, S6K inhibition in tumor, and PBMC) indicated that the everolimus dosage regimen had a greater influence on tumor S6K activity than on PBMC S6K activity, with daily everolimus dosing giving more complete and sustained inhibition of S6K than once-weekly administration [26]. Additional modeling of rat (and mouse) PK also suggested that significant tumor accumulation of everolimus of approximately eight-fold in rats and two-fold in mice would occur with daily dosing but none with weekly dosing [14]. Extending the model to include data from cancer patients [28] indicated that, other than correcting for the differences in PK properties of everolimus between humans and rats, the model also accurately fits the inhibition of S6K1 in the PBMC of cancer patients. This provided evidence that there is little difference between tumor-bearing rats and cancer patients regarding the concentration effect of everolimus and its PD effect on signal transduction proteins. However, it is important to note that this may not be the case for brain tumors, where weekly dosing may be preferred, because everolimus only significantly accumulated in rat brain at higher doses [14].
PD Studies with the Rapalog Temsirolimus
A similar set of studies in PBMC was performed with temsirolimus (i.e., CCI-779) [29]. These studies demonstrated a linear dose response for rapamycin-dependent inhibition of S6K in lymphoid cells and reduced S6K activity in the PBMCs and subcutaneous (s.c.) tumors of treated mice. Furthermore, in PBMCs from human volunteers, S6K activity was stable for 8 days incubated ex vivo, although substantial variation in the levels occurred. Lastly, in PBMCs taken from a small group of advanced renal cancer patients treated with temsirolimus, there was a strong impairment of PMBC S6K activity that was protracted and showed a linear relationship between the extent of decrease in S6K activity and time to treatment failure. Overall, these studies support the use of PBMC S6K activity as a marker for the effects of rapalog-based therapy for cancer.
Clinical Pharmacology and Efficacy of Everolimus as a Single Agent
The PK/PD model described [26] was used and further refined in a phase 1 study [28], with the PD marker being PBMC S6K1 activities. The study continued with a safety evaluation of everolimus, with a secondary evaluation being efficacy. The PK of everolimus in cancer patients (n = 4–6 per dose) with a once-weekly administration demonstrated minimal plasma accumulation, and steady state was obtained after two treatments. TheCmax at steady state increased in a dose-dependent manner at 5, 10, and 20 mg and was less than dose-proportional at 30, 50, and 70 mg; AUC, however, remained dose-proportional. The elimination half-life was approximately 30 hours at all doses.
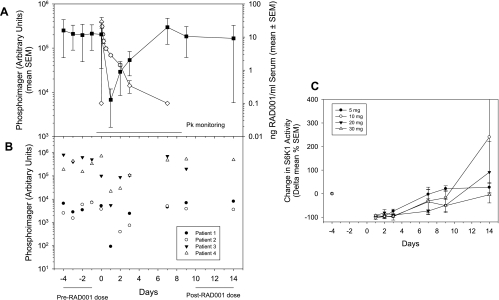
The PBMC sampling strategy used several samples before treatment to establish a baseline of S6K1 activity. An example of the data obtained from single doses of everolimus is presented in Figure 6B, which shows that the interpatient and intrapatient PBMC S6K1 levels varied considerably. However, the levels of PBMC S6K1 were uniformly dramatically reduced during the time of high blood levels of everolimus and increased again when everolimus levels dropped below ca. 2 ng/ml (i.e., 2 nM; Figure 6A). This recovery of S6K1 was apparently partially dose-dependent (Figure 6C). Continuous suppression of pS6 levels in PBMC was achieved with doses greater than 20 mg (once weekly) and at both 5- and 10-mg doses when administered once daily. These clinical data are in congruence with the predictions from the PK/PD model.
Figure 6.
Data in panels (A) and (B) are from the patient group dosed with 5 mg of everolimus as described by O'Donnell et al. [28]. (A) Everolimus blood PK (open symbols) with the PD readout of S6K activity (closed symbols) in circulating PBMCs from treated patients. (B) Time-dependent PBMC S6K levels in four individual patients. (C) Recovery of S6K acitivity in PBMC from patients receiving various doses (5–30 mg) of everolimus [28].
In general, the safety profile in this phase 1 trial was considered good (92 patients), with only two patients showing dose-limiting tolerability issues. Thus, the maximum tolerated dose was not reached, although almost every patient had some adverse event related to drug dosing. Furthermore, partial responses were seen in four patients, and 12 patients were progression-free for greater than 6 months, including 5 of 10 patients with RCCs. Overall, it was concluded that on the basis of these PK and PD readouts, the minimum doses of 5 mg/d or 20 mg/week should be used in further clinical development [28].
A concomitant phase 1 study also studied dose and schedule with PK and more extensive PD readouts [30]. They determined pretreatment and on-treatment steady-state tumor and skin biopsies for a variety of markers (both total and phosphorylated state) using immunohistochemistry (IHC) for S6K1 elF-4E, 4E-BP1, p4E-BP1-Thr70, eIF-4G, AKT, and Ki-67 expression. During the first 4 weeks of treatment, irrespective of the administration regimen, plasma trough levels of everolimus were determined once per week before dosing.
Trough levels of everolimus were, respectively, 8.5 and 17 ng/ml for 5 and 10 mg administered daily, whereas weekly administration of 20, 50, or 70 mg produced trough levels of 0.1, 1.0, and 4.2 ng/ml, respectively. There was almost complete and sustained inhibition of pS6 and p-eIF-4G in both tumor and skin at the trough concentration of 10 mg daily and more than 50 mg weekly. Strangely, this did not occur with the 5-mg daily dosage, although trough levels were higher than the 50- and 70-mg weekly regimens. Significant inverse correlations occurred between the daily dose of everolimus and p-eIF-4G and pS6 levels, but sustained reduction occurred only at 50- and 70-mg doses. Although not observed in all patients, everolimus caused dramatic reductions in skin p-4E-BP1, but the effect on tumor was less profound; however, when inhibition occurred, it was sustained. There was an increase in pAKT (Ser473) in tumor and skin in 50% of the patients, and in some cases, this was sustained. Cellular proliferation, as indicated by Ki-67 nuclear staining, was variably reduced in both tumor and skin by everolimus treatment. An analysis linking trough levels of everolimus with the PD readouts indicated a significant correlation between PK and p-4E-BP1, a trend for a correlation with p-eIF-4G, but no correlation between pAKT and trough levels; the near-complete reduction of pS6 at all doses prohibited any correlation analyses. Importantly, there was no change in the total levels of S6, 4E-BP1, eIF-4G, or AKT, suggesting that the observed changes in phosphorylated levels of these proteins were due to the inhibition of mTOR-dependent signal transduction and protein activation as opposed to the changes in protein levels.
