Abstract
The protective antigen component of anthrax toxin binds the I domain of the anthrax toxin receptors, ANTXR1 and ANTXR2, in a manner akin to how integrins bind their ligands. The I domains of integrins and ANTXR1 both have high and low affinity conformations and the cytosolic tails of these receptors associate with the actin cytoskeleton. The association of ANTXR1 with the cytoskeleton correlates with diminished binding to PA, although a mechanistic explanation for this observation is lacking. Here, we identified a segment in the cytoplasmic tail of ANTXR1 required for its association with the cytoskeleton. We synthesized a 60-mer peptide based on this segment and demonstrated a direct interaction between the peptide and β-actin, indicating that in contrast to integrins, ANTXR1 does not use an adaptor to bind the cytoskeleton. This peptide orders actin filaments into arrays, demonstrating an actin-bundling activity that is novel for a membrane protein.
Keywords: ANTXR1, anthrax toxin, cytoskeleton, bundling
Anthrax toxin consists of protective antigen (PA), edema factor (EF), and lethal factor (LF). PA binds the anthrax toxin receptors, ANTXR1 and ANTXR2, and delivers the enzymatic components of the toxin, EF and LF, into the cell where they exert their toxic activities (1). EF is an adenylate cyclase and LF is a zinc metalloprotease that cleaves most mitogen-activated protein kinase kinases (MAPKKs) (2, 3).
ANTXR1 and ANTXR2 are Type I membrane proteins that exhibit a high degree of similarity (1). PA binds the extracellular von Willebrand Factor type A or integrin inserted (VWA/I) domain of each receptor. The I domains also bind the natural ligands of the receptors: collagen type I and VI for ANTXR1, laminin and collagen IV for ANTXR2 (4-6). The receptors are expressed widely, although their physiological functions have not been fully elucidated. Recently, however, it has been shown that ANTXR1 and ANTXR2 knock-out female mice are unable to reproduce (7). Another study showed that ANTXR1 knock-out mice accumulate excessive extracellular matrix in several tissues, including the ovaries and uterus (8). Mutations in ANTXR2 are associated with the human diseases juvenile hyaline fibromatosis and infantile systemic hyalinosis, which are characterized by the accumulation of extracellular matrix and contractures of the joints (9). Cumulatively, these results suggest that the receptors are cell adhesion molecules involved in extracellular matrix homeostasis.
ANTXR1 has been shown to mediate cell spreading on collagen I by a process that is dependent on an interaction between its cytoplasmic tail and the actin cytoskeleton (10). We previously demonstrated that the linkage between the cytoplasmic domain of ANTXR1 and the actin cytoskeleton affected the binding of PA (11). Cells that expressed a splice variant of ANTXR1 that associated with actin (splice variant 1, sv1) bound markedly less PA than cells that expressed a shorter variant that did not associate with actin (sv2) (11). In addition, introduction of the Y383C point mutation into the cytoplasmic tail of ANTXR1-sv1, which is synonymous to a disease-causing mutation in ANTXR2, disrupted receptor-cytoskeleton association and increased binding of PA (11). These results suggest that ANTXR1 possesses different affinity states that may be modulated by cytoplasmic interactions. This notion is reminiscent of what has been observed for integrins, which can shift from low to high affinity conformations as a result of how adaptor proteins interact with their cytoplasmic tails (12).
Here we have delimited a short region within the ANTXR1 cytoplasmic domain required for its association with the cytoskeleton. We synthesized a peptide based on this region and found that it interacted with β-actin in HeLa cells. Furthermore, this peptide bound to purified monomeric β-actin in vitro, indicating that the interaction between ANTXR1 and the actin cytoskeleton is direct. In addition, we show that ANTXR1 is capable of organizing actin filaments into bundles.
Experimental Procedures
Cell culture
HeLa cells were maintained in HG-DME medium supplemented with 10% fetal bovine serum (Invitrogen) and 1× penicillin-streptomycin. HeLa cells were transfected with polyethyleneimine (PEI) at a 5 to 1 PEI:DNA ratio in HG-DME medium.
