Abstract
In carcinogenesis, determination of gene and protein expression profiles is important for prevention and treatment. Caffeic acid phenethyl ester (CAPE) in a single dose administered before carcinogenic initiation induced by diethylnitrosamine (DEN) prevents the appearance of preneoplastic lesions. On the basis of this approach, the main purpose of this work was to compare the gene expression profiles induced by DEN or a previously administered single dose of CAPE. Using a modified hepatocarcinogenesis-resistant hepatocyte model, male Fischer-344 rats were administered with one intraperitoneal dose of CAPE (20 mg/kg) 12 hours before DEN administration (200 mg/kg). Livers were removed and processed for microarray analysis and reverse transcription-polymerase chain reaction 12 hours after CAPE dosing and 24 hours after DEN administration with or without CAPE. CAPE alone did not alter the expression profile. DEN treatment modified the expression of 665 genes, and CAPE plus DEN induced changes in 1371 genes. DEN treatment increased the expression of genes associated with oxidative stress such as glutathione reductase, genes involved in cell cycle regulation including p53, and modified cytochrome P450. CAPE plus DEN diminished the expression of cytochrome involved in DEN bioactivation such as CYP2B1 as well as the expression of regulators of oxidative stress such as glutathione reductase, GST-κ and GST-θ, and cell cycle regulators such as p53. Using CAPE as a tool, we uncovered new approaches for studying the altered expression of reactive genes and identifying proteins that will help to propose well-sustained and concrete hypothesis of DEN mechanism of hepatocarcinogenesis initiation.
Introduction
Exposure to chemical carcinogens is one of the most studied areas in carcinogenesis, and three major stages in carcinogenesis have been recognized: initiation, when mutations occur and initiated cells are generated; promotion, when clonal expansion of initiated cells takes place and forms preneoplastic lesions; and progression, when preneoplastic lesions become tumors through the gain of additional genetic and metabolic alterations [1].
Diethylnitrosamine (DEN), an environmental carcinogen, has been used in several experimental models [2]. Specifically, it has been used as an initiator in the modified resistant hepatocyte model [3,4] because initiation is necessary for carcinogenesis [5]. This effect has not yet been completely studied, with only a few elements of the pathway having been identified thus far. As early as 3 hours after DEN administration, lipid peroxidation can be detected [6], as well as overexpression of glutathione-S-transferase Pi (GST-p) at 4 hours; this is considered a marker of initiation in chemical carcinogenesis and tumors [7]. In addition, the formation of DNA adducts have been detected as early as 3 hours after administration [8], and generalized necrosis clearly seen after 24 hours is a sign of subsequent damage [5,9].
In the modified resistant hepatocyte model of hepatocarcinogenesis, it has been demonstrated that oxidative stress is necessary for initiation. When N-ethyl-N-nitrosourea is used as the initiator carcinogen instead of DEN, it generates the same adducts as DEN, but without an important oxidative stress. When N-ethyl-N-nitrosourea is used, preneoplastic lesions are not observed at 25 days, in contrast to the high number that occurs when oxidative stress and adducts are produced by DEN. This finding is also supported by the decreased induction of preneoplastic lesions by the antioxidant quercetin [6]. Oxidative stress generated during the initiation stage is mainly due to DEN metabolism by cytochrome P450 (CYP). DEN must be metabolized by several CYP family members to produce the active procarcinogen, and when the isoforms related to this bioactivation are inhibited by a chemoprotector such as caffeic acid phenethyl ester (CAPE), the lesions observed in the model are drastically reduced [5,10].
As previously shown in the modified resistance hepatocyte model, three stages are necessary to produce preneoplastic lesions at 25 days and liver cancer after 7 months of initiation [5,11]. As such, all components of this model (DEN, 2-acetyl aminofluorene administration and partial hepatectomy) are necessary, and it is implied that each has an individual pathological mechanism; however, the specific mechanisms of each step in carcinogenesis process remain unclear.
Gene expression profile (GEP) of the progression stage has shown important genetic changes related to the evolution of liver preneoplastic lesions toward neoplastic lesions and tumors [11]. However, GEP and possible expression modifications by anticancer agents have not been extensively studied.
The chemoprotectors quercetin, celecoxib, and CAPE are inhibitors of the initiation stage and prevent preneoplastic lesions; CAPE is one of the most efficient inhibitors. When administered 12 hours before DEN, this natural component of propolis diminished tissue damage produced 24 hours after DEN administration, as well as lipid peroxidation and modification of CYP isoforms 1A1/2 and 2B1/2 related to DEN bioactivation. All of these alterations are thought to be related to the reduced incidence of preneoplastic lesions at 25 days after DEN treatment in CAPE-pretreated animals [5,11].