Comparing the two schedules, mTOR pathway inhibition was more profound (and maintained) with the daily schedule. Although PBMC samples were not analyzed in this clinical study [30], the PD effects of everolimus on pS6 levels in patients' tumor and skin paralleled the observations made in CA20498 pancreatic tumor-bearing rats [13]; this effect was also observed using KB-31 tumor-bearing nude mice (A. Boulay, PhD, T. O'Reilly, PhD, and H.A. Lane, PhD, unpublished observations). Thus, in congruence with the PK/PD modeling of animal data [26], the clinical data indicated a difference in the PD effects caused by everolimus depending on the regimen, with more sustained inhibition of mTOR signaling components with daily or high-dose once-weekly dosing, whereas lower doses given once weekly could not achieve durable inhibition of downstream target effectors.
As in the clinical study of O'Donnell, all patients in the Tabernero study showed some adverse event, but only 5 of 55 patients demonstrated dose-limiting toxicities. Four patients demonstrated clinical benefit (all receiving weekly dosages of 20–70 mg weekly), including one partial response in a heavily pretreated patient with metastatic colorectal cancer treated who received 20 mg of everolimus once weekly. Three patients showed disease stabilization for more than 5 months, including one with advanced renal cell cancer and two with advanced breast cancer [30].
These initial clinical responses in the phase 1 studies described, plus impressive response rates in another phase 2 study for RCC only recently reported [31], encouraged a phase 3 registration trial in RCC, known as RECORD-1 [32]. An update of this trial showed an impressive extension of progression-free survival from 1.9 months in the placebo arm to 4.9 to 5.5 months in the everolimus arm, despite a relatively low (partial) response rate of just 2% [33]. Such data emphasize that, for a cytostatic agent such as everolimus, clinical benefit may not necessarily be detected using RECIST criteria but rather may require suitable biomarkers for early detection of response.
Limitations of Currently Used Biomarkers for Rapalogs
Biomarkers are objectively determined indicators of normal or pathologic biologic processes or a response to a pharmacological agent [34]. They are categorized as markers of 1) natural history (type 0), indicating disease progression or resolution, independent of any particular therapy, and are often used for patient stratification in clinical trials; 2) drug activity (type 1), indicating response to drug therapy that can be used in optimizing therapeutic intervention; or 3) surrogate (type 2), which is intended to substitute for clinical end points.
S6K activity is elevated in many cancer cells, but this seems to be a consequence of alterations at other points in the PI3K/AKT/mTOR pathway, as opposed to being tumorigenic in itself. Furthermore, there is little information that would securely suggest that reduction of S6K activity would be directly indicative of a clinical outcome. Accordingly, S6K activity used as a biomarker would be classified at present as a type 1 marker; however, it cannot be considered a specific marker form TOR inhibition or for the detection of the “sensitivity” toward rapamycin derivatives because both “sensitive” and “insensitive” cells demonstrated reduction of S6K activity, as described above [9]. However, S6K may indeed be a possible marker for clinical outcome because overexpression of S6K has been shown to be related to increased in vitro sensitivity toward rapamycins in a series of breast cancer cell lines [35,36] and because higher levels of S6K expression are associated with increased local recurrence in breast cancer and lower levels are associated with reduced disease-free interval in both breast cancer [37] and renal cancer patients [29].
Although clearly proven useful in determining optimal monotherapy regimens for clinical evaluation of everolimus, use of S6K1 activity as a type 1 biomarker will be difficult in the situations in combination with other anticancer agents. This is because cytotoxic agents can modulate components of the mTOR signaling pathway, e.g., etoposide, mitomycin C, and cisplatin all reduce the activity of S6K1, with concomitant reduction in the phosphorylation status [38]. Reduction of the phosphorylation of 4E-BP1 also occurred and increased its binding to eIF-4E, which would prevent the binding of eIF-4E to the 5′-cap region of mRNA to allow translation. Conversely, paclitaxel causes the phosphorylation of S6K at Thr421 and Ser424 in breast and ovarian cell lines [39]. Furthermore, hypophosphorylated S6 protein seems to be a mediator of TRAIL-associated apoptosis induction, which occurs with a variety of agents including tumor necrosis factor, cycloheximide, etoposide, doxorubicin, tunicamycin, and staurosporine [40]. Consequently, the use of these markers as indicators of the activity of rapalogs when used in combination with cytotoxic agents would be complicated. Therefore, unless new markers of sensitivity to the combination partner can be developed, use of “target-independent” tumor response measurement methods (e.g., noninvasive imaging) may be required (see below).
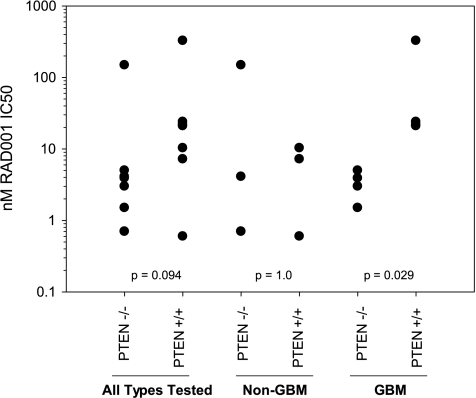
Alternatively, these markers could be used as type 0 biomarkers. Thus, basal levels of markers of mTOR pathway could, however, still be useful in determining the potential sensitivity of the tumor, especially if several different markers are combined. A recent report studying 13 different human tumor cell lines showed a significant correlation between basal levels of pAKT (Ser473) and sensitivity to everolimus as measured by IC50 values (r = 0.73) and similar levels of correlation between pGSK3 (Ser9) or pTSC2 (Thr1462) and IC50 [41]. PTEN status was not examined in this report, whose loss of function has been thought to render cells more dependent on the mTOR signaling cascade and, accordingly, more sensitive to mTOR inhibition. However, our observations suggest that PTEN status does not predict well the in vitro sensitivity to everolimus with the exception of glioblastoma multiforme cell lines (Figure 7). However, PTEN status was used in clinical trials of the rapalog temsirolimus. Glioma and melanoma are often PTEN-mutated, but clinical trials of temsirolimus showed a low activity [42]. In contrast, endometrial cancer (also often PTEN-mutated) patients demonstrated a much better outcome when treated with temsirolimus. Although many explanations could be offered, one proposal [42] is that the time of PTEN mutation (late in melanoma and glioma; early in endometrial) may dictate sensitivity to the inhibition of mTORC1-independent pathways by rapalogs.