Plasmids
The ANTXR1-sv1-HA and ANTXR1-sv2-HA plasmids have been described previously (11). QuickChange site-directed mutagenesis (Stratagene) was used to introduce triple alanine mutations between amino acids 370-420 of ANTXR1-sv1 (Supplementary Information Table 1). The pEGFP-ANTXR1360-420 and pEGFP-ANTXR1360-420(Y383C) plasmids were made by amplifying amino acids 360-420 from the previously described plasmids, ANTXR1-sv1-HA and ANTXR1-sv1-Y383C-HA (11), respectively, using the forward primer 5′-GCGAAGCTTCGGAGGAGAG TGAGGAAGAAG-3′ and the reverse primer 5′-CAAAGAATGCAAGAGTCAAGATGTAGGCATCC-3′. The PCR products were digested with HindIII and BamHI, and ligated into pEGFP-C1 (BD Biosciences Clontech).
Actin association assays
HeLa cells transiently transfected with wildtype ANTXR1 (sv1 and sv2) and ANTXR1 mutants were washed and scraped in PHEM buffer (60mM PIPES, 25mM HEPES, 10mM EGTA, and 2mM MgCl2: pH 6.9). Cells were lysed in PHEM buffer + 0.15% Triton X-100 for 12 minutes at 4° C. Following lysis, cells were centrifuged at 16,000 g for 30 min at 4° C. The soluble fraction was collected, and the pellet (insoluble fraction) was re-suspended in equal volumes of PHEM+0.15% Triton X-100 buffer. SDS sample buffer was added to both fractions, boiled, and equal volumes of each were subjected to SDS-PAGE and Western blotting. Blots were probed with anti-HA antibodies (Santa Cruz Cat# E1608) and visualized using a Kodak Image Station 400MM Pro. The intensity of the bands were quantified and the ANTXR1 ratio in the pellet to supernatant was determined.
Fluorescence microscopy
HeLa cells were seeded on coverslips and transfected with pEGFP-ANTXR1360-420, pEGFP-ANTXR1360-420(Y383C), or pEGFP plasmids. Cells were washed twice in phosphate buffered saline (PBS), and fixed in 4% paraformaldehyde in PBS for 15 min at room temperature. Cells were then washed 2 more times with PBS, permeabilized with 0.2% Triton-X 100 in PBS for 5 min, washed 4 times with PBS, and blocked in 5% BSA in PBS for 30 min at room temperature. Cells were stained with 5 units/mL Alexa555-Phalloidin (Molecular Probes) in 5% BSA for 1 h in the dark at room temperature. Following staining, cells were washed 3 times with PBS and mounted on glass slides. To visualize actin and EGFP signals, conventional fluorescence microscopy was performed on Zeiss Axioplan 2 and the images compiled using AxioVision LE software.
ANTXR1 peptides and actin preparations
The peptide TAIL360-420 containing amino acids 360-420 of human ANTXR1-sv1 and an amino-terminal biotin tag was synthesized (AnaSpec Inc). TAIL360-420(Y383A) is identical to TAIL360-420 except for a tyrosine to alanine substitution corresponding to ANTXR1-sv1 position 383. TAIL360-420 and TAIL360-420(Y383A) were incubated with streptavidin agarose resin at a concentration of 4 mg peptide/1 mL resin and rotated for 30 min at room temperature in EBC buffer (50 mM Tris pH 8, 120mM NaCl, 0.5% (v/v) NP-40, 50 μg/mL phenylmethylsulfonyl fluoride). Peptide-bound streptavidin agarose beads were centrifuged at 1500 g for 5 min and resuspended in equal volumes of EBC buffer.
For ANXTR1 peptide experiments and native gel electrophoresis, purified human β-actin (Cytoskeleton Inc., Cat. # APHL95) was diluted and depolymerized in general actin buffer (0.2 mM CaCl2, 5 mM Tris pH 8, supplemented with 0.2 mM ATP) at a concentration of 0.5 mg/mL for 45 min at room temperature, and centrifuged at 16,000 g for 30 minutes to remove residual nucleating centers. For electron microscopy experiments, 0.5 mg/mL β-actin in general actin buffer was depolymerized as above, followed by addition of 10× actin polymerization buffer (Cytoskeleton Inc., Cat. # BSA02: 500mM KCl, 20mM MgCl2, 10mM ATP) diluted to a 1× concentration, and incubated for 1 h at room temperature to stimulate filament formation.