Thus, the aim of this work was to analyze the initiation stage using microarrays to contrast the changes in GEP produced by DEN alone with those produced when CAPE was administered before DEN. We have hypothesized that the comparison of the carcinogenic treatment versus the chemoprotector plus carcinogen will show relevant changes in gene expression pertaining to the initiation of carcinogenesis.
Results indicated that there is a differential effect on GEP induced by CAPE with or without DEN. When CAPE was administered alone, it did not modify GEP. After DEN administration, 665 genes were modulated and CAPE plus DEN modified surprisingly 1371 genes. CAPE modulated several cell physiological aspects, such as DEN metabolism, regulation of oxidative stress, and others such as proliferation, DNA replication, and DNA repair. These results proposed new routes to study the principal events performed in the initiation of hepatocarcinogenesis process and in the chemoprotector action mechanism of CAPE.
Materials and Methods
Animals
Male Fischer-344 rats (180–200 g) were obtained from the Unit for Production of Experimental Laboratory Animals (UPEAL Cinvestav, Mexico City, Mexico). Animals had access to food (PMI Feeds, Inc, Laboratories Diet, Richmond, IN) and water ad libitum, with each rat consuming approximately 12 to 15 g of food and 10 to 15 ml of water per day. Animals were maintained in a holding room under controlled conditions of 12-hour light/dark cycles, 50% relative humidity, and 21°C. Animal care followed institutional guidelines for the use of laboratory animals.
Experimental Protocol
Animals were administered 200 mg/kg DEN (Sigma-Aldrich, St Louis, MO) as in the modified resistant hepatocyte model of Semple-Robert [3]. A group of rats was pretreated with a 20-mg/kg single dose of CAPE (kindly provided by Dr. Javier Hernández-Martínez, CIAD, Hermosillo, Mexico) 12 hours before DEN treatment [10]. Animals were killed 12 hours after CAPE alone and 24 hours after DEN administration with and without CAPE. Animals were killed by exsanguination. Livers were excised, washed in physiological saline solution, frozen in 2-methyl butane with liquid nitrogen, or immersed in RNA later (Sigma) and stored at -80°C. Frozen livers sections were used to complementary DNA (cDNA) microarray and reverse transcription-polymerase chain reaction (RT-PCR) assays (n = 4 in each group).
cDNA Microarray Slides
Microarrays were assayed at the Biochips Platform of Genopole, University of Toulouse, INSA, UPS, INP & INRA (Toulouse, France). cDNA hybridization was performed on slides with 28,800 spots using 27,004 oligonucleotides of 22,012 rat genes, including 5000 genes with known function in the rat. Data were analyzed with the Web service Bioplot, Bioclust, and Venn diagrams from Transcriptome-Biochips Platform of Genopole Toulouse Midi-Pyrenees (http://biopuce.insa-toulouse.fr). Altered genes were selected using a ratio of up-regulation threshold greater than 1.5, downregulation threshold less than 0.6, and Student's t test with P < .05 considered statistically significant. The generated data set has been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/index.cgi) database (GSE12030).
Relative Quantitative RT-PCR
Total RNA extraction was performed using Tripure Isolation Reagent (Roche Diagnostics, Indianapolis, IN). DNAse treatment was performed using RNase-free DNAse I (Roche Diagnostics), and cDNA synthesis was performed using SuperScript Reverse Transcriptase (Invitrogen, Carlsbad, CA). Real-time PCR was performed in a LightCycler Carousel-Based System Instrument 2.0 (Roche Diagnostics) using LightCycler FastStart DNA MasterPLUS SYBR Green I (Roche Diagnostics), and primers were synthesized by Invitrogen (Table 1). RT-PCR was performed in quadruplicate, and additional reactions were performed without reverse transcriptase to verify the absence of DNA contamination. Gene expression quantification was performed using a derivation of the 2-ΔΔCT method, 2-ΔC′T [12]. Student's t test was used as statistical analysis.
Table 1.
Primers Used in RT-PCR.
| Symbol | Gene ID | Forward | Reverse | Size (bp) |
| CYP1A2 | NM_012541 | AGGGACACCTCACTGAATGG | CCGAAGAGCATCACCTTCTC | 182 |
| CYP2A2 | NM_012693 | ATCCAGATGTGGAAGCCAAG | CCACGGAAGGTTGTGTTCTT | 187 |
| CYP2B1 | NM_001134844 | GGAAGCTCTGGTTGCTTGAC | CAAAGAAGTGCAGACCGACA | 206 |
| CYP2E1 | NM_031543 | CCTACATGGATGCTGTGGTG | CTGGAAACTCATGGCTGTCA | 171 |
| GST-θ | NM_012796 | ATGGCATTCCCTTTCAGTTG | GTGGTCTGCCACCTGGTACT | 179 |
| GST-κ | NM_181371 | CACGGAGTCCCAGAACATTT | CGGTCAGACCCAAATAGCAT | 210 |
| p53 | NM_030989 | TCTCCCAGCAAAAGAAAAA | CTTCGGGTAGCTGGAGTGAG | 168 |
| p38 | NM_031020 | AGCGATACCAGAACCTGTCC | GAACGTGGTCATCGGTAAGC | 318 |
| YY1 | NM_173290 | GACGACGACTACATCGAGCA | TTGATCTGCACCTGCTTCTG | 209 |
| DNApolδ | NM_021662 | GACGACGACTACATCGAGCA | AGCGGGAGGAGAAGAGTAGG | 217 |
| GLUTATHION REDUCTASE | NM_053906 | CCATGTGGTTACTGCACTTCC | GTTCCTTTCCTTCTCCTGAGC | 171 |
These genes were selected from microarrays analysis and validated by RT-PCR.
