Abstract
The transcription factor, nuclear factor κB (NF-κB), plays a central role as a key mediator of cell survival and proliferation, and its activation may confer increased tumor chemoresistance. Curcumin, an orally available naturally occurring compound, has been shown to inhibit NF-κB and has a potential role in cancer chemoprevention. We investigated the effects of curcumin on NF-κB activity, on cell viability, and as a chemosensitizing agent with 5-fluorouracil (5-FU) or cisplatin (CDDP) in esophageal adenocarcinoma (EAC). Oligonucleotide microarray analysis of 46 cases, consisting of Barrett metaplasia, low-grade dysplasia, high-grade dysplasia and EAC, showed increased expression of NF-κB and IκB kinase subunits and decreased effector caspase expression in EAC compared with Barrett metaplasia. Stromal expression of both IκB and phospho-IκB was detected in several EAC samples by tissue microarray analysis. Curcumin alone inhibited NF-κB activity and induced apoptosis in both Flo-1 and OE33 EAC cell lines as determined by Western blot analysis, NF-κB reporter assays, and Caspase-Glo 3/7 assays. It also increased 5-FU- and CDDP-induced apoptosis in both cell lines. These data suggest that activation of NF-κB and inhibition of apoptosis may play a role in the progression from Barrett metaplasia to EAC. In addition, curcumin, a well-known inhibitor of NF-κB activity, was shown to increase apoptosis and enhance both 5-FU- and CDDP-mediated chemosensitivity, suggesting that it may have potential application in the therapy of patients with EAC.
Introduction
The incidence of esophageal adenocarcinoma (EAC) has increased significantly, especially in western countries. Surveillance, Epidemiology, and End Results (SEER) registry data indicate a three- to four-fold increase in incidence during the past 30 years [1], with current estimates of approximately 7000 new cases per year in the United States alone. EAC is generally diagnosed at a late stage and has a poor prognosis, with a 5-year survival of less than 10%. Although the current treatment includes chemotherapy, radiation therapy, and, if possible, esophagogastric resection, many patients with EAC experience progression of disease despite such treatment, suggesting that such tumors are resistant to chemotherapy.
Nuclear factor κB (NF-κB) is a transcription factor that is associated with tumorigenesis, and its increased activity has been associated with evasion of apoptosis, malignant transformation, sustained cell proliferation, metastasis, and angiogenesis [2]. NF-κB is a protein complex composed of several subunits including p50, p52, RelA (p65), RelB, and c-Rel that dimerize, with the most common form being the p50/RelA heterodimer. Inactive NF-κB is retained in the cytoplasm by its interaction with inhibitors of κB (IκBα, IκBβ, or IκBɛ) [3]. Activation of extrinsic pathway-mediated apoptosis is initiated by extracellular signaling such as that mediated by tumor necrosis factor-α (TNFα) [4]. Resultant phosphorylation of IκB, its subsequent ubiquitination and proteasome-mediated degradation releases NF-κB, which then translocates to the nucleus [2].
Activation of NF-κB has been reported in several epithelial cancers, including breast [5–7], pancreas [8], oropharynx [9], lung [10], and esophagus [11]. Increased bile acid exposure and an acidic environment have been shown to induce NF-κB in dysplastic Barrett esophagus, the precursor to EAC [12]. With its central role as a transcription factor in a number of malignancies, NF-κB is a target for ongoing development of novel targeted pharmacotherapy.
Curcumin, a phytopolyphenolic pigment derived from turmeric (Curcuma longa), has been shown to have multiple anticancer effects, including inhibition of proliferation, induction of apoptosis, inhibition of angiogenesis, and inhibition of DNA topoisomerase II. A variety of mechanisms has been implicated as mediators of these effects. Specifically, inhibition of activator protein 1 (AP-1), c-Jun N-terminal kinase, Akt, and NF-κB signaling pathways has been described [13,14]. Curcumin has been reported to inhibit NF-κB activation by suppressing the IκB kinases (IKKs), thereby resulting in decreased proliferation and increased apoptosis [15,16]. It seems to suppress constitutively active NF-κB in a variety of cancers [17]. Although several clinical studies are in progress to evaluate the chemotherapeutic activity of curcumin in various cancers (www.cancer.gov/clinicaltrials, accessed November 21, 2009), none have investigated the effectiveness of curcumin in the treatment of EAC. Its pharmacology has been extensively studied, and data support the feasibility of evaluating the efficacy of its oral administration in further clinical trials for the treatment of gastrointestinal tract lesions [18].
We report in the present study that there is increased gene expression of NF-κB and IKK subunits and decreased expression of apoptosis-effector genes in primary EAC samples compared with Barrett metaplasia. We demonstrate that curcumin inhibits NF-κB activity and promotes apoptosis in EAC cell lines, as has been demonstrated in other types of epithelial malignancies [8,9,13,17]. We also show that curcumin can enhance the in vitro cytotoxicity of 5-fluorouracil (5-FU) and cisplatin (CDDP), two first-line chemotherapeutic agents used in the treatment of EAC.
Materials and Methods
Patients and Tissues
After obtaining informed consent, tissues were obtained from patients undergoing esophagectomy for adenocarcinoma at the University of Michigan Medical Center (Ann Arbor, MI) and transported to the laboratory in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) on ice. A portion of each sample was embedded in OCT compound (Miles, Inc, Elkhart, IN) and frozen in isopentane cooled in liquid nitrogen for cryostat sectioning. The remainder was frozen in liquid nitrogen and stored at -80°C. Metaplastic or dysplastic mucosa and tumor samples with at least 70% cellularity were identified using hematoxylin and eosin-stained frozen sections, and 2-mm3 samples were obtained for RNA and protein isolation. The sections were then examined by two pathologists to confirm the histopathologic diagnosis of EAC, high-grade or low-grade dysplasia, Barrett metaplasia, or normal esophageal mucosa. None of the patients had received previous chemotherapy or radiotherapy.