ANTXR1 peptide experiments
HeLa cells were washed three times and scraped in PBS. Cells were lysed in EBC buffer by rotating for 1 hour at 4°C, and lysates were cleared by centrifugation, followed by determination of protein concentrations by the Bradford assay. Either 50 μL of TAIL360-420 or TAIL360-420(Y383A) peptide-bound strepatividin agarose (50% slurry) was added to 2 mg of cell lysates and rotated for 2 h at 4°C. After incubation, beads were washed three times in EBC buffer, and proteins were eluted using SDS loading dye. Eluted proteins were subjected to SDS-PAGE and Western blotting. Blots were probed for β-actin (Sigma Cat# A5441). To ensure equal levels of each peptide were used, blots were stripped and re-probed with anti-streptavidin HRP conjugated antibodies (Pierce Cat# 21126) to detect the biotin-labeled peptides.
To investigate direct binding between ANTXR1 peptides and β-actin, 50 μL peptide-bound streptavidin agarose was incubated with 5 μg of monomeric β-actin and rotated for 2 h at 4°C. Beads were then washed three times with EBC buffer and proteins were eluted and subjected to SDS-PAGE and Western blotting for β-actin as described above.
Native gel electrophoresis
Monomeric β-actin (10 μM) was incubated alone or with 2 μM, 10 μM, or 50 μM TAIL360-420 or TAIL360-420(Y383A) for 15 min in a total volume of 12 μL. The mixtures were run on a 4-20% polyacrylamide gel (BioRad) and proteins were visualized by SimplyBlue SafeStain (Invitrogen).
Electron microscopy
β-actin filaments polymerized from 10 μM monomeric actin were incubated with or without 10 μM TAIL360-420 or TAIL360-420(Y383A) in general actin buffer for 30 min at room temperature. After incubation, samples were applied to carbon-coated 0.25% formvar films (in dichloroethane), and negatively stained with 2% phosphotungstate acid (PTA) in dH20, pH 6.9. Electron micrographs were obtained using a Hitachi H-7000 TEM in the Microscopy Imaging Laboratory at the University of Toronto.
Results
Triple alanine substitutions were introduced into ANTXR1-sv1 within a 50 amino acid cytoplasmic region, which is conserved between ANTXR1-sv1 and ANTXR2, to define the region within the cytoplasmic tail of ANTXR1-sv1 that mediates its interaction with the actin cytoskeleton. HeLa cells were transiently transfected with wild-type and mutant ANXTR1 HA-tagged fusion proteins. The cells were lysed and sedimented such that cytoskeleton-associated receptors were predominantly pelleted and receptors not associated with the cytoskeleton remained in the supernatant. We have used this assay previously to demonstrate that ANTXR1-sv1 associates with the actin cytoskeleton, whereas ANTXR1-sv2 does not (11). ANTXR1-sv1 mutants containing triple alanine substitutions between amino acids 379-411 were found predominantly in the supernatant, suggesting that this 33 amino acid region is necessary for the association of ANTXR1-sv1 with the actin cytoskeleton (Fig. 1). Substitution of amino acids 373-375 or 376-378 with alanines had a more modest effect – the two mutant receptors were almost equally partitioned between the pellet and supernatant fractions. In contrast, flanking mutations (370-372, 412-414, 415-417, 418-420) did not alter the sedimentation properties of the receptor.
Figure 1.
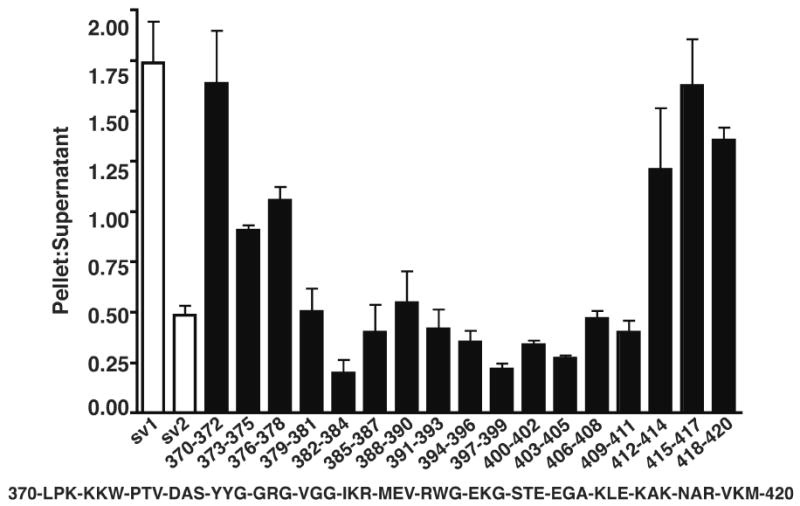
Mutations in the cytoplasmic domain of ANTXR1-sv1 impair cytoskeleton association. ANTXR1-sv1-HA and ANTXR1-sv2-HA (white bars) and ANTXR1-sv1-HA triple alanine mutants (black bars) were individually expressed in Hela cells. The cells were lysed and the cellular lysates were centrifuged for 30 min. The ratio of the amount of receptor that sedimented to the amount that remained in the supernatant was quantified. The sequence of a segment of the ANTXR1-sv1 cytoplasmic tail is indicated. Error bars indicate standard error of the mean of three independent experiments.