Results
Differential GEPs in Liver Cancer Initiation
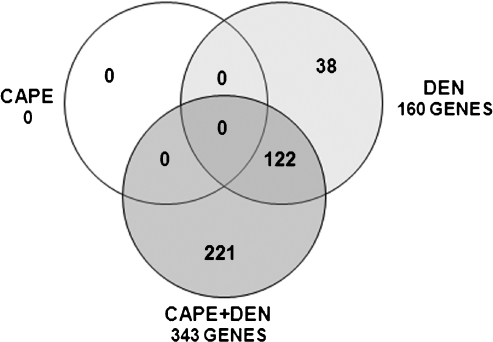
Considering that omission of the carcinogen initiator is sufficient to avoid preneoplastic lesions and cancer induction [5], molecular alterations at the initiation stage are likely necessary for cancer to occur. To characterize the GEP modifications that occur at the initiation stage, microarray analyses were performed to determine genes differentially expressed on DEN, CAPE, or CAPE plus DEN administration. At 24 hours after DEN administration, 665 genes were transcriptionally modified; of these, 160 had known function in the rat, with 91 upregulated and 69 downregulated. The effect of CAPE was analyzed at 12 hours after administration; this elapsed period before DEN administration was used to determine GEP changes produced by the chemoprotector alone. As such, CAPE administration did not induce modifications in the GEP. When CAPE was administered 12 hours before DEN and its effect evaluated 24 hours after DEN treatment, the expression of 1371 genes was changed; 343 of these have known function in the rat. Of these genes, 100 were upregulated and 243 were downregulated. Of the 160 altered genes with known function in the rat, 38 were restored to normal levels, 122 remained altered with DEN treatment alone, and the expression of 221 was newly modified in the CAPE plus DEN group (Figure 1). It is tempting to speculate that genes whose expression was initially modified by DEN were restored by CAPE treatment thus describing a chemoprotective mechanism for CAPE.
Figure 1.
Venn diagramof transcriptionally altered genes. Data were analyzed using the Venn diagram program from http://biopuce.insa-toulouse.fr. Genes were identified as upregulated (threshold > 1.5) or downregulated (threshold < 0.6) with P < .05 considered statistically significant with Student's t test (n = 4 in each group).
Upregulated Genes
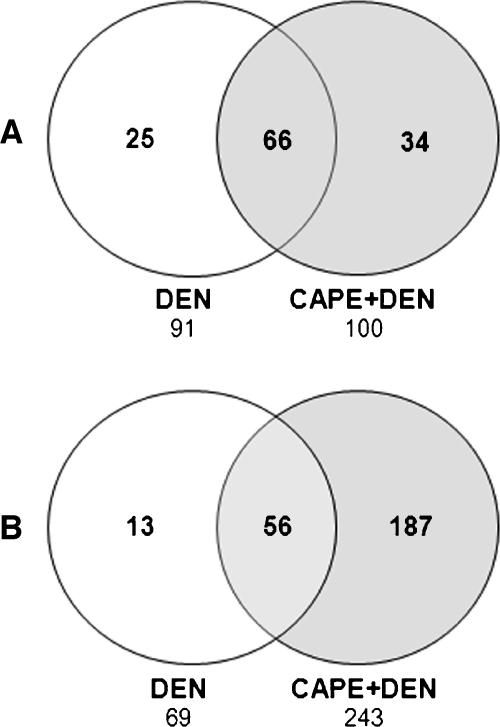
A general analysis of GEP was the starting point for this study. In the DEN-treated group, 91 genes were upregulated; of these, 66 remained modified after CAPE treatment (Table W1), whereas the expression of 25 was restored to untreated levels (Table W2). The 91 genes modified by DEN included several ribosomal proteins, CYP, heat shock proteins, and genes such as GAPDH and GST-κ, suggesting an important alteration of protein synthesis, metabolism, and oxidative stress, which are all involved the initiation stage (Table W1). In the CAPE plus DEN group, 100 genes were upregulated; of these, 66 were upregulated after DEN treatment (Table W1) and 34 were only upregulated after CAPE plus DEN administration (Table W3). These 34 genes encode ribosomal proteins as well as heat shock proteins and CYP, among others. In line with chemoprotective effects of CAPE, there were differences between in GEPs between both groups (Figure 2A).