Figure 7.
Role of PTEN in sensitivity of tumor cell lines in vitro to everolimus. Results show the individual IC50 values for different human tumor cell lines defined as glioblastoma multiforme (GBM) or not, according to presence of PTEN mutation (P values from a 2-tailed t test).
Finally, other potential biomarkers should also be considered as potential stratifiers that are not strictly part of the PI3K/AKT/mTOR pathway. For example, it was reported that loss of the tumor-suppressor protein merlin and activation of mTORC1 are correlated in mesothelioma and that merlin-negative lines are sensitive to the growth-inhibitory effect of rapamycin, suggesting that loss of merlin may be a marker for rapalog sensitivity in mesothelioma [43]. Also, those functions further downstream of control of protein translation such as the effectors themselves such as HIF-1 and the many proteins that this transcription factor regulates could also be stratifiers. Thus, rates of glycolysis and the degree of vascularization maybe factors governing overall response to rapalogs, although this has not yet been reported (also discussed below).
Combinations of Biomarkers
Single markers for stratification of the population, i.e., preselection of the patients most likely to respond to chemotherapy are ideal but may not suffice, and thus, the use of combinations of biomarkers may be more appropriate. It has long been known that use of several markers increases sensitivity and specificity of the prediction of therapeutic outcome [44]. Using a total of 7 markers (6 in urine and 1 in plasma), both as absolute values and as ratios, stepwise multiple regression techniques demonstrated that a maximal of 15 markers (measured before and after treatment and marker ratios) was required to obtain a maximal predictive power of therapy for breast carcinoma patients; a 2.4-fold improvement over the best single marker.
As an example of what might be achieved in the clinic using a combination of biomarkers, we have applied a number of different statistical approaches to a data set of 21 cell lines of diverse tumor origins with 1) known sensitivity to everolimus (IC50) and 2) quantified levels (Western blot with quantitation of light-emitting paper-bound immune complexes) of several of the members of the AKT/mTOR pathway (total S6 levels, p235-S6, p240-S6, pEIF4, Rictor, Raptor, total AKT, and pAKT). In addition, the ratios of pS6/total S6, pAKT/AKT, and Rictor/Raptor were determined (M. Klopfenstein, BSc, M. Breuleux, PhD, and H.A. Lane, PhD, unpublished observations).
Univariate correlation analyses (Table 2) demonstrated significant correlations between sensitivity to everolimus with the markers total S6, p235-S6, total AKT, and pAKT by applying the Spearman rank correlation test. Note that different answers were obtained depending on whether IC50, log IC50, or rank IC50 were used, which seemed to be due to the lack of normal distribution and the extent of kurtosis. In the case of p235-S6/total S6, combining two markers as a ratio dramatically improved both the correlation coefficient and the statistical significance compared with use of a single marker. Perhaps interestingly, use of the pAKT/total AKT ratio was uncorrelated to everolimus sensitivity.
Table 2.
Univariate Correlation of Key mTOR Pathway Components and Sensitivity to Everolimus On the Basis of Rank IC50.
| Spearman Rank Correlation Coefficient | ||
| r | P | |
| pS6 240/244 | 0.06 | 0.80 |
| pS6 235/236 | 0.53 | 0.01 |
| Total S6 | -0.51 | 0.02 |
| pEIF4 | -0.06 | 0.80 |
| Raptor | -0.01 | 0.98 |
| Rictor | -0.16 | 0.50 |
| Total AKT | 0.37 | 0.10 |
| pAKT 473 | 0.66 | 0.001 |
| 235/total S6 | 0.76 | <0.0001 |
| 240/total S6 | 0.30 | 0.18 |
| 235 + 240/total S6 | 0.52 | 0.01 |
| Raptor/Rictor | 0.19 | 0.40 |
| pAKT/total AKT | 0.41 | 0.07 |
These markers were then used individually in linear regression models (Table 3). All models passed goodness-of-fit tests but varied dramatically in their regression parameters, with p235-S6/total S6 providing the best fit (r = 0.77), followed by pAKT (r = 0.69), p235-S6 (r = 0.51), total S6 (r = 0.48), and total AKT (r = 0.45). All of the measured data and their ratios were then combined in multiple regression to search for an improved predictive ability. Use of backward stepwise regression eliminated two of the eight measured markers, and their ratios on the basis of their lack of significance contribute to a regression function predicting rank IC50. The regression yielded a highly significant fit (r = 0.98, ANOVA P < .0001, each parameter fit P < .04; see Table 3). These data demonstrate that the use of multiple biomarkers can lead to dramatic improvement in predicting tumor cell sensitivity to everolimus. However, the cost and time required to measure all of the parameters maybe prohibitive. Use of just the ratio of p235-S6/total-S6 and pAKT (the two factors correlating best with rank IC50) still provided a significant fit (r = 0.79, ANOVA P = .0001), with the p235-S6/total-S6 ratio alone providing the major contribution (P = .0113) and pAKT providing much less (P = .14).
Table 3.
Univariate and Multiple Linear Regression Using Rank IC50 Values.
| Regressor | r | ANOVA Results, P |
| Univariate linear regression | ||
| Total S6 | 0.48 | .026 |
| p235–240 S6 | 0.51 | .017 |
| p235/total S6 | 0.77 | .001 |
| pAkt | 0.69 | .006 |
| Total AKT | 0.45 | .043 |
| pAKT/total AKT | 0.33 | .14 |
| Multivariate linear regression | ||
| 11 parameters* | 0.98 | <.0001 |
| p235/total S6 and pAKT | 0.79 | .0001 |
Backward stepwise regression selected the following parameters to be used in multiple least squares regression: total S6 levels, p235-S6, p240-S6, pEIF4, Rictor, Raptor, total AKT, pAKT, and the ratios p235-S6/total S6, pAKT/AKT, and Rictor/Raptor.