Cell Lines
OE33 (European Collection of Cell Cultures, Sigma-Aldrich, St Louis, MO) and Flo-1 cell lines [19,20] were derived from EACs and grown in RPMI or Dulbecco's modified Eagle medium(Invitrogen), respectively, supplemented with 10% fetal bovine serum (Atlanta Biologicals, Norcross, GA) and 1% penicillin/streptomycin/fungizone (Invitrogen) and incubated at 37°C in 5% CO2/95% air.
Reagents
Curcumin was obtained from Sigma-Aldrich (catalog no. C7727), diluted in dimethyl sulfoxide (DMSO, catalog no. D4540; Sigma-Aldrich) to 100 mM and kept at -20°C. TNFα (catalog no. 2169; Cell Signaling, Danvers, MA) was used to stimulate NF-κB. CDDP (catalog no. P4394; Sigma-Aldrich) and 5-FU (catalog no. F6627; Sigma-Aldrich) were used in chemosensitivity experiments.
Oligonucleotide Microarray
Oligonucleotide expression microarray analysis was performed as previously described [21]. Briefly, total RNA was isolated from 46 esophageal samples using TRIzol (Invitrogen) and purified with RNeasy spin columns (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA quality was assessed by 1% agarose gel electrophoresis and A260/A280 spectrophotometer ratios. RNA quality was reassessed with Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA) at intermediate steps after double-stranded complementary DNA and RNA (cDNA and cRNA, respectively) syntheses. cDNA synthesis, cRNA amplification, hybridization, and washing of HGU133A gene chips (Affymetrix, Santa Clara, CA) were performed by the University of Michigan Cancer Center Microarray Core according to the manufacturer's instructions.
To normalize microarray data, a summary statistic was calculated using 11 probe pairs for each gene and the robust multichip average method as implemented in the Affymetrix Library of Bioconductor (version 1.3, www.bioconductor.org), which provides adjustment, quantile normalization, and summarization. Expression values for each sample were then compared with the mean expression value for the seven Barrett metaplasia samples [22]. Hierarchical cluster analysis and gene expression “heat maps” were generated using the method of Eisen et al. [23].
Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA from treated cells was isolated and column purified using the RNeasy Mini Kit (Qiagen) as per the manufacturer's instructions and was then reverse-transcribed with 250 µM dNTPs, 12.5 ng/µl oligo(dT)12–18 primer, 75 ng/µl random primers, 10 mM dithiothreitol, 1 U/µl RNaseOUT, and 5 U/µl SuperScript II reverse transcriptase (Invitrogen). Real-time polymerase chain reaction (PCR) amplification using 20 ng worth of total RNA, 1 x Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen), and 0.2 µM both forward and reverse primers was performed on the Corbett Rotor-Gene 6000 (Qiagen). The following cycling parameters were used: 50°C hold for 2 minutes; 95°C hold for 2 minutes; 40 cycles of 95°C for 10 seconds, annealing for 15 seconds, and 72°C for 20 seconds. Significant differences of relative quantification were determined using the 2(-ΔΔC(T)) method [24]. These data are represented as the fold increase in gene expression normalized to the endogenous reference gene, β2-microglobulin (B2M).
Primer sets (Invitrogen) were designed using DNASTAR software (Madison, WI).
Casp7: forward, 5′-TTCCGAAGCCTGGGTTTTGACG-3′; and reverse, 5′-GCGAAGCAGGCGGCATTTGTA-3′
RelA: forward, 5′-GACCCCGGCCATGGACGAAC-3′; and reverse, 5′-CCGCTGCTTGGGCTGCTCA-3′.
B2M: forward, 5′-GCTGTGCTCGCGCTACTCTC-3′; and reverse, 5′-CAATGTCGGATGGATGAAACC-3′.
Immunohistochemistry and Tissue Microarray
The expression of IKK and Ser-32 phosphorylated IKK was determined using an esophageal tissue microarray (TMA) [25,26] from 73 patients including 64 tumor, 8 lymph node metastases, 8 dysplastic Barrett mucosa, and 11 nondysplastic Barrett metaplasia samples. Normal esophagus was also included. Immunohistochemical staining was performed on the DAKO Autostainer and the DAKO EnVision+ System, Peroxidase (DAKO, Carpinteria, CA). Microwave epitope retrieval in 1 mM EDTA (pH 8) was performed for 20 minutes. Dewaxed and rehydrated sections of the TMA at 4-mm thickness were labeled with IκB (1:100, catalog no. 9242; Cell Signaling) or phosphorylated IκB (Ser 32, 1:100, catalog no. 9241; Cell Signaling). Slides were lightly counterstained with hematoxylin.
TNFα-Mediated Stimulation of NF-κB
To determine the optimal dosage for TNFα-mediated stimulation of NF-κB, Flo-1 and OE33 cells were treated with TNFα at 0.1, 0.5, 1, 5, 7.5, and 10 ng/ml for 5, 15, 30, 60, 120, 240, and 480 minutes.
Cellular Proliferation and Chemosensitivity Assay
Cellular proliferation was determined by WST-1 assay (Roche Diagnostics, Indianapolis, IN), in accordance with the manufacturer's directions, with varying concentrations of curcumin, 5-FU, or CDDP.
Western Blot Analysis for Apoptosis and NF-κB Activity
Cells were plated at a density of 1.4 x 106 cells/60-mm plate and allowed to adhere for 24 hours. Cells were treated with curcumin at 6.25, 12.5, 25, 50, or 100 µM for 48 hours. Untreated and DMSO (100 µM)-treated cells were used as controls. In addition, cells observed for NF-κB activity were treated with curcumin for 48 hours at varying doses and then stimulated by TNFα at 10 ng/ml for 5 minutes. In separate experiments, Flo-1 and OE33 cell lines were treated with combinations of curcumin and either 5-FU or CDDP.
Total cellular protein was extracted in lysis buffer (150 mM NaCl, 20 mM Tris, pH 7.5, 1 mM EDTA, 1 mM EGTA, 2.5 mM Na4P2O7, 1 mM β-glycerol phosphate, 1 mM Na3VO4, 1 µg/ml leupeptin, and 1% Triton X-100) supplemented with additional protease inhibitor cocktail (catalog no. P8340; Sigma-Aldrich). Lysates (40 µg) were separated by electrophoresis in an 8% to 16% gradient Tris-glycine gel (Invitrogen), then transferred to Immobilon-P nylon membranes (Millipore, Bedford, MA). Membranes were blocked with 5% evaporated milk in 1 x TBS/0.1% Tween 20 (TBST) for 1 hour at room temperature.