Figure 2.
Comparison of Venn diagrams for genes upregulated or downregulated after DEN or DEN plus CAPE treatment. (A) Upregulated genes, threshold greater than 1.5 with P < .05 by Student's t test. (B) Downregulated genes, threshold less than 0.6 with P < .05 by Student's t test. Data were analyzed using the Venn diagram program from http://biopuce.insa-toulouse.fr (n = 4 in each group).
Downregulated Genes
After DEN treatment, 69 genes were downregulated, and 56 genes remained downregulated after CAPE treatment (Table W4). Of these, the expression of 13 was restored to control levels by CAPE administration before DEN (Table W5). Within the set of downregulated genes, there were several CYP isoforms as well as microglobulins, macroglobulins, VEGFR1, and APC. In general, gene down-regulation was a hallmark of CAPE pretreatment because CAPE plus DEN treatment downregulated 243 genes, of which 56 were common to the DEN alone condition and 187 genes were downregulated independently of DEN administration (Table W6), including ribosomal proteins, CYP, globulins, p53, p38, transcription factor YY1, GST-θ, and others. It should be noted that some CYP family members were downregulated in the CAPE plus DEN group, including bioactivators of DEN such as CYP2E1, CYP2B1, CYP1A2; this suggests that CAPE treatment might affect DEN bioactivation. In addition, CAPE might act by modifying the cell cycle, oxidative stress, and other pathways involved in cell regulation (Figure 2B).
PCR Validation
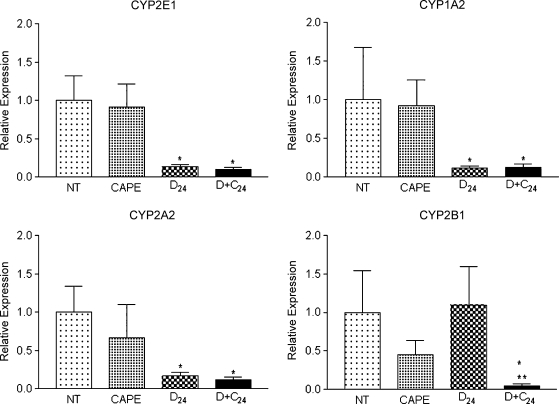
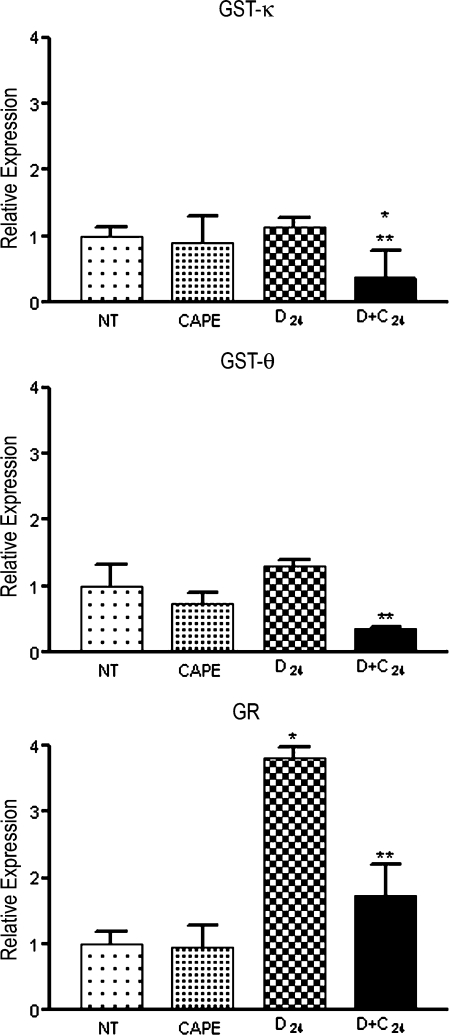
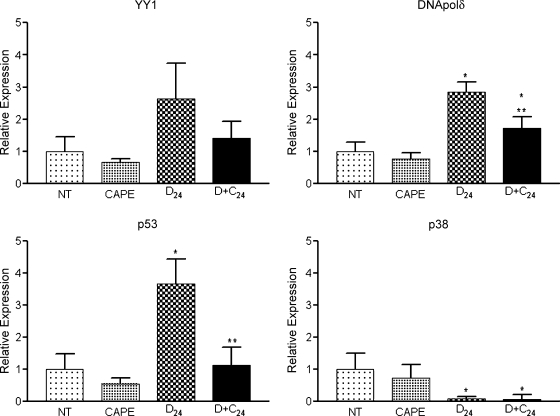
The aim of using microarrays was to explore the GEP to generate hypothesis about events involved in tumor initiation. Microarrays are completely reliable because the genes are not individually evaluated. Thus, microarray results must be validated using a quantitative method. For RT-PCR validation, we selected 11 genes (Table 2): four related to DEN metabolism (CYP2E1, 2B1, 1A2, and 2A2), three related to oxidative stress regulation (GST-κ, GST-θ, and glutathione reductase), and four related to cell regulation (transcription factor YY1, DNA polymerase δ [DNA polδ], p53, and p38). We found that DEN administration decreased CYP2E1, 1A2, and 2A2 expression, but CAPE administration did not ameliorate this effect. Conversely, CYP2B1 expression was not altered by DEN, but at 24 hours after DEN treatment, CAPE administration significantly decreased the expression of this isoform (Figure 3). The expression of GST-κ, GST-θ, and glutathione reductase genes increased with DEN administration and decreased in the CAPE plus DEN group (Figure 4). DEN treatment increased YY1, p53, and DNA polδ expression; this effect decreased with CAPE pretreatment. DEN reduced the expression of p38 and CAPE had no effect (Figure 5). This suggests that CAPE modulates the alterations produced by DEN at different levels. Furthermore, these results confirm the importance of microarray validation, because on RT-PCR analysis, the expression of some genes was not in complete agreement with microarray data. We also observed expression changes that were not observed when using the microarray, such as increased expression of YY1, p53, and DNA polδ genes.