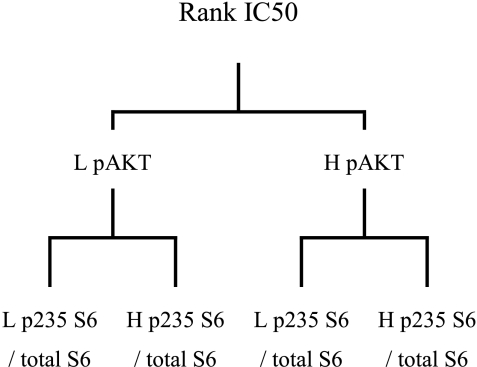
In clinical decision making, discreet criteria (e.g., high vs low) are preferable than continuous data, and an attempt was made to “convert” the continuous data of the present data set. Recursive partitioning is a technique that finds partitions of factors within a data set grouped by a particular response [45]. Use of recursive partitioning to select which markers best separated the cell lines when grouped according to rank IC50 indicated that pAKT and the ratio p235-S6/total S6 significantly segregated the 21 cell lines, with no other markers being identified (Figure 8). Recursive partitioning suffers from being sensitive to changes in the database, and particularly in the present case, a large database is required for obtaining accurate results [45,46]. Therefore, the results obtained with the present database must be considered preliminary in nature.
Figure 8.
Recursive partitioning of the sensitivity of the 21 tumor cell lines to everolimus activity in vitro. H, high level expression; L, low level expression.
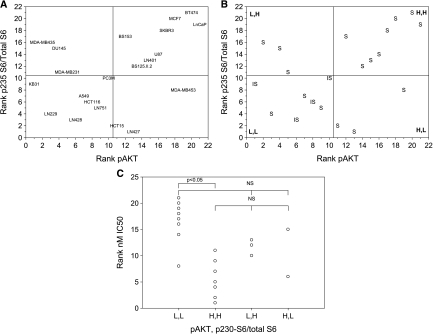
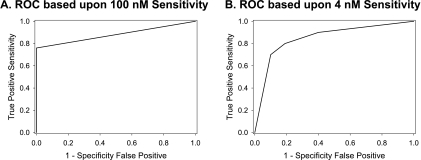
On the basis of this partitioning analysis, the cell lines were ranked according to expression levels of pAKT and p235-S6/total S6 and were plotted (Figure 9). Midpoints of the ranks were used to segregate the lines into four categories on the basis of expression levels of the two markers (Figure 9B). Finally, comparing the in vitro IC50 “cutoff” of 100 nM, which seems to segregate the cell lines into sensitive and insensitive groups (see above), largely matched the categories of marker expression level, with a high level expression of both pAKT and p235-S6/total S6 representing the most sensitive tumor cell line phenotype (Figure 9B). However, Figure 9C demonstrates that these categories indeed overlap with respect to everolimus sensitivity, with only the high pAKT and high p235-S6/total S6 and low pAKT and low p235-S6/total S6 categories being statistically different. The use of other “cutoff levels” for everolimus sensitivity, namely 20 and 4 nM (the approximate Cmax and Cmin, respectively, on the basis of the administration of 10 mg to cancer patients [28]) accordingly gave slightly different results. Lastly, nominal logistic regression was performed using everolimus sensitivity as the outcome categories (sensitive [S] or insensitive [IS]) and pAKT and p235-S6/total S6 expression as the regressor categories (high pAKT and high p235-S6/total S6, high pAKT and low p235-S6/total S6, low pAKT and high p235-S6/total S6, and low pAKT and low p235-S6/total S6). The models compared sensitivity criteria of IC50 < 100 nM (an apparently naturally occurring breakpoint) and IC50 < 4 nM (the Cmin unbound levels at steady state after 10-mg dosing [28]. With the 100-nM breakpoint, 13 cell lines were considered sensitive and 8 were insensitive. The regression produced an adequate fit (r2 = 0.46, χ2 P = .025, with Wald test for goodness-of-fit passed), providing the resulting receiver operating characteristic (ROC) curve (Figure 10A). The AUC of this curve (0.88) suggests that the use of these two parameters provides a good indicator of everolimus sensitivity [47].Using the categories high pAKT and high p235-S6/total S6 (a predictor of sensitivity) and low pAKT and low p235-S6/total S6 (a predictor of insensitivity), the sensitivity is 73% and the specificity is 100%. With IC50 < 4 nM as the sensitivity cutoff, 10 cell lines were considered sensitive and 11 were insensitive. The resulting model fitted well (r2 = 0.36, χ2 P = .015, Wald test passed, AUC = 0.86), and the ROC curve is presented in Figure 10B. At this lower cutoff, the sensitivity and specificity are 88% and 86%, respectively. It must be emphasized that with the low number of cell lines tested and few biomarkers analyzed, these results should be viewed as illustrative and not definitive.
Figure 9.
Segregation of 21 tumor cell lines according to levels (A) or categories (B, C) of p-AKT and the ratio of p235-S6/total S6. L,H, low pAKT expression and high p235 S6/total S6 expression; H,H, high pAKT expression and high p235 S6/total S6 expression; H,L, high pAKT expression and low p235 S6/total S6 expression; L,L, low pAKT expression and low p235 S6/total S6 expression. Sensitive (S): IC50 < 100 nM; insensitive (IS): IC50 > 100 nM.
Figure 10.
ROC curve of pAKT and p235-S6/total S6 levels as predictors of sensitivity of 21 tumor cell lines to everolimus. Nominal logistic regression was performed using sensitivity to everolimus at the 100 nM (A) and 4 nM (B) cutoff limits and pAKT and p235 S6/total S6 expression categories (see legend to Figure 9) and the ROCs derived from the regression are presented.
It could be envisioned that such an approach may individualize cancer chemotherapy with everolimus, if tumor biopsies could be obtained before treatment and assessed for pAKT and p235-S6/total S6 levels (using a suitable set of reference controls), and determine the expression category for each marker. One difficulty is the relation of IHC data (typically H-scores) with the continuous data provided by quantitative Western blot or ELISA used preclinically or in clinical trials. H-scores do not provide a unique number describing the marker expression level, and it will have to be clarified if a very high level marker expression by a few cells within a tumor is therapeutically equivalent to a low-level expression by most tumor cells; both scenarios could give similar H-scores. Once the PK properties of everolimus in that patient are determined, comparing the marker expression level with the achieved everolimus blood level will provide relevant decision-making criteria regarding the continuance of everolimus therapy and the likelihood of clinical success. However, this marker set cannot be used to monitor response to treatment because insensitive cells show a decrease in p235-S6 levels, which is not correlated to the eventual antitumor effect preclinically (see earlier section). Furthermore, if rapalogs are used in combination, these other agents can also induce significant effects on pAKT and pS6 (see earlier section).