To determine whether this region of ANTXR1-sv1 was sufficient to interact with the actin cytoskeleton, we transiently expressed in HeLa cells the EGFP fusion proteins EGFP-ANTXR1360-420 and EGFP-ANTXR1360-420(Y383C) and stained the cells with Alexa555-Phalloidin to visualize actin. EGFP-ANTXR1360-420 localized to actin-rich regions of the cell including protrusions that resembled filopodia (Fig. 2). In contrast, EGFP and EGFP-ANTXR1360-420(Y383C), which contains a mutation that disrupts the interaction between ANTXR1-sv1 and the cytoskeleton (11), did not localize to the actin-rich periphery of the cell. These results indicate that amino acids 360-420 of ANTXR1-sv1 are sufficient to bind the cytoskeleton.
Figure 2.
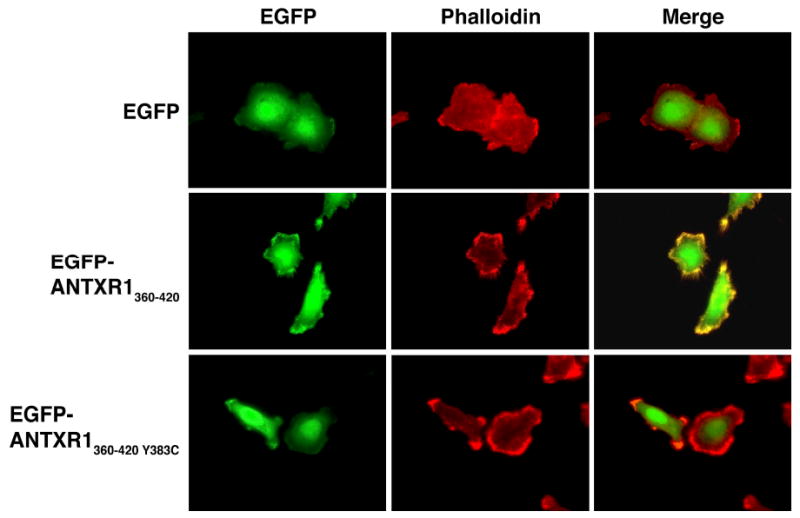
ANTXR1360-420 localizes to actin-rich regions of the cell. HeLa cells were transfected with pEGFP, pEGFP-ANTXR1360-420, or pEGFP-ANTXR1360-420(Y383C) (left panels, green) and stained with Alexa555-Phalloidin to visualize actin (middle panels, red). Right panels show merged images with colocalization indicated in yellow. Images are representative of ∼25 cells from four independent experiments.
We next synthesized peptides based on this region, TAIL360-420, and TAIL360-420(Y383A). A biotin tag was incorporated at the N-terminus of the peptides to facilitate binding to streptavidin agarose resin. The peptide-bound resin was incubated with HeLa cell lysates and the associated proteins were subjected to SDS-PAGE and Western blotting with an anti-β-actin antibody. We observed that TAIL360-420 precipitated β-actin from cellular lysates, whereas TAIL360-420(Y383A) did not (Fig. 3A).
Figure 3.