Table 2.
Genes Validated by RT-PCR for Microarray Results.
| Gen | Function |
| CYP2B1* | |
| CYP1A2* | DEN metabolism |
| CYP2A2* | |
| CYP2E1 | |
| GST-κ† | |
| GST-θ* | Oxidative stress regulation |
| GLUTATIÓN REDUCTASE* | |
| YY1* | |
| DNA polδ* | Replication |
| p53* | DNA reparation |
| p38* | Cell signaling process. |
Genes downregulated by CAPE plus DEN as determined by microarray.
Genes upregulated by DEN as determined by microarray.
Figure 3.
RT-PCR validation of CYP expression levels. Samples were analyzed by RT-PCR using primers described in Table 1. CAPE indicates rats killed 12 hours after CAPE dosing; D + C24, killed 24 hours after DEN dosing with CAPE pretreatment 12 hours before DEN (n = 4 in each group); D24, killed 24 hours after DEN dosing; NT, untreated. *P < .05 compared with NT, **P < .05 compared with D24 by Student's t test.
Figure 4.
RT-PCR validation of GST-κ, GST-θ, and glutathione reductase (GR) expression levels. Samples were analyzed by RT-PCR using primers from Table 1. CAPE indicates rats killed 12 hours after CAPE dosing; D + C24, killed 24 hours after DEN dosing with CAPE pretreatment 12 hours before DEN (n = 4 in each group); D24, killed 24 hours after DEN dosing; NT, untreated. *P < .05 compared with NT, **P < .05 compared with D24 by Student's t test.
Figure 5.
RT-PCR validation of YY1, DNA polδ, p53, and p38 gene expression levels. Samples were analyzed by RT-PCR using primers from Table 1. CAPE indicates rats killed 12 hours after CAPE dosing; D + C24, killed 24 hours after DEN dosing with CAPE pretreatment 12 hours before DEN (n = 4 in each group); D24, killed 24 hours after DEN dosing; NT, untreated. *P < .05 compared with NT, **P < .05 compared with D24 by Student's t test.
Discussion
Chemoprevention is helpful for preventing or even curing cancer in any of the three stages of carcinogenesis: initiation, promotion, or progression. To this end, CAPE has been studied for the past 20 years, and it has shown chemoprotective properties in several types of cancer. In different experimental models, the mechanism of action differs depending on the situation tested [4,5,10,12–19]. In the modified resistant hepatocyte model of liver cancer, the chemoprotective mechanism of action of CAPE in the initiation stage is not completely understood, but it seems to be related to the inhibition of DEN bioactivation [5].
The initiation stage is a necessary step in the modified resistant hepatocyte model. Initiation is caused by DEN administration. The metabolites produced during bioactivation produce mutations in DNA, generate oxidative stress, and, finally, initiate cells toward transformation. Furthermore, oxidative stress and DNA alkylation have both been suggested to be necessary for initiation [6]. During the first 24 hours after DEN administration, the expression of the tumor marker GST-p is observed, and lipid peroxidation, adduct formation, CYP modifications, and tissue damage are also detected [5–8]. These alterations thus represent a phenotype characteristic of initiated hepatocytes. However, it is important to identify the immediate early changes in hepatocarcinogenesis to uncover specific molecular targets for detection and chemoprevention.