Nevertheless, the use of initial levels of pAKT and pS6 does seem to have some predictive power for clinical outcome. For example, in RCC, an initial high-level expression of pAKT and pS6 correlated with outcome to treatment with the rapalog temsirolimus [48]. Also, in RCC, there was a positive and statistically significant correlation of pS6 levels, and a trend for an association with a high-level pAKT expression, with temsirolimus response [49]. In breast cancer, PTEN, pAKT, and pS6K1 levels were associated with rapamycin sensitivity, and pS6K1-positive tumors were associated with a worse prognosis compared with pS6K1-negative tumors [50]. Furthermore, pS6 expression has been linked to sensitivity to the rapalog AP23573 where a high-level pS6 expression (≥20% of tumor cells staining) was associated with a clinical response among metastatic sarcoma patients, whereas a low expression (<10% of tumor cells) was not [51].
Whatever molecular approach is used, there persists several important issues: 1) biopsies are not always readily obtained because of a) access and b) consent; 2) a biopsy is only ever a small sample from a very heterogeneous target tissue; and 3) in particular, for IHC, the samples need to be carefully preserved because they may degrade. Consequently, alternatives such as noninvasive imaging, which can sample the whole tissue, are potentially very attractive.
Alternatives to Molecular Stratification
Given the complexity of the mTOR pathway, and the issues described above, it may be that other generic nonmolecular approaches to determine tumor response before, or even soon after treatment, may also prove beneficial. In this respect, one should consider the possibilities of functional imaging using dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) or ultrasound (US) and positron emission tomography (PET). In principle, these methods can measure aspects of tumor biology relevant to the downstream effects of everolimus: DCE-MRI/US for antivascular/antiangiogenic effects and 18FDG- and 18FLT-PET for measuring effects on glycolysis and cell proliferation, respectively. At present, preclinical data using these methods are scarce, and there is even less in the clinic.
With respect to antivascular effects, DCE-US has shown that everolimus (5 mg/kg for 3 weeks) significantly decreased perfusion of tumors growing s.c. in rats, and this paralleled growth inhibition [52]. Similar reductions in tumor blood flow were observed using a Doppler method in C57BL/6 mice bearing GL261 tumors [53]. In contrast, DCE-MRI was unable to detect any significant changes in tumor vessel permeability (Ktrans) or blood volume in murine B16/BL6 melanoma and rat breast BN472 tumors [9]. The absence of an effect on Ktrans was consistent with the lack of effect on Evans Blue measurement of permeability in the B16/BL6 model, and this suggests that, although everolimus does have antiangiogenic activity, this is manifested differently in comparison to VEGF-R or PI3K inhibitors [54]. Everolimus has been reported to rapidly reduce both FDG and FLT uptake in xenograft models that are classified as sensitive but not in the insensitive models [55]. Similar data were shown for rapamycin in human tumor xenografts where reduced tracer uptake was linked to decreased activity of hexokinase for FDG and thymidine kinase 1 for FLT [56]. Everolimus was shown to dose-dependently reduce FDG uptake in a gastric xenograft model, with the effect showing a plateau at 5 to 15 mg/kg daily, consistent with effects on pS6 in the same model [57]. These effects also paralleled those on growth inhibition, and thus, this study emphasized the potential power of FDG-PET to obtain an OBD for clinical application. In contrast, the only full clinical report for the effect of rapalogs on imaging indicated that, although rapamycin reduced FDG uptake in 17 of 34 patients, 46% of these patients progressed, suggesting that the PET method was not of value in predicting response to rapamycin at the dose and schedule used in that study [58].
Lastly, it is possible that imaging could also be used for stratification, in particular, if one considers the antivascular/antiangiogenic characteristics of mTORi. For example, if the PI3K/AKT/mTOR is highly activated in a particular tumor, then this should lead to an increased expression of HIF-1 and, consequently, increased expression of VEGF and platelet-derived growth factor and increased glucose transport and glycolysis (Figure 1B). Thus, it may be that sensitive tumors have, at baseline, higher rates of glucose uptake and/or vascularity; the latter manifested as a higher vessel density or increased blood volume, permeability, or flow. These characteristics are all detectable by the different noninvasive imaging methods described, i.e., FDG-PET, DCE-MRI, and also arterial spin labeling (ASL) for blood flow. Indeed, in RCC clinical trials, the VEGF-R inhibitor sorafenib showed significantly increased activity in tumors with a higher permeability (Ktrans) than in those with a lower permeability [59]. Recently, a similar approach has been followed in experimental RCC models where tumor blood flow was measured by ASL at baseline and in response to sorafenib, which showed increased sensitivity in tumors with higher blood flow [60]. Thus, where imaging methods have been set up to monitor early response, retrospective analysis may indicate cutoffs in vascularity or glycolysis that can assign tumors into sensitive or insensitive groups, perhaps eventually providing the opportunity for stratification on the basis of noninvasive imaging.
Conclusions
Through a concentrated research effort acquiring a solid understanding of the molecular targets and affected pathways of everolimus (both in vitro and in vivo), along with a clear definition of more classic pharmacological aspects (dose-response, PK/PD relationship, therapeutic window, etc.), principles were obtained, which were used to formulate clinically testable questions that guided initial clinical trials. This endeavor reduced the clinical efforts needed to decide on the optimal dose for further testing, allowing a rapid progression from phase 1 trials to successful phase 3 trials. This effort also demonstrates that the appropriate use of animal models, on the basis of a thorough knowledge of the properties of the model and restricting the questions asked to those that the model is capable of answering, does indeed provide a predictive system from which to plan clinical trials.
Acknowledgments
O'Reilly and McSheehy are employees of Novartis Pharma AG. The authors thank M. Breuleux, D. Fabbro, M. Klopfenstein, H.A. Lane, and T. Mayer for access to unpublished data.