Primary antibodies against polyadenosine-5′-diphosphate-ribose polymerase 1 (PARP) (1:1000, catalog no. 9542; Cell Signaling), IκBα (1:1000, catalog no. 4812; Cell Signaling), phospho-IκBα (1:1000, catalog no. 9246; Cell Signaling), or β-actin (1:10,000, catalog no. ab6276; Abcam, Inc, Cambridge, MA) were incubated with membranes overnight at 4°C. Antirabbit HRP-conjugated secondary antibodies (1:5000, catalog no. PI-1000, Vector; Burlingame, CA) or antimouse HRP-conjugated secondary antibodies (1:5000, catalog no. 1010-05; Southern Biotech; Birmingham, AL) in 5% evaporated milk/TBST were incubated with membranes for 1 hour at room temperature. Membranes were washed five times for 5 minutes in TBST after each antibody incubation. Protein bands were visualized using the Pierce ECL kit (Thermo Fisher Scientific, Rockford, IL).
Apoptosis Assay
Cells were plated in white-walled 96-well tissue culture plates at a density of 5 x 103 cells per well in 100 µl of medium and allowed to adhere for 24 hours. Cells were treated in triplicate for 8 to 48 hours with curcumin at 12.5, 25, and 50 µM with DMSO as the vehicle control. In separate experiments, Flo-1 and OE33 cell lines were treated with a combination of curcumin and either 5-FU or CDDP. Caspase-3 and -7 activity was determined at 8, 24, and 48 hours using the Caspase-Glo 3/7 assay (catalog no. G8091; Promega, Madison, WI) as directed by the manufacturer. Briefly, Caspase-Glo 3/7 reagent was added to each well in a 1:1 ratio and incubated for 30 minutes before measuring luminescence as relative light units (RLUs) using a Xenogen IVIS 100 luminometer (Xenogen, Corp, Almeda, CA). Caspase activity was normalized to the cell number using the WST-1 reagent in side-by-side assays performed under the same plating density and conditions.
NF-κB Luciferase Reporter Assay
Flo-1 and OE33 cells were plated at a density of 125,000 cells/well and transfected with NF-κB-Luc reporter plasmid pNifty-Luc (InvivoGen, San Diego, CA) using FuGENE 6 (Roche Diagnostics) at a reagent-plasmid ratio of 16 µl:4 µg in accordance with the manufacturer's directions. Twenty-four hours after transfection, cells were treated as described above with curcumin 10 to 100 µM for 48 hours and 10 ng/ml of TNFα for 5 minutes. At the end of treatment, 1 µl of 40 mg/ml of d-luciferin (Promega) was added to cells, and bioluminescence was monitored using the Xenogen IVIS 100. A grayscale image was collected followed by luminescence acquisition, which was overlaid as a pseudocolor image representing the spatial distribution of the detected photons emitted from cells. This analysis was performed using Living Image software (Xenogen). Signal intensity was quantified as the sum of all detected photon counts within a uniform region of interest manually placed over the wells during data postprocessing. The total numbers of cells in each well were counted, and NF-κB activity was normalized to cell count before the fold increase was determined.
Statistical Analysis
Data points are reported as experimental averages and error bars represent SD. Comparisons between experimental groups used the two-sided Student's t test, with P < .05 considered statistically significant.
Results
NF-κB Subunit Gene Expression Increases and Apoptosis-Related Gene Expression Decreases in EAC Compared with Barrett Metaplasia
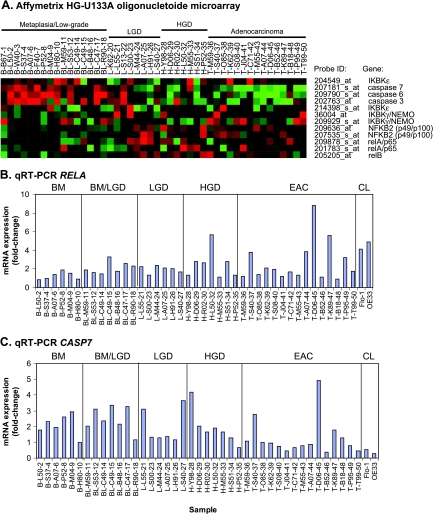
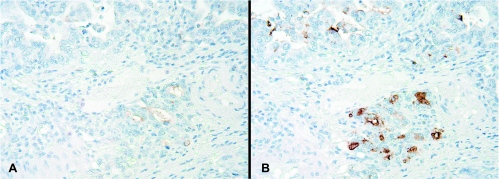
Oligonucleotide expression microarrays were used to evaluate a cohort of 46 esophageal tissue samples and revealed a 1.5-fold increase or higher in REL (33%), RELA (40%), RELB (40%), and NFκB2 (47%) messenger RNA (mRNA) expression in EAC compared with Barrett metaplasia (P < .05; Figure 1A). In addition, we observed overexpression of IKK β, γ, and ɛ subunits, components of the upstream NF-κB activation pathway [27,28], with 9% to 21% greater mean expression in adenocarcinoma compared with metaplastic Barrett esophagus (P < .01). The oligonucleotide microarray analysis also revealed a two-fold or more decrease in CASP3 (30%), CASP6 (47%), and CASP7 (60%) mRNA expression in EAC compared with Barrett metaplasia (P < .05; Figure 1A). Reverse transcription-polymerase chain reaction analysis of RELA and CASP7 confirmed the overall expression patterns observed in the oligonucleotide microarray data set. In addition, the EAC cell lines demonstrated similar expression patterns for RELA (Figure 1B) and CASP7 (Figure 1C) when compared with Barrett metaplasia. To validate that NFκB is activated in EAC, we performed immunohistochemical analysis of a TMA composed of tissue samples of normal, dysplastic, and EAC for the expression of IκB and its phosphorylated product, P-IκB. Stromal expression of IκB was observed in 9 (12.3%) of 73 patients, and P-IκB expression was observed in 5 (6.8%) of 73 patients. Representative photomicrographs from one subject (T-M59-36) indicate focal expression of both IκB and P-IκB in tumor glands (Figure 2), indicating activation of NF-κB in this patient's adenocarcinoma tissue sample.