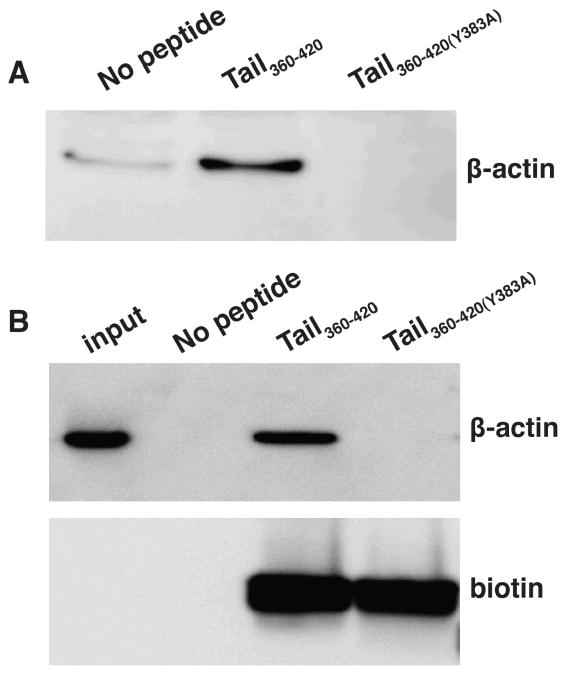
A segment of the ANTXR1-sv1 cytoplasmic domain binds actin. Peptides TAIL360-420 and TAIL360-420(Y383A) were bound to streptavidin beads and incubated with (A) HeLa cell lysates or (B) with purified β-actin. The beads were pelleted and the associated proteins were subjected to SDS-PAGE and tranferred to nitrocellulose. Blots were probed for biotin, to detect the peptides, or for β-actin. Blots are representative of three independent experiments.
We next tested whether TAIL360-420 bound actin directly by incubating the peptide with purified human β-actin. This mixture was centrifuged and the precipitated proteins were subjected to SDS-PAGE. We observed that TAIL360-420, but not TAIL360-420(Y383A), bound actin (Fig. 3B). To further establish the direct interaction between actin and this cytoplasmic region of ANTXR1, we performed native gel electrophoresis using various concentrations of the ANTXR1 peptides incubated in the presence or absence of purified human β-actin. β-actin alone was visualized as a smear in the gel, which progressively decreased in intensity when actin was incubated with increasing concentrations of TAIL360-420 (Fig. 4). The decrease in the actin smear was accompanied by the appearance of a band that only just entered the gel, which could represent a large protein complex. In contrast, no detectable change in the mobility of actin was observed when actin was incubated with TAIL360-420(Y383A) (Fig. 4).
Figure 4.
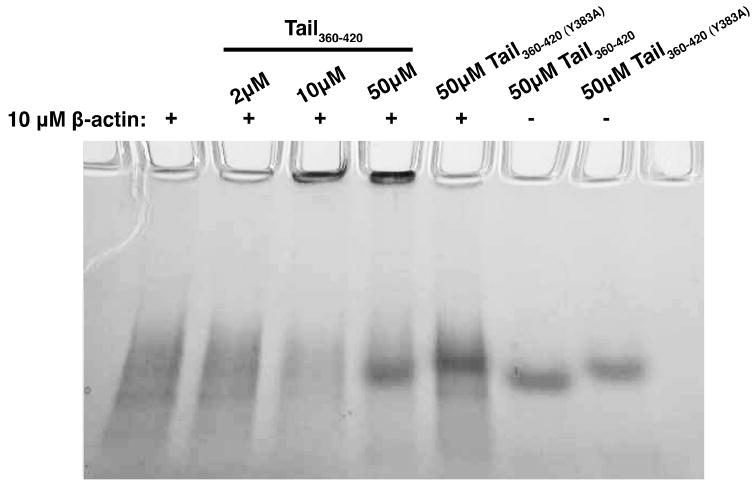
Association of β-actin with TAIL360-420. Combinations of β-actin, TAIL360-420 and TAIL360-420(Y383A) were subjected to native gel electrophoresis. Proteins were visualized with SimplyBlue SafeStain. The gel is representative of thee independent experiments.
As the native gel electrophoresis data suggested large complexes could be forming between actin and TAIL360-420, we were interested in whether this peptide promoted actin filament polymerization. As determined by transmission electron microscropy, incubation of monomeric β-actin with TAIL360-420 led to the formation of disorganized actin aggregates, which were absent when β-actin was incubated in the presence of TAIL360-420(Y383A) (data not shown). This observation suggested that TAIL360-420 does not stimulate polymerization, but might crosslink actin monomers.