Microarray analysis is a powerful technique for genetic and genomic investigation, making it possible to simultaneously analyze gene expression of thousand of genes [20]. This is important, considering that gene expression variations are associated with common diseases and do not lead directly to disease, but instead generate an intermediary molecular phenotype. Therefore, it is important to identify the molecular phenotypes associated with changes in disease state. In this manner, molecular networks that involve changes in disease can be elucidated [21].
We used microarray slides with 28,800 spots, allowing for a large and complex data set to be analyzed. It was necessary to further select a small group of genes to validate. We started with genes with well-known function in rats. From the analysis of 28,800 spots, we identified genes expressed differentially in each treatment; 160 were modified by DEN, 343 by DEN plus CAPE, and none by CAPE alone before DEN administration.
To validate microarrays by RT-PCR, we selected three groups of genes related to specific functions in the rat liver that could be involved in the initiation process. Genes that 1) might be involved in the initiation of carcinogenesis, 2) are directly involved in the initial oxidative stimulus that modifies the phenotype of hepatocytes, and 3) activate possible proliferation signals through DNA alkylation were selected. The first genes selected were those related to DEN bioactivation because alteration of DEN metabolism should result in altered initiation. Gene expressions of 1A2, 2E1, and 2A2 isoforms were diminished by DEN treatment and were not modified by CAPE pretreatment. The expression of 2B1 was not affected by DEN but was significantly reduced by pretreatment with CAPE alone and by administration of both CAPE and DEN. Because CYP2B1 is an important bioactivator of DEN, we propose that DEN bioactivation by CYP2B1 is diminished by CAPE, as previously shown enzymatically [5].
The second group of genes selected were those involved in oxidative stress regulation including GST-κ, a mitochondrial GST isoform, GST-θ, a cytosolic isoform, and glutathione reductase [22]. These genes were upregulated by DEN treatment in agreement with the previous report of altered redox state [6]. The messenger RNA up-regulation of these genes was not present with CAPE pretreatment alone or in the CAPE plus DEN group. This suggests that pretreatment-derived counteraction of oxidative stress is likely related to the decreased bioactivation of DEN; this effect is likely derived from the reduced generation of oxidative stress by CYP together with the direct antioxidant activity of CAPE [10]. Both mechanisms deserve further analysis because there is a need to identify new candidates as possible initiation markers during the early stages of carcinogenesis, such as GST-p [7].
The third group of selected genes was related to cell regulation processes such as proliferation and DNA repair. The transcription factor YY1 participates in many aspects of cell regulation including activation, repression, and modification of growth, development, and differentiation GEPs [23]. DNA polδ participates in DNA replication and DNA repair processes including base pair excision, nucleotide excision, and mismatch repair [24]. p53 protects the genomic integrity by participating in cell cycle arrest, DNA repair, and apoptosis, and it has been found to be overexpressed in hepatocellular carcinoma [25]. After validating the GEPs, we observed that p38 expression, a mitogen-activated protein kinase, was diminished with DEN treatment and was not restored by CAPE [26]. The expression of p53, transcription factor YY1, and DNA polδ was increased by DEN treat ment, suggesting altered DNA repair and replication. Up-regulation of these genes was avoided by CAPE pretreatment. After DEN bioactivation, DNA damage is followed by DNA repair and replication; however, if DEN is not completely metabolized, the DNA damage is decreased. These effects are likely mediated directly by CAPE, with subsequently reduced expression of YY1 and p53; the transcription factor YY1 regulates the transcription of p53 [23]. Further investigation is required to determine the participation of these genes and their protein products during carcinogenesis initiation.
There are many more genes that could be selected for further analysis from our microarray; thus, our analysis only represents an initial effort to identify the genes participating in the initiation stage of this model of chemical hepatocarcinogenesis.
Other groups of genes relevant for future validation and investigation include those encoding heat shock proteins, which are molecular chaperones induced by several pathological stimuli. Alterations in processing proteins have been related to the carcinogenesis process; these proteins have been found to be overexpressed in gastric cancer, and they serve as possible markers of tumorigenesis, malignant phenotype, tumor immunity, resistance to apoptosis, and poor prognosis [27]. Several CYP family members seemed to be downregulated after DEN administration; these include 3A1 and 2C11, which have been previously reported to be downregulated most likely as the result of tissue damage produced by DEN [28]. CYP down-regulation is expected in the response to inflammation and infection. As consequence of generalized necrosis observed at 24 hours after DEN, the inflammation may have contributed to CYP down-regulation, as previously reported for CYP isoforms such as 2C11, 3A2, and 2E1 [29,30]. In addition, several microglobulins and macroglobulins were downregulated, specifically α-2µ-globulins that might be involved in increased DEN-mediated proliferation as previously reported [31].