Figure 1.
NFκB (relA, relB, and NFκB2), IκB kinase (γ, ɛ), and effector-caspase (3, 6, 7) mRNA expression in the progression from metaplastic Barrett esophagus to dysplasia and EAC. (A) HG-U133A (Affymetrix) oligonucleotide microarray analysis of primary tissue samples from patients with Barrett metaplasia (Barrett, n = 9), low-grade dysplasia (LGD, n = 15), high-grade dysplasia (HGD, n = 7), and EAC (EAC, n = 15). Individual samples are labeled by tissue type (B, BL, L, H, or T), subject identifier code, and sample number for the purpose of cross-comparison. Gene expression is labeled as increased (red) or decreased (green). Quantitative real-time PCR analysis of RELA (B) and CASP7 (C) expression in a panel of primary tissue samples from the oligonucleotide microarray analysis compared with normal intestinal epithelium. Significant differences of relative quantification were determined using the 2(-ΔΔC(T)) method.
Figure 2.
Immunohistochemical analysis of (A) IκB and (B) phospho (Ser-32)-IκB in EAC tissues. Paraffin-fixed tissue cores were prepared in a TMA. Representative sections from subject T-M59-36 (original magnification, x200) are displayed and indicate focal expression in tumor glands. This subject also had increased mRNA expression of IKKγ and relB (Figure 1A), consistent with increased NF-κB signaling.
Curcumin Promotes Apoptosis in EAC Cell Lines
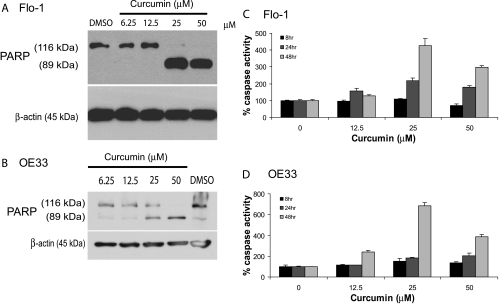
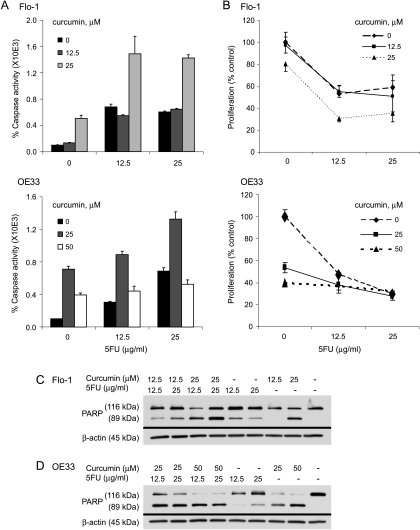
To determine whether caspase-dependent apoptotic activity could be modulated by curcumin, EAC cell lines Flo-1 and OE33 were treated with increasing doses of curcumin for 48 hours. Levels of apoptosis, as determined by immunoblot analysis of PARP expression and cleavage [29,30], were increased in a dose-dependent fashion up to 50 µM curcumin for both Flo-1 (Figure 3A) and OE33 (Figure 3B). We confirmed this dose-dependent apoptotic response by luminescent-based caspase 3/7 assays, which showed more than a four-fold and six-fold increase in caspase 3/7 activity with a 48-hour treatment with 25 µM curcumin in Flo-1 (Figure 3C) and OE33 (Figure 3D; P < .05) cells, respectively.
Figure 3.
Curcumin dose-response and time course to determine the levels of apoptosis in the EAC cell lines Flo-1 and OE33. Western blot analysis of PARP cleavage (89-kDa subunit) after a 48-hour treatment with increasing doses of curcumin in both (A) Flo-1 and (B) OE33 cells. Levels of caspase 3/7 activity as determined by Caspase-Glo 3/7 luminescent-based assays after curcumin treatment at the indicated doses and with increasing duration of incubation in (C) Flo-1 and (D) OE33 cells. Caspase 3/7 activity was normalized to cell number as determined by WST-1 proliferation assay. These numbers were then compared with the level of vehicle control at each time point and expressed as percentage caspase activity (mean % ± SD). Each data point represented three individual wells. Independently repeated experiments were performed to ensure reproducibility. DMSO was used as the vehicle control in all experiments.
Curcumin Inhibits TNFα-Mediated NF-κB Activation in EAC Cell Lines
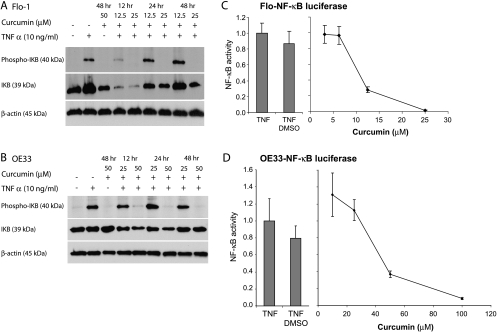
To determine whether the inhibitory effects of curcumin could be mediated through the NF-κB pathway, we examined IκBα and phospho-IκBα expression levels and NF-κB luciferase reporter assay activity after curcumin treatment in the setting of TNFα stimulation. A 5-minute incubation with 10 ng/ml TNFα induced maximal NF-κB activity as demonstrated by IκBα phosphorylation (data not shown). Curcumin inhibited TNFα-mediated phosphorylation of IκB at 25 µM in Flo-1 and at 50 µM in OE33 cell lines, respectively (Figure 4, A and B). Curcumin inhibited NF-κB reporter gene activity in a dose-dependent fashion in both Flo-1 (90% at 25 µM) and OE33 (50% at 50 µM) cell lines (P < .05; Figure 4, C and D). In Flo-1 cells, maximal caspase 3/7 activity and PARP cleavage was detected with 25 µM curcumin and correlated with almost complete inhibition of NF-κB activity. The curcumin dose required for 50% inhibition (45 µM) in OE33 cells was higher than in Flo-1 cells, and doses as high as 100 µM were required for 90% inhibition. Curcumin at 25 µM did not inhibit NF-κB activity, yet it induced the maximum caspase 3/7 activity, suggesting that this induction of apoptosis was not through NF-κB modulation.