To test the crosslinking ability of the peptides, we next incubated β-actin filaments (Fig. 5A) with TAIL360-420 and observed by electron microscopy that the filaments became organized into side-by-side arrays or bundles (Fig. 5B); we observed no effect on the arrangement of filaments incubated with TAIL360-420(Y383A) (Fig. 5C). These data suggest this region of the cytoplasmic tail of ANTXR1 bundles filamentous actin.
Figure 5.
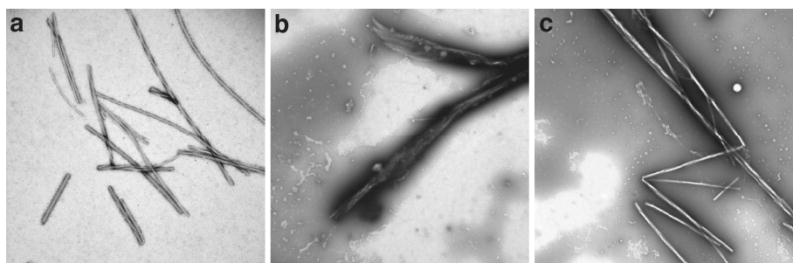
The peptide TAIL360-420 bundles actin filaments. Electron micrographs of β-actin filaments incubated alone (a), or with TAIL360-420 (b), or TAIL360-420(Y383A) (c). Images were taken at 20,000 times magnification, and scale bar represents 500 nm. Micrographs are representative of three independent experiments.
Discussion
Here we provide evidence for a direct interaction between the anthrax toxin receptor ANTXR1-sv1 and the actin cytoskeleton. There have been few reports demonstrating direct interactions between the cytoskeleton and single transmembrane domain receptors. The epidermal growth factor receptor (EGFR) and the Coxsackie and adenovirus receptor (CAR) are notable exceptions (13, 14). It appears, however, that most membrane proteins that bind the cytoskeleton directly are ion channels. The epithelial cell ClC-2 chloride channel is directly linked to the actin cytoskeleton and this linkage inhibits channel activity (15). Binding of the epithelial sodium channel, ENaC, to the cytoskeleton activates the channel to allow Na2+ influx into the cell; elevated intracellular Na2+ levels then trigger actin to dissociate from ENaC, thereby downregulating channel activity (16).
Our previous work revealed an inverse correlation between the binding of ANTXR1 to the cytoskeleton and the amount of PA bound to receptors on cells, which suggested that the association of the cytoplasmic tail with the cytoskeleton converts the extracellular I domain into a closed conformation. This process might occur in a manner similar to how signals from the cytoplasm are transmitted to the I domains of integrins: the cytoplasmic domains of integrin heterodimers are pulled apart to disrupt the interaction between the two transmembrane domains, which serves to transmit the signal across the plasma membrane (17). A difference between the ANTXR1 and integrin activation models appears to be that integrins do not bind the cytoskeleton directly and rely on adaptor proteins to mediate inside-out signaling. Whether it is the binding of the cytoplasmic tail of ANTXR1 to actin filaments or bundles that affects I domain conformation is not clear.
Actin bundling proteins organize actin filaments into ordered arrays that play roles in cell shape, adhesion, and motility (18). For example, fascin organizes actin filaments to form cell-protruding structures called filopodia and α-actinin increases the stiffness of stress fibres by bundling filaments (19). ANTXR1 and ANTXR2, being ubiquitous membrane proteins, may have distinct activities compared to previously described bundling proteins.
Proteins that organize actin into bundles can possess two distinct actin binding domains or contain a single actin binding domain and a dimerization domain (18). We performed sedimentation equilibrium ultracentrifugation to determine the molecular weights of TAIL360-420 and TAIL360-420(Y383A) and found that the molecular weight of each peptide was within 1% of their predicted monomeric molecular weights (data not shown). This result suggests that the TAIL360-420 peptide does not form a dimer, so it may have two distinct actin-binding surfaces that facilitate actin crosslinking. It should be noted, however, that monomeric proteins possessing a single actin-binding domain could induce bundles by facilitating packing of actin filaments (20). Further work is needed to determine how the ANTXR1-sv1 cytoplasmic tail bundles actin.
Supplementary Material
Acknowledgments
This research was supported by NIH grant RO1 AI067683.