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer chemotherapy. Nat Rev Cancer. 2004;4:335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 3.Lane HA, Breuleux M. Optimal targeting of the mTORC1 kinase in human cancer. Curr Opin Cell Biol. 2009;21:219–229. doi: 10.1016/j.ceb.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol. 2008;8:393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Thomas GV, Tran C, Mellinghoff IK, Welsbie DS, Chan E, Fueger B, Czernin J, Sawyers CL. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 7.Mabuchi S, Altomare DA, Cheung M, Zhang L, Poulikakos PI, Hensley HH, Schilder RJ, Ozols RF, Testa JR. RAD001 inhibits human ovarian cancer cell proliferation, enhances cisplatin-induced apoptosis, and prolongs survival in an ovarian cancer model. Clin Cancer Res. 2007;13:4261–4270. doi: 10.1158/1078-0432.CCR-06-2770. [DOI] [PubMed] [Google Scholar]
- 8.Manegold PC, Paringer C, Kulka U, Krimmel K, Eichhorn ME, Wilkowski R, Jauch KW, Guba M, Bruns CJ. Antiangiogenic therapy with mammalian target of rapamycin inhibitor RAD001 (everolimus) increases radiosensitivity in solid cancer. Clin Cancer Res. 2008;14:892–900. doi: 10.1158/1078-0432.CCR-07-0955. [DOI] [PubMed] [Google Scholar]
- 9.Lane HA, Wood JM, McSheehy PM, Allegrini PR, Boulay A, Brueggen J, Littlewood-Evans A, Maira SM, Martiny-Baron G, Schnell CR, et al. mTOR inhibitor RAD001 (everolimus) has antiangiogenic/vascular properties distinct from a VEGFR tyrosine kinase inhibitor. Clin Cancer Res. 2009;15:1612–1622. doi: 10.1158/1078-0432.CCR-08-2057. [DOI] [PubMed] [Google Scholar]
- 10.Dormond O, Contreras AG, Meijer E, Datta D, Flynn E, Pal S, Briscoe DM. CD40-induced signaling in human endothelial cells results in mTORC2- and Akt-dependent expression of vascular endothelial growth factor in vitro and in vivo. J Immunol. 2008;181:8088–8095. doi: 10.4049/jimmunol.181.11.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boernsen KO, Egge-Jacobsen W, Inverardi B, Strom T, Streit F, Schiebel HM, Benet LZ, Christians U. Assessment and validation of the MS/MS fragmentation patterns of the macrolide immunosuppressant everolimus. J Mass Spectrom. 2007;42:793–802. doi: 10.1002/jms.1215. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn B, Jacobsen W, Christians U, Benet LZ, Kollman PA. Metabolism of sirolimus and its derivative everolimus by cytochrome P450 3A4: insights from docking, molecular dynamics, and quantum chemical calculations. J Med Chem. 2001;44:2027–2034. doi: 10.1021/jm010079y. [DOI] [PubMed] [Google Scholar]
- 13.Boulay A, Zumstein-Mecker S, Stephan C, Beuvink I, Zilbermann F, Haller R, Tobler S, Heusser C, O'Reilly T, Stolz B, et al. Antitumor efficacy of intermittent treatment schedules with the rapamycin derivative RAD001 correlates with prolonged inactivation of ribosomal protein S6 kinase 1 in peripheral blood mononuclear cells. Cancer Res. 2004;64:252–261. doi: 10.1158/0008-5472.can-3554-2. [DOI] [PubMed] [Google Scholar]
- 14.O'Reilly T, McSheehy P, Kawai R, Kretz O, McMahon L, Brueggen J, Bruelisauer A, Gschwind H, Allegrini P, Lane H. Comparative pharmacokinetics of RAD001 (everolimus) in normal and tumor-bearing rodents. Cancer Chemother Pharm. 2010;65:625–639. doi: 10.1007/s00280-009-1068-8. [DOI] [PubMed] [Google Scholar]
- 15.Mabuchi S, Altomare DA, Connolly DC, Klein-Szanto A, Litwin S, Hoelzle MK, Hensley HH, Hamilton TC, Testa JR. RAD001 (everolimus) delays tumor onset and progression in a transgenic mouse model of ovarian cancer. Cancer Res. 2007;67:2408–2413. doi: 10.1158/0008-5472.CAN-06-4490. [DOI] [PubMed] [Google Scholar]
- 16.Bianco R, Garofalo S, Rosa R, Damiano V, Gelardi T, Daniele G, Marciano R, Ciardiello F, Tortora G. Inhibition of mTOR pathway by everolimus cooperates with EGFR inhibitors in human tumours sensitive and resistant to anti-EGFR drugs. Br J Cancer. 2008;98:923–930. doi: 10.1038/sj.bjc.6604269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan TM, Pitts TE, Gross TS, Poliachik SL, Vessella RL, Corey E. RAD001 (everolimus) inhibits growth of prostate cancer in the bone and the inhibitory effects are increased by combination with docetaxel and zoledronic acid. Prostate. 2008;68:861–871. doi: 10.1002/pros.20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernando E, Charytonowicz E, Dudas ME, Menendez S, Matushansky I, Mills J, Socci ND, Behrendt N, Ma L, Maki RG, et al. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748–753. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- 19.Rossi F, Ehlers I, Agosti V, Socci ND, Viale A, Sommer G, Yozgat Y, Manova K, Antonescu CR, Besmer P. Oncogenic kit signaling and therapeutic intervention in a mouse model of gastrointestinal stromal tumor. Proc Natl Acad Sci USA. 2006;103:12843–12848. doi: 10.1073/pnas.0511076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milam MR, Celestino J, Wu W, Broaddus RR, Schmeler KM, Slomovitz BM, Soliman PT, Gershenson DM, Wang H, Ellenson LH, et al. Reduced progression of endometrial hyperplasia with oral mTOR inhibition in the Pten heterozygote murine model. Am J Obstet Gynecol. 2007;196:247.e1–247.e5. doi: 10.1016/j.ajog.2006.10.872. [DOI] [PubMed] [Google Scholar]
- 21.Law M, Forrester E, Chytil A, Corsino P, Green G, Davis B, Rowe T, Law B. Rapamycin disrupts cyclin/cyclin-dependent kinase/p21/proliferating cell nuclear antigen complexes and cyclin D1 reverses rapamycin action by stabilizing these complexes. Cancer Res. 2006;66:1070–1080. doi: 10.1158/0008-5472.CAN-05-1672. [DOI] [PubMed] [Google Scholar]