Figure 4.
Inhibition of TNFα-induced activation (5 minutes) of NF-κB by curcumin. Flo-1 and OE33 EAC cells were treated with increasing doses of curcumin that were administered for 12, 24, and 48 hours. TNFα was then added to the medium for 5 minutes followed by protein isolation for immunoblot analysis of IκB and phospho-IκB in both (A) Flo-1 and (B) OE33 cells. Actin was used as a loading control. A TNFα-inducible NF-κB-luciferase reporter was transfected into (C) Flo-1 and (D) OE33 cell lines. After treatments with increasing doses of curcumin for 48 hours and TNFα for 5 minutes, d-luciferin was added, and bioluminescence was monitored. Luciferase reporter experiments were performed in triplicate and expressed as mean expression ± SD relative to TNFα stimulation alone.
Chemosensitivity to 5-FU and CDDP Increases with Curcumin
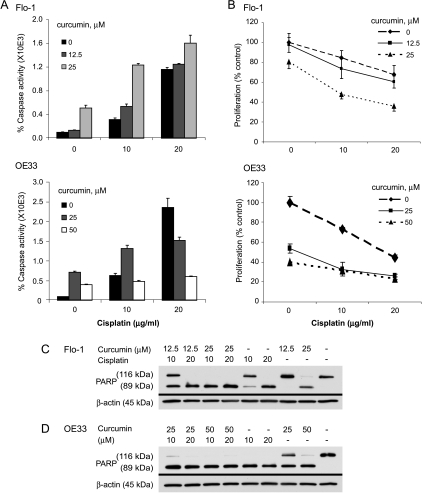
The lethal dose, 50% (LD50) values for Flo-1 and OE33 cell lines were 25 µg/ml for 5-FU and 20 µg/ml for CDDP (data not shown). Curcumin enhanced apoptosis in both Flo-1 and OE33 EAC cell lines when administered in combination with 5-FU, as determined by both greater caspase-3/7 activity and PARP cleavage (Figure 5). 5-FU doses of 12.5 µg/ml or greater seemed to have similar levels of cytotoxicity and caspase 3/7 activity in Flo-1 cells. Although a cytotoxic plateau effect was also noted in OE33 cells, a dose-dependent response to 5-FU was observed in both apoptotic assays. Both cell lines seemed to be sensitive to CDDP treatment in a dose-dependent fashion, with curcumin having an additive effect for both caspase activity and cellular proliferation (Figure 6). The decrease in caspase 3/7 activation observed in OE33 cells in response to 20 µg/ml CDDP/curcumin combinations and high doses of curcumin alone may be due to shifts toward nonapoptotic mechanisms of cell death such as mitotic catastrophe, as has been recently reported [31].
Figure 5.
Effect of curcumin on 5-FU chemosensitivity in the EAC cell lines Flo-1 and OE33. (A) Levels of apoptosis induced by increasing doses of 5-FU and/or curcumin treatment for 48 hours in Flo-1 and OE33 cells were determined using the Caspase-Glo 3/7 activity assay. (B) Caspase 3/7 activity was normalized to cell number as determined by WST-1 proliferation assay. These numbers were then compared with the level of vehicle control and expressed as percentage caspase activity (mean % ± SD). Each data point represented three individual wells. Immunoblot analyses of PARP cleavage were used to assess apoptosis in (C) Flo-1 cells and (D) OE33 cells that were treated with increasing doses of 5-FU and/or curcumin for 48 hours. β-Actin was used as a loading control. DMSO was used as the vehicle control in all experiments. Independently repeated experiments were performed to ensure reproducibility.
Figure 6.
Effect of curcumin on CDDP chemosensitivity in the EAC cell lines Flo-1 and OE33. (A) Levels of apoptosis induced by increasing doses of CDDP (24-hour treatment) and/or curcumin (48-hour treatment) in Flo-1 and OE33 cells were determined using the Caspase-Glo 3/7 activity assay. (B) Caspase 3/7 activity was normalized to cell number as determined by WST-1 proliferation assay. These numbers were then compared with the level of vehicle control and expressed as percentage caspase activity (mean % ± SD). Each data point represented three individual wells. Immunoblot analyses of PARP cleavage were used to assess apoptosis in (C) Flo-1 cells and (D) OE33 cells that were treated with increasing doses of CDDP (24 hours) and/or curcumin (48 hours). β-Actin was used as a loading control. DMSO was used as the vehicle control in all experiments. Independently repeated experiments were performed to ensure reproducibility.
Discussion
Oligonucleotide expression microarray evaluation of a panel of tissue samples suggested that activation of NF-κB occurs in the neoplastic progression from Barrett metaplasia to preneoplastic high-grade dysplasia and adenocarcinoma. We also observed a concomitant decline in caspase-3, -6, and -7 gene expression. Taken together, these findings suggest that NF-κB could be an important mediator of tumor cell survival in EAC, possibly through its inhibitory influence on apoptosis. In addition, activation of NF-κB might be an important mechanism of chemoresistance in the treatment of EAC.
The oligonucleotide expression data reported in this series of tissue samples are consistent with previous reports of increasing frequency of NF-κB activation in the neoplastic progression from Barrett metaplasia to adenocarcinoma [32,33]. The impact of NF-κB inhibition in EAC has not been widely reported. Our data demonstrated that activation of NF-κB occurred not only in EAC but also in its preneoplastic lesion, Barrett esophagus with high-grade dysplasia (Figures 1 and 2). Recently, investigators have demonstrated that NF-κB activation increases the resistance of Barrett epithelial cells to UV-B-mediated apoptotic signaling [34], suggesting one mechanism for increased survival of these precursor lesions as they progress to adenocarcinoma. There is additional indirect evidence regarding the role for NF-κB activation as a mediator of cellular proliferation. Indeed, inhibitory RNA to the NF-κB p50 subunit has been shown to reduce cyclooxygenase 2-dependent cellular proliferation in an EAC cell line [35]. NF-κB activation may also have a role in epithelial tumor chemoresistance [36,37], possibly through its induction of cell survival genes including the inhibitors of apoptosis (IAPs) such as survivin and X-linked IAP (XIAP) [38].