When CAPE was administered alone, it did not modify gene expression; but when DEN was administered after CAPE, a dramatic response was obtained when compared with that produced by the carcinogen alone. This induction of a protective phenotype by CAPE is similar to a previously observed differentiation of effects between normal and altered cells, where CAPE produced radiosensitization in lung cancer [12], CT26 colorectal adenocarcinoma cells [13], and preferential cytotoxicity in several cancer cells but not in normal ones [15]. This intriguing effect of CAPE should be further investigated.
It would seem that tumor initiation by DEN induces changes in GEP. These changes probably influence protein modifications. CAPE given before DEN interferes with DEN-mediated expression changes in genes involved in DEN bioactivation, oxidative stress regulation, DNA repair and replication, among others. The effect of CAPE on this process can be used as a tool to suggest new mechanisms for DEN initiation process in the modified resistant hepatocyte model as well as to uncover possible molecular targets for liver cancer chemoprotection.
Supplementary Material
Acknowledgments
The authors thank the technical support of Jorge T. Ayala-Sumuano and Jaime Arellanes-Robledo, animal technical support of UPEAL-Cinvestav from M.V.Z María Antonieta López-López, Rafael Leyva-Muñoz, Manuel Flores-Cano, Ricardo Gaxiola-Centeno, and UPEAL Chairman Jorge Fernández.
Footnotes
The authors thank CONACYT for funding (39525-M) and a scholarship (OBR 185604).
This article refers to supplementary materials, which are designated by Tables W1 to W6 and are available online at www.transonc.com.
References
- 1.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 2.Verna L, Whysner J, Williams GM. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol Ther. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 3.Semple-Roberts E, Hayes MA, Armstrong D, Becker RA, Racz WJ, Farber E. Alternative methods of selecting rat hepatocellular nodules resistant to 2-acetylaminofluorene. Int J Cancer. 1987;40:643–645. doi: 10.1002/ijc.2910400512. [DOI] [PubMed] [Google Scholar]
- 4.Carrasco-Legleu CE, Marquez-Rosado L, Fattel-Fazenda S, Arce-Popoca E, Perez-Carreon JI, Villa-Trevino S. Chemoprotective effect of caffeic acid phenethyl ester on promotion in a medium-term rat hepatocarcinogenesis assay. Int J Cancer. 2004;108:488–492. doi: 10.1002/ijc.11595. [DOI] [PubMed] [Google Scholar]
- 5.Beltran-Ramirez O, Aleman-Lazarini L, Salcido-Neyoy M, Hernandez-Garcia S, Fattel-Fazenda S, Arce-Popoca E, Arellanes-Robledo J, García-Román R, Vázquez-Vázquez P, Sierra-Santoyo A, et al. Evidence that the anticarcinogenic effect of caffeic acid phenethyl ester in the resistant hepatocyte model involves modifications of cytochrome P450. Toxicol Sci. 2008;104:100–106. doi: 10.1093/toxsci/kfn071. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Perez Y, Carrasco-Legleu C, Garcia-Cuellar C, Perez-Carreon J, Hernandez-Garcia S, Salcido-Neyoy M, Alemán-Lazarini L, Villa-Treviño S. Oxidative stress in carcinogenesis. Correlation between lipid peroxidation and induction of preneoplastic lesions in rat hepatocarcinogenesis. Cancer Lett. 2005;217:25–32. doi: 10.1016/j.canlet.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 7.Cameron RG. Identification of the putative first cellular step of chemical hepatocarcinogenesis. Cancer Lett. 1989;47:163–167. doi: 10.1016/0304-3835(89)90086-4. [DOI] [PubMed] [Google Scholar]
- 8.Becker RA, Lu SS, Bresil H, Shank RC, Montesano R. DNA ethylation in target and non-target organs of hamsters and rats treated with diethylnitrosamine. Cancer Lett. 1985;26:17–24. doi: 10.1016/0304-3835(85)90168-5. [DOI] [PubMed] [Google Scholar]
- 9.Ying TS, Sarma DS, Farber E. Role of acute hepatic necrosis in the induction of early steps in liver carcinogenesis by diethylnitrosamine. Cancer Res. 1981;41:2096–2102. [PubMed] [Google Scholar]