Abbreviations
- ANTXR
anthrax toxin receptor
- PA
protective antigen
- sv1
splice variant 1
Footnotes
Supporting Information Available: Supporting information contains the sequences of the oligonucleotides used for site directed mutagenesis. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Young JA, Collier RJ. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 2.Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci U S A. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 4.Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA, Davis GE. Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J Cell Sci. 2001;114:2755–2773. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]
- 5.Hotchkiss KA, Basile CM, Spring SC, Bonuccelli G, Lisanti MP, Terman BI. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell-matrix interactions on collagen. Exp Cell Res. 2005;305:133–144. doi: 10.1016/j.yexcr.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 6.Nanda A, Carson-Walter EB, Seaman S, Barber TD, Stampfl J, Singh S, Vogelstein B, Kinzler KW, St Croix B. TEM8 interacts with the cleaved C5 domain of collagen alpha 3(VI) Cancer Res. 2004;64:817–820. doi: 10.1158/0008-5472.can-03-2408. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Crown D, Miller-Randolph S, Moayeri M, Wang H, Hu H, Morley T, Leppla SH. Capillary morphogenesis protein-2 is the major receptor mediating lethality of anthrax toxin in vivo. Proc Natl Acad Sci U S A. 2009;106:12424–12429. doi: 10.1073/pnas.0905409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen M, Seaman S, Chaudhary A, Yang MY, Hilton MB, Logsdon D, Haines DC, Tessarollo L, St Croix B. Host-derived tumor endothelial marker 8 promotes the growth of melanoma. Cancer Res. 2009;69:6021–6026. doi: 10.1158/0008-5472.CAN-09-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanks S, Adams S, Douglas J, Arbour L, Atherton DJ, Balci S, Bode H, Campbell ME, Feingold M, Keser G, Kleijer W, Mancini G, McGrath JA, Muntoni F, Nanda A, Teare MD, Warman M, Pope FM, Superti-Furga A, Futreal PA, Rahman N. Mutations in the gene encoding capillary morphogenesis protein 2 cause juvenile hyaline fibromatosis and infantile systemic hyalinosis. Am J Hum Genet. 2003;73:791–800. doi: 10.1086/378418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner E, Kowalczyk AP, Faundez V. Anthrax toxin receptor 1/tumor endothelium marker 8 mediates cell spreading by coupling extracellular ligands to the actin cytoskeleton. J Biol Chem. 2006;281:23227–23236. doi: 10.1074/jbc.M603676200. [DOI] [PubMed] [Google Scholar]
- 11.Go MY, Chow EM, Mogridge J. The cytoplasmic domain of anthrax toxin receptor 1 affects binding of the protective antigen. Infect Immun. 2009;77:52–59. doi: 10.1128/IAI.01073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 13.den Hartigh JC, van Bergen en Henegouwen PM, Verkleij AJ, Boonstra J. The EGF receptor is an actin-binding protein. J Cell Biol. 1992;119:349–355. doi: 10.1083/jcb.119.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang KC, Yasruel Z, Guerin C, Holland PC, Nalbantoglu J. Interaction of the Coxsackie and adenovirus receptor (CAR) with the cytoskeleton: binding to actin. FEBS Lett. 2007;581:2702–2708. doi: 10.1016/j.febslet.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed N, Ramjeesingh M, Wong S, Varga A, Garami E, Bear CE. Chloride channel activity of ClC-2 is modified by the actin cytoskeleton. Biochem J. 2000;352(Pt 3):789–794. [PMC free article] [PubMed] [Google Scholar]
- 16.Mazzochi C, Benos DJ, Smith PR. Interaction of epithelial ion channels with the actin-based cytoskeleton. Am J Physiol Renal Physiol. 2006;291:F1113–1122. doi: 10.1152/ajprenal.00195.2006. [DOI] [PubMed] [Google Scholar]
- 17.Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Winder SJ, Ayscough KR. Actin-binding proteins. J Cell Sci. 2005;118:651–654. doi: 10.1242/jcs.01670. [DOI] [PubMed] [Google Scholar]
- 19.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 20.Tang JX, Ito T, Tao T, Traub P, Janmey PA. Opposite effects of electrostatics and steric exclusion on bundle formation by F-actin and other filamentous polyelectrolytes. Biochemistry. 1997;36:12600–12607. doi: 10.1021/bi9711386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