Pretreatment NF-κB status correlated significantly with overall survival, disease-free survival, and complete pathologic response, as reported in one retrospective study, in which pretreatment NF-κB activation was detected in 78% of patients who failed to achieve complete pathologic response and 51% of patients who developed metastatic esophageal carcinoma [39]. Whether manipulation of NF-κB activation will be of clinical significance remains to be determined.
A number of pharmacologic agents directed at inhibiting NF-κB activation are under investigation including curcumin, a naturally occurring phytopolyphenol pigment that is considered the most active component of turmeric [14]. Curcumin has been shown to inhibit NF-κB activation during tumor promotion [40] mediated by phorbol ester, TNF-α, hydrogen peroxide, and cigarette smoke. It also suppressed cellular proliferation and induced apoptosis through the inhibition of constitutively activated NF-κB in pancreatic cancer [8]. In human melanoma cells, curcumin arrested cell cycle progression at the G2/M phase and induced apoptosis by inhibiting NF-κB activation [41].
Given these cell cycle effects, curcumin could potentially synergize with chemotherapeutic agents, inhibiting tumor growth and metastasis. In bladder cancer models, curcumin seemed to potentiate the apoptotic effects of the chemotherapeutic agents, gemcitabine and paclitaxel [42]. Furthermore, curcumin suppressed the paclitaxel-induced NF-κB pathway in breast cancer cells and inhibited metastasis of human breast cancer to the lungs in nude mice [17]. Although curcumin has limited oral bioavailability as a result of hepatic clearance by glucuronidation and sulfation [43,44], it also has a low toxicity profile [14,45] and might be suitable for treatment of upper gastrointestinal malignancies, including esophageal cancers.
Curcumin dose-dependently inhibited NF-κB activity, reduced cell viability and/or proliferation, and promoted apoptosis in EAC cell lines. We, therefore, hypothesized that curcumin might sensitize EAC cell lines to antiproliferative agents such as 5-FU or CDDP, two “first-line” chemotherapeutic agents for the treatment of EAC. We observed an enhanced apoptotic response to these chemotherapeutic agents when either Flo-1 or OE33 cells were concomitantly treated with curcumin. Curcumin may have enhanced the proapoptotic effects of these chemotherapeutic agents through the inhibition of NF-κB activation, or functioned in concert with CDDP, which has been shown to inhibit NF-κB-dependent transcription, leading to the down-regulation of XIAP and, in turn, releasing caspase 3 inhibition [46]. In addition, it may attenuate CDDP-induced NF-κB activation, as has been shown in several chemoresistant carcinoma cell lines [47–49]. Whereas curcumin likely has several modes of action, our data suggest that the combination of curcumin and “first-line” chemotherapeutic agents increased apoptosis, as demonstrated by increased caspase activity and PARP cleavage. Although we have shown that curcumin inhibits NF-κB in EAC cells, other cell signaling pathways including Akt, AP-1, or c-Jun N-terminal kinase [14] might also be significant mediators of this curcumin-induced apoptotic activity. In fact, the lack of NF-κB inhibition with the dose of curcumin that resulted in maximal caspase 3/7 activity in OE33 cells suggested that other signaling pathways were also involved.
It has been reported that curcumin can reduce cell survival in a p53- and caspase-independent manner, possibly through inhibition of both the NF-κB and AP-1 signaling pathways. O'Sullivan-Coyne et al. recently showed that curcumin induced apoptosis-independent cell death in esophageal cancer cells [31]. Specifically, OE33 cells treated with 15 µM curcumin for 24 hours resulted in 13.4% apoptosis and 22.2% mitotic catastrophe. Therefore, the decrease in caspase activity we observed with 50 µM compared with 25 µM curcumin may reflect a shift toward mitotic catastrophe at higher doses of curcumin in both OE33 and Flo-1 cells.
In summary, our findings provide further support for the investigation of curcuminoids in the treatment of patients with esophageal cancer or those with its neoplastic precursor, dysplastic Barrett esophagus. Although efforts to increase systemic bioavailability will likely be necessary for the treatment of more advanced esophageal cancer, proximal digestive tract lesions such as Barrett esophagus might be more amenable to oral-based chemoprevention with curcuminoids.
Footnotes
This study was supported by the National Institutes of Health through 5T32CA009672 (W.H.), 5R01CA071606 (D.G.B.), 5P50CADE97248 (M.S.B.), R01CA129623 (A.R.), 5P30CA046592 (T.J.G.), and 1K08CA127212 (A.C.C.) and by the Thoracic Surgery Foundation for Research and Education (A.C.C.).
