Abstract
Atypical meningiomas exhibit heterogeneous clinical outcomes. It is unclear which atypical meningiomas require aggressive multimodality treatment with surgery and radiation therapy versus surgery alone to prevent recurrence. Detailed molecular-genetic characterization of these neoplasms is necessary to better understand their pathogenesis and to identify genetic markers. Oligonucleotide array comparative genomic hybridization was used to identify frequent genetic alterations in 47 primary atypical meningiomas resected at Massachusetts General Hospital between August 1987 and September 2006. Eighty five percent of samples exhibited loss of 22q, including the NF2 gene. The second most frequent regions of loss were confined to the short arm of chromosome 1, particularly 1p33-p36.2 (70%) and 1p13.2 (64%). Other frequent regions of loss, detected in more than 50% of samples, included 14q, 10q, 8q, 7p, 21q, 19, 9q34, and 4p16. Frequent regions of gain were detected along 1q (59%), 17q (44%), 9q34 (30%) and 7q36 (26%). Univariate marker-by-marker analysis of all frequently identified copy number alterations showed potential correlation between gain of 1q and shorter progression free survival. Given the heterogeneous treatment outcomes of atypical meningioma, investigation of large-scale and focal genomic alterations in multi-institutional efforts may help clarify molecular-genetic signatures of clinical utility.
Keywords: atypical meningioma, array comparative genomic hybridization
Introduction
Meningiomas comprise up to 30% of intracranial neoplasms (1–3). Approximately 10–40% of meningiomas correspond to atypical (WHO grade II) and anaplastic (grade III) subtypes, which are associated with less favorable clinical outcomes (4–6). These non-benign subtypes have been associated with two-fold increased relative risk of local failure and four-fold relative excess risk of death (7, 8). Among them, the atypical subtype (grade II) comprises up to 35% of meningiomas (3, 4, 9, 10). By the World Health Organization (WHO) criteria, grade II atypical meningiomas exhibit four or more mitotic figures per ten high-power fields or have at least three other histologic features associated with higher grade (architectural sheeting, necrosis, prominent nucleoli, hypercellularity and high nuclear:cytoplasmic ratio). WHO grade III malignant meningiomas exhibit frank histological malignancy or twenty mitotic figures per 10 high-power fields. According to the 2000 and 2007 WHO classifications, brain invasion does not necessarily imply WHO grade III meningioma; in the absence of frank anaplasia, meningiomas exhibiting brain invasion behave most like atypical meningiomas (11) and correspond to WHO grade II (3). These changes in the WHO grading systems have resulted in increased diagnosis of atypical meningioma (4, 9, 11, 12