- 22.Jundt F, Raetzel N, Müller C, Calkhoven CF, Kley K, Mathas S, Lietz A, Leutz A, Dórken B. A rapamycin derivative (everolimus) controls proliferation through down-regulation of truncated CCAAT enhancer binding protein {beta} and NF-{kappa}B activity in Hodgkin and anaplastic large cell lymphomas. Blood. 2005;106:1801–1807. doi: 10.1182/blood-2004-11-4513. [DOI] [PubMed] [Google Scholar]
- 23.Weisberg E, Banerji L, Wright RD, Barrett R, Ray A, Moreno D, Catley L, Jiang J, Hall-Meyers E, Sauveur-Michel M, et al. Potentiation of antileukemic therapies by the dual PI3K/PDK-1 inhibitor, BAG956: effects on BCR-ABL- and mutant FLT3-expressing cells. Blood. 2008;111:3723–3734. doi: 10.1182/blood-2007-09-114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeager N, Brewer C, Cai KQ, Xu XX, Di Cristofano A. Mammalian target of rapamycin is the key effector of phosphatidylinositol-3-OH-initiated proliferative signals in the thyroid follicular epithelium. Cancer Res. 2008;68:444–449. doi: 10.1158/0008-5472.CAN-07-3030. [DOI] [PubMed] [Google Scholar]
- 25.André F, Campone M, Hurvitz SA, Vittori L, Pylvaenaeinen I, Sahmoud T, O'Regan RM. Multicenter phase I clinical trial of daily and weekly RAD001 in combination with weekly paclitaxel and trastuzumab in patients with HER2-overexpressing metastatic breast cancer with prior resistance to trastuzumab. J Clin Oncol. 2008;26 Abstract 1003. [Google Scholar]
- 26.Tanaka C, O'Reilly T, Kovarik JM, Shand N, Hazell K, Judson I, Raymond E, Zumstein-Mecker S, Stephan C, Boulay A, et al. Identifying optimal biologic doses of everolimus (RAD001) in patients with cancer based on the modeling of preclinical and clinical pharmacokinetic and pharmacodynamic data. J Clin Oncol. 2008;26:1596–1602. doi: 10.1200/JCO.2007.14.1127. [DOI] [PubMed] [Google Scholar]
- 27.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Donnell A, Faivre S, Burris HA, III, Rea D, Papadimitrakopoulou V, Shand N, Lane HA, Hazell K, Zoellner U, Kovarik JM, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral mammalian target of rapamycin inhibitor everolimus in patients with advanced solid tumors. J Clin Oncol. 2008;26:1588–1595. doi: 10.1200/JCO.2007.14.0988. [DOI] [PubMed] [Google Scholar]
- 29.Peralba JM, DeGraffenried L, Friedrichs W, Fulcher L, Grunwald V, Weiss G, Hidalgo M. Pharmacodynamic evaluation of CCI-779, an inhibitor of mTOR, in cancer patients. Clin Cancer Res. 2003;9:2887–2892. [PubMed] [Google Scholar]
- 30.Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, Martinelli E, Ramon y Cajal S, Jones S, Vidal L, et al. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol. 2008;26:1576–1578. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 31.Amato RJ, Jac J, Giessinger S, Saxena S, Willis JP. A phase 2 study with a daily regimen of the oral mTOR inhibitor RAD001 (everolimus) in patients with metastatic clear cell renal cell cancer. Cancer. 2009;115:2438–2446. doi: 10.1002/cncr.24280. [DOI] [PubMed] [Google Scholar]
- 32.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA, Hollaender N, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 33.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA, Osanto S, et al. Phase 3 trial of everolimus for metastatic renal cell carcinoma: final results and analysis of prognostic factors. Cancer. doi: 10.1002/cncr.25219. (in press) [DOI] [PubMed] [Google Scholar]
- 34.Rigatto C, Barrett BJ. Biomarkers and surrogates in clinical studies. In: Parfrey P, Barrett B, editors. Methods of Molecular Biology, Clinical Epidemiology. Totowa, NJ: Humana Press; 2009. pp. 137–154. [DOI] [PubMed] [Google Scholar]
- 35.Noh WC, Mondesire WH, Peng J, Jian W, Zhang H, Dong J, Mills GB, Hung MC, Meric-Bernstam F. Determinants of rapamycin sensitivity in breast cancers cells. Clin Cancer Res. 2004;10:1013–1023. doi: 10.1158/1078-0432.ccr-03-0043. [DOI] [PubMed] [Google Scholar]
- 36.Yamnik RL, Digilova A, Davis DC, Brodt ZN, Murphy CJ, Holz MK. S6 kinase 1 regulates estrogen receptor in control of breast cancer cell proliferation. J Biol Chem. 2009;284:6361–6369. doi: 10.1074/jbc.M807532200. [DOI] [PubMed] [Google Scholar]
- 37.van der Hage JA, van den Broek LJ, Legrand C, Clahsen PC, Bosch CJ, Robanus-Maandag EC, van de Velde CJ, van de Vijver MJ. Overexpression of p70 S6 kinase protein is associated with increased risk of locoregional recurrence in node-negative premenopausal early breast cancer patients. Br J Cancer. 2004;90:1543–1550. doi: 10.1038/sj.bjc.6601741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tee AR, Proud CG. DNA-damaging agents cause inactivation of translational regulators linked to mTOR signalling. Oncogene. 2000;19:3021–3031. doi: 10.1038/sj.onc.1203622. [DOI] [PubMed] [Google Scholar]
- 39.Le XF, Hittelman WN, Liu J, McWatters A, Li C, Mills GB, Bast RC., Jr Paclitaxel induces inactivation of p70 S6 kinase and phosphorylation of Thr421 and Ser424 via multiple signaling pathways in mitosis. Oncogene. 2003;22:484–497. doi: 10.1038/sj.onc.1206175. [DOI] [PubMed] [Google Scholar]
- 40.Jeon YJ, Kim IK, Hong SH, Nan H, Kim HJ, Lee HJ, Masuda ES, Meyuhas O, Oh BH, Jung YK. Ribosomal protein S6 is a selective mediator of TRAIL-apoptotic signaling. Oncogene. 2008;27:4344–4352. doi: 10.1038/onc.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Breuleux M, Klopfenstein M, Stephan C, Doughty CA, Barys L, Maira SM, Kwiatkowski D, Lane HA. Increased AKT S473 phosphorylation after mTORC1 inhibition is rictor dependent and does not predict tumor cell response to PI3K/mTOR inhibition. Mol Cancer Ther. 2009;8:742–753. doi: 10.1158/1535-7163.MCT-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abraham RT, Eng CH. Role of mTOR in solid tumor systems: a therapeutical target against primary tumor growth, metastases, and angiogenesis. Expert Opin Ther Targets. 2008;12:209–222. [Google Scholar]
- 43.López-Lago MA, Okada T, Murillo MM, Socci N, Giancotti FG. Loss of the tumor suppressor gene NF2, encoding merlin, constitutively activates integrin-dependent mTORC1 signaling. Mol Cell Biol. 2009;29:4235–4249. doi: 10.1128/MCB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woo KB, Waalkes TP, Ahmann DL, Tormey DC, Gehrke CW, Oliverio VT. A quantitative approach to determining disease response during therapy using multiple biologic markers: application to carcinoma of the breast. Cancer. 1978;41:1685–1703. doi: 10.1002/1097-0142(197805)41:5<1685::aid-cncr2820410507>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 45.Looney SW. Statistical methods for assessing biomarkers. In: Looney SW, editor. Methods in Molecular Biology. Vol 184: Biostatistical Methods. Totowa, NJ: Humana Press; 2001. pp. 81–109. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Yu CY, Singer B. Cell and tumor classification using gene expression data: construction of forests. Proc Natl Acad Sci USA. 2003;100:4168–4172. doi: 10.1073/pnas.0230559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fardy JM. Evaluation of diagnostic tests. In: Parfrey P, Barrett B, editors. Methods of Molecular Biology, Clinical Epidemiology. Totowa, NJ: Humana Press; 2009. pp. 127–136. [Google Scholar]
- 48.Atkins MB, Choueiri TK, Cho D, Regan M, Signoretti S. Treatment selection for patients with metastatic renal cell carcinoma. Cancer. 2009;115(10 suppl):2327–2333. doi: 10.1002/cncr.24231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho D, Signoretti S, Dabora S, Regan M, Seeley A, Mariotti M, Youmans A, Polivy A, Mandato L, McDermott D, et al. Potential histologic and molecular predictors of response to temsirolimus in patients with advanced renal cell carcinoma. Clin Genitourin Cancer. 2007;5:379–385. doi: 10.3816/CGC.2007.n.020. [DOI] [PubMed] [Google Scholar]
- 50.Noh WC, Kim YH, Kim MS, Koh JS, Kim HA, Moon NM, Paik NS. Activation of the mTOR signaling pathway in breast cancer and its correlation with the clinicopathologic variables. Breast Cancer Res Treat. 2008;110:477–483. doi: 10.1007/s10549-007-9746-x. [DOI] [PubMed] [Google Scholar]
- 51.Iwenofu OH, Lackman RD, Staddon AP, Goodwin DG, Haupt HM, Brooks JS. Phospho-S6 ribosomal protein: a potential new predictive sarcoma marker for targeted mTOR therapy. Mod Pathol. 2008;21:231–237. doi: 10.1038/modpathol.3800995. [DOI] [PubMed] [Google Scholar]
- 52.Broillet A, Hantson J, Ruegg C, Messager T, Schneider M. Assessment of microvascular perfusion changes in a rat breast tumor model using SonoVue to monitor the effects of different anti-angiogenic therapies. Acad Radiol. 2005;12(suppl 1):S28–S33. doi: 10.1016/j.acra.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 53.Shinohara ET, Cao C, Niermann K, Mu Y, Zeng F, Hallahan DE, Lu B. Enhanced radiation damage of tumor vasculature by mTOR inhibitors. Oncogene. 2005;24:5414–5422. doi: 10.1038/sj.onc.1208715. [DOI] [PubMed] [Google Scholar]
- 54.Schnell CR, Stauffer F, Allegrini PR, O'Reilly T, McSheehy PM, Dartois C, Stumm M, Cozens R, Littlewood-Evans A, García-Echeverría C, et al. Effects of the dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235 on the tumor vasculature: implications for clinical imaging. Cancer Res. 2008;68:6598–6607. doi: 10.1158/0008-5472.CAN-08-1044. [DOI] [PubMed] [Google Scholar]
- 55.McSheehy PM, Allegrini PR, Honer M, Ebenhan T, Ametaby S, Schubiger P, Schnell C, Stumm M, O'Reilly TM, Lane HA. Monitoring the activity of the mTOR pathway inhibitor RAD001 (everolimus) non-invasively by functional imaging. Target Oncol. 2007;(suppl):S45–S46. [Google Scholar]
- 56.Wei LH, Su H, Hildebrandt IJ, Phelps ME, Czernin J, Weber WA. Changes in tumor metabolism as readout for mammalian target of rapamycin kinase inhibition by rapamycin in glioblastoma. Clin Cancer Res. 2008;14:3416–3426. doi: 10.1158/1078-0432.CCR-07-1824. [DOI] [PubMed] [Google Scholar]
- 57.Cejka D, Kuntner C, Preusser M, Fritzer-Szekeres M, Fueger BJ, Strommer S, Werzowa J, Fuereder T, Wanek T, Zsebedics M, et al. FDG uptake is a surrogate marker for defining the optimal biological dose of the mTOR inhibitor everolimus in vivo. Br J Cancer. 2009;100:1739–1745. doi: 10.1038/sj.bjc.6605076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma WW, Jacene H, Song D, Vilardell F, Messersmith WA, Laheru D, Wahl R, Endres C, Jimeno A, Pomper MG, et al. [18F]fluorodeoxyglucose positron emission tomography correlates with Akt pathway activity but is not predictive of clinical outcome during mTOR inhibitor therapy. J Clin Oncol. 2009;27:2697–2704. doi: 10.1200/JCO.2008.18.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flaherty KT, Rosen MA, Heitjan DF, Gallagher ML, Schwartz B, Schnall MD, O'Dwyer PJ. Pilot study of DCE-MRI to predict progression-free survival with sorafenib therapy in renal cell carcinoma. Cancer Biol Ther. 2008;7:496–501. doi: 10.4161/cbt.7.4.5624. [DOI] [PubMed] [Google Scholar]
- 60.Schor-Bardach R, Alsop DC, Pedrosa I, Solazzo SA, Wang X, Marquis RP, Atkins MB, Regan M, Signoretti S, Lenkinski RE, et al. Does arterial spin-labeling MR imaging-measured tumor perfusion correlate with renal cell cancer response to antiangiogenic therapy in a mouse model? Radiology. 2009;251:731–742. doi: 10.1148/radiol.2521081059. [DOI] [PMC free article] [PubMed] [Google Scholar]