References
- 1.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 2.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 3.Dejardin E, Deregowski V, Chapelier M, Jacobs N, Gielen J, Merville MP, Bours V. Regulation of NF-κB activity by I κB-related proteins in adenocarcinoma cells. Oncogene. 1999;18:2567–2577. doi: 10.1038/sj.onc.1202599. [DOI] [PubMed] [Google Scholar]
- 4.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 5.Kim DW, Gazourian L, Quadri SA, Romieu-Mourez R, Sherr DH, Sonenshein GE. The RelA NF-κB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells. Oncogene. 2000;19:5498–5506. doi: 10.1038/sj.onc.1203945. [DOI] [PubMed] [Google Scholar]
- 6.Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ, Jr, Sledge GW., Jr Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol Cell Biol. 1997;17:3629–3639. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sovak MA, Bellas RE, Kim DW, Zanieski GJ, Rogers AE, Traish AM, Sonenshein GE. Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J Clin Invest. 1997;100:2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Aggarwal BB, Shishodia S, Abbruzzese J, Kurzrock R. Nuclear factor-κB and IκB kinase are constitutively active in human pancreatic cells, and their down-regulation by curcumin (diferuloylmethane) is associated with the suppression of proliferation and the induction of apoptosis. Cancer. 2004;101:2351–2362. doi: 10.1002/cncr.20605. [DOI] [PubMed] [Google Scholar]
- 9.Aggarwal S, Takada Y, Singh S, Myers JN, Aggarwal BB. Inhibition of growth and survival of human head and neck squamous cell carcinoma cells by curcumin via modulation of nuclear factor-κB signaling. Int J Cancer. 2004;111:679–692. doi: 10.1002/ijc.20333. [DOI] [PubMed] [Google Scholar]
- 10.Chen G, Bhojani MS, Heaford AC, Chang DC, Laxman B, Thomas DG, Griffin LB, Yu J, Coppola JM, Giordano TJ, et al. Phosphorylated FADD induces NF-κB, perturbs cell cycle, and is associated with poor outcome in lung adenocarcinomas. Proc Natl Acad Sci USA. 2005;102:12507–12512. doi: 10.1073/pnas.0500397102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Latif MM, O'Riordan J, Windle HJ, Carton E, Ravi N, Kelleher D, Reynolds JV. NF-κB activation in esophageal adenocarcinoma: relationship to Barrett's metaplasia, survival, and response to neoadjuvant chemoradiotherapy. Ann Surg. 2004;239:491–500. doi: 10.1097/01.sla.0000118751.95179.c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald RC, Omary MB, Triadafilopoulos G. Dynamic effects of acid on Barrett's esophagus. An ex vivo proliferation and differentiation model. J Clin Invest. 1996;98:2120–2128. doi: 10.1172/JCI119018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Zhang ZS, Zhang YL, Zhou DY. Curcumin inhibits cell proliferation by interfering with the cell cycle and inducing apoptosis in colon carcinoma cells. Anticancer Res. 1999;19:3675–3680. [PubMed] [Google Scholar]
- 14.Sharma RA, Gescher AJ, Steward WP. Curcumin: the story so far. Eur J Cancer. 2005;41:1955–1968. doi: 10.1016/j.ejca.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Jobin C, Bradham CA, Russo MP, Juma B, Narula AS, Brenner DA, Sartor RB. Curcumin blocks cytokine-mediated NF-κB activation and proinflammatory gene expression by inhibiting inhibitory factor I-κB kinase activity. J Immunol. 1999;163:3474–3483. [PubMed] [Google Scholar]
- 16.Plummer SM, Holloway KA, Manson MM, Munks RJ, Kaptein A, Farrow S, Howells L. Inhibition of cyclo-oxygenase 2 expression in colon cells by the chemopreventive agent curcumin involves inhibition of NF-κB activation via the NIK/IKK signalling complex. Oncogene. 1999;18:6013–6020. doi: 10.1038/sj.onc.1202980. [DOI] [PubMed] [Google Scholar]
- 17.Aggarwal BB, Shishodia S, Takada Y, Banerjee S, Newman RA, Bueso-Ramos CE, Price JE. Curcumin suppresses the paclitaxel-induced nuclear factor-κB pathway in breast cancer cells and inhibits lung metastasis of human breast cancer in nude mice. Clin Cancer Res. 2005;11:7490–7498. doi: 10.1158/1078-0432.CCR-05-1192. [DOI] [PubMed] [Google Scholar]
- 18.Sharma RA, Euden SA, Platton SL, Cooke DN, Shafayat A, Hewitt HR, Marczylo TH, Morgan B, Hemingway D, Plummer SM, et al. Phase I clinical trial of oral curcumin. Clin Cancer Res. 2004;10:6847–6854. doi: 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 19.Stoner G, Kaighn M, Reddel R, Resau J, Bowman D, Naito Z, Matsukura N, You M, Galati A, Harris C. Establishment and characterization of SV40 T-antigen immortalized human esophageal epithelial cells. Cancer Res. 1991;51:365–371. [PubMed] [Google Scholar]
- 20.Hughes S, Nambu Y, Soldes O, Hamstra D, Rehemtulla A, Iannettoni M, Orringer M, Beer D. Fas/APO-1 (CD95) is not translocated to the cell membrane in esophageal adenocarcinoma. Cancer Res. 1997;57:5571–5578. [PubMed] [Google Scholar]
- 21.Beer DG, Kardia SLR, Huang C-C, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 22.Risinger JI, Maxwell GL, Chandramouli GV, Jazaeri A, Aprelikova O, Patterson T, Berchuck A, Barrett JC. Microarray analysis reveals distinct gene expression profiles among different histologic types of endometrial cancer. Cancer Res. 2003;63:6–11. [PubMed] [Google Scholar]
- 23.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Raoof DA, Wang Z, Lin MY, Thomas DG, Greenson JK, Giordano TJ, Orringer MB, Chang AC, Beer DG, et al. Expression and effect of inhibition of the ubiquitin-conjugating enzyme E2C on esophageal adenocarcinoma. Neoplasia. 2006;8:1062–1071. doi: 10.1593/neo.05832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kononen J, Bubendorf L, Kallionimeni A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallionimeni O-P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 27.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li JW, Young DB, Barbosa M, Mann M, Manning A, et al. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 28.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 29.Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 30.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 31.O'Sullivan-Coyne G, O'Sullivan GC, O'Donovan TR, Piwocka K, McKenna SL. Curcumin induces apoptosis-independent death in oesophageal cancer cells. Br J Cancer. 2009;101:1585–1595. doi: 10.1038/sj.bjc.6605308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konturek PC, Nikiforuk A, Kania J, Raithel M, Hahn EG, Muhldorfer S. Activation of NFκB represents the central event in the neoplastic progression associated with Barrett's esophagus: a possible link to the inflammation and overexpression of COX-2, PPARγ and growth factors. Dig Dis Sci. 2004;49:1075–1083. doi: 10.1023/b:ddas.0000037790.11724.70. [DOI] [PubMed] [Google Scholar]
- 33.O'Riordan JM, Abdel-latif MM, Ravi N, McNamara D, Byrne PJ, McDonald GS, Keeling PW, Kelleher D, Reynolds JV. Proinflammatory cytokine and nuclear factor κ-B expression along the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Gastroenterol. 2005;100:1257–1264. doi: 10.1111/j.1572-0241.2005.41338.x. [DOI] [PubMed] [Google Scholar]
- 34.Hormi-Carver K, Zhang X, Zhang HY, Whitehead RH, Terada LS, Spechler SJ, Souza RF. Unlike esophageal squamous cells, Barrett's epithelial cells resist apoptosis by activating the nuclear factor-κB pathway. Cancer Res. 2009;69:672–677. doi: 10.1158/0008-5472.CAN-08-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Si J, Fu X, Behar J, Wands J, Beer DG, Souza RF, Spechler SJ, Lambeth D, Cao W. NADPH oxidase NOX5-S mediates acid-induced cyclooxygenase-2 expression via activation of NF-κB in Barrett's esophageal adenocarcinoma cells. J Biol Chem. 2007;282:16244–16255. doi: 10.1074/jbc.M700297200. [DOI] [PubMed] [Google Scholar]
- 36.Zong W-X, Edelstein LC, Chen C, Bash J, Galinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNFα-induced apoptosis. Genes Dev. 1999;13:382–387. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grumont RJ, Rourke IJ, Gerondakis S. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 1999;13:400–411. doi: 10.1101/gad.13.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huerta S, Heinzerling JH, Anguiano-Hernandez Y-M, Huerta-Yepez S, Lin J, Chen D, Bonavida B, Livingston EH. Modification of gene products involved in resistance to apoptosis in metastatic colon cancer cells: roles of Fas, Apaf-1, NFκβ, IAPs, Smac/DIABLO, and AIF. J Surg Res. 2007;142:184–194. doi: 10.1016/j.jss.2006.12.551. [DOI] [PubMed] [Google Scholar]
- 39.Izzo JG, Correa AM, Wu T-T, Malhotra U, Chao CKS, Luthra R, Ensor J, Dekovich A, Liao Z, Hittelman WN, et al. Pretherapy nuclear factor-κB status, chemoradiation resistance, and metastatic progression in esophageal carcinoma. Mol Cancer Ther. 2006;5:2844–2850. doi: 10.1158/1535-7163.MCT-06-0351. [DOI] [PubMed] [Google Scholar]
- 40.Singh S, Aggarwal BB. Activation of transcription factor NF-κB is suppressed by curcumin (diferuloylmethane) J Biol Chem. 1995;270:24995–25000. doi: 10.1074/jbc.270.42.24995. [DOI] [PubMed] [Google Scholar]
- 41.Zheng M, Ekmekcioglu S, Walch ET, Tang CH, Grimm EA. Inhibition of nuclear factor-κB and nitric oxide by curcumin induces G2/M cell cycle arrest and apoptosis in human melanoma cells. Melanoma Res. 2004;14:165–171. doi: 10.1097/01.cmr.0000129374.76399.19. [DOI] [PubMed] [Google Scholar]
- 42.Kamat AM, Sethi G, Aggarwal BB. Curcumin potentiates the apoptotic effects of chemotherapeutic agents and cytokines through down-regulation of nuclear factor-κB and nuclear factor-κB regulated gene products in IFNα-sensitive and IFNα-resistant human bladder cancer cells. Mol Cancer Ther. 2007;6:1022–1030. doi: 10.1158/1535-7163.MCT-06-0545. [DOI] [PubMed] [Google Scholar]
- 43.Cheng AL, Hsu CH, Lin JK, Hsu MM, Ho YF, Shen TS, Ko JY, Lin JT, Lin BR, Ming-Shiang W, et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001;21:2895–2900. [PubMed] [Google Scholar]
- 44.Garcea G, Berry DP, Jones DJ, Singh R, Dennison AR, Farmer PB, Sharma RA, Steward WP, Gescher AJ. Consumption of the putative chemopreventive agent curcumin by cancer patients: assessment of curcumin levels in the colorectum and their pharmacodynamic consequences. Cancer Epidemiol Biomarkers Prev. 2005;14:120–125. [PubMed] [Google Scholar]
- 45.Dhandapani KM, Mahesh VB, Brann DW. Curcumin suppresses growth and chemoresistance of human glioblastoma cells via AP-1 and NFκB transcription factors. J Neurochem. 2007;102:522–538. doi: 10.1111/j.1471-4159.2007.04633.x. [DOI] [PubMed] [Google Scholar]
- 46.Shen Y, Wang J, Yang T, Li Y, Jiang W, Guan Z, Wang Z, Tan J, Wu J, Li G, et al. Platinums sensitize human epithelial tumor cells to lymphotoxin α by inhibiting NFκB-dependent transcription. Cancer Biol Ther. 2008;7:1407–1414. doi: 10.4161/cbt.7.9.6429. [DOI] [PubMed] [Google Scholar]
- 47.Notarbartolo M, Poma P, Perri D, Dusonchet L, Cervello M, D'Alessandro N. Antitumor effects of curcumin, alone or in combination with cisplatin or doxorubicin, on human hepatic cancer cells. Analysis of their possible relationship to changes in NF-κB activation levels and in IAP gene expression. Cancer Lett. 2005;224:53–65. doi: 10.1016/j.canlet.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 48.Chuang SE, Yeh PY, Lu YS, Lai GM, Liao CM, Gao M, Cheng AL. Basal levels and patterns of anticancer drug-induced activation of nuclear factor-κB (NF-κB), and its attenuation by tamoxifen, dexamethasone, and curcumin in carcinoma cells. Biochem Pharmacol. 2002;63:1709–1716. doi: 10.1016/s0006-2952(02)00931-0. [DOI] [PubMed] [Google Scholar]
- 49.Yeh PY, Chuang SE, Yeh KH, Song YC, Ea CK, Cheng AL. Increase of the resistance of human cervical carcinoma cells to cisplatin by inhibition of the MEK to ERK signaling pathway partly via enhancement of anticancer drug-induced NF κB activation. Biochem Pharmacol. 2002;63:1423–1430. doi: 10.1016/s0006-2952(02)00908-5. [DOI] [PubMed] [Google Scholar]