- 10.Carrasco-Legleu CE, Sanchez-Perez Y, Marquez-Rosado L, Fattel-Fazenda S, Arce-Popoca E, Hernandez-Garcia S, Villa-Treviño S. A single dose of caffeic acid phenethyl ester prevents initiation in a medium-term rat hepatocarcinogenesis model. World J Gastroenterol. 2006;12:6779–6785. doi: 10.3748/wjg.v12.i42.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez-Carreon JI, Lopez-Garcia C, Fattel-Fazenda S, Arce-Popoca E, Aleman-Lazarini L, Hernandez-Garcia S, Le Berre V, Sokol S, Francois JM, Villa-Treviño S. Gene expression profile related to the progression of preneoplastic nodules toward hepatocellular carcinoma in rats. Neoplasia. 2006;8:373–383. doi: 10.1593/neo.05841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Chen MF, Wu CT, Chen YJ, Keng PC, Chen WC. Cell killing and radiosensitization by caffeic acid phenethyl ester (CAPE) in lung cancer cells. J Radiat Res (Tokyo) 2004;45:253–260. doi: 10.1269/jrr.45.253. [DOI] [PubMed] [Google Scholar]
- 14.Chen YJ, Liao HF, Tsai TH, Wang SY, Shiao MS. Caffeic acid phenethyl ester preferentially sensitizes CT26 colorectal adenocarcinoma to ionizing radiation without affecting bone marrow radioresponse. Int J Radiat Oncol Biol Phys. 2005;63:1252–1261. doi: 10.1016/j.ijrobp.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Chen YJ, Shiao MS, Hsu ML, Tsai TH, Wang SY. Effect of caffeic acid phenethyl ester, an antioxidant from propolis, on inducing apoptosis in human leukemic HL-60 cells. J Agric Food Chem. 2001;49:5615–5619. doi: 10.1021/jf0107252. [DOI] [PubMed] [Google Scholar]
- 16.Grunberger D, Banerjee R, Eisinger K, Oltz EM, Efros L, Caldwell M, Estevez V, Nakanishi K. Preferential cytotoxicity on tumor cells by caffeic acid phenethyl ester isolated from propolis. Experientia. 1988;44:230–232. doi: 10.1007/BF01941717. [DOI] [PubMed] [Google Scholar]
- 17.Na HK, Wilson MR, Kang KS, Chang CC, Grunberger D, Trosko JE. Restoration of gap junctional intercellular communication by caffeic acid phenethyl ester (CAPE) in a ras-transformed rat liver epithelial cell line. Cancer Lett. 2000;157:31–38. doi: 10.1016/s0304-3835(00)00470-5. [DOI] [PubMed] [Google Scholar]
- 18.Natarajan K, Singh S, Burke TR, Jr, Grunberger D, Aggarwal BB. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-κB. Proc Natl Acad Sci USA. 1996;93:9090–9095. doi: 10.1073/pnas.93.17.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Son S, Lewis BA. Free radical scavenging and antioxidative activity of caffeic acid amide and ester analogues: structure-activity relationship. J Agric Food Chem. 2002;50:468–472. doi: 10.1021/jf010830b. [DOI] [PubMed] [Google Scholar]
- 20.Song YS, Park EH, Hur GM, Ryu YS, Lee YS, Lee JY, Kim YM, Jin C. Caffeic acid phenethyl ester inhibits nitric oxide synthase gene expression and enzyme activity. Cancer Lett. 2002;175:53–61. doi: 10.1016/s0304-3835(01)00787-x. [DOI] [PubMed] [Google Scholar]
- 21.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 22.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 24.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 25.Maga G, Villani G, Tillement V, Stucki M, Locatelli GA, Frouin I, Spadari S, Hübscher U. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc Natl Acad Sci USA. 2001;98:14298–14303. doi: 10.1073/pnas.251193198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueda H, Ullrich SJ, Gangemi JD, Kappel CA, Ngo L, Feitelson MA, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41–47. doi: 10.1038/ng0195-41. [DOI] [PubMed] [Google Scholar]
- 27.Loesch M, Chen G. The p38 MAPK stress pathway as a tumor suppressor or more? Front Biosci. 2008;13:3581–3593. doi: 10.2741/2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao ZG, Shen WL. Heat shock protein 70 antisense oligonucleotide inhibits cell growth and induces apoptosis in human gastric cancer cell line SGC-7901. World J Gastroenterol. 2005;11:73–78. doi: 10.3748/wjg.v11.i1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riddick DS, Lee C, Bhathena A, Timsit YE, Cheng PY, Morgan ET, Prough RA, Ripp SL, Miller KK, Jahan A, et al. Transcriptional suppression of cytochrome P450 genes by endogenous and exogenous chemicals. Drug Metab Dispos. 2004;32:367–375. doi: 10.1124/dmd.32.4.367. [DOI] [PubMed] [Google Scholar]
- 30.Cheng PY, Wang M, Morgan ET. Rapid transcriptional suppression of rat cytochrome P450 genes by endotoxin treatment and its inhibition by curcumin. J Pharmacol Exp Ther. 2003;307:1205–1212. doi: 10.1124/jpet.103.057174. [DOI] [PubMed] [Google Scholar]
- 31.Matuoka K, Markus I, Wong A, Smith GJ. Diethylnitrosamine- and partial hepatectomy-induced decrease in α 2µ-globulin mRNA level in the rat liver. J Cancer Res Clin Oncol. 1993;119:572–575. doi: 10.1007/BF01372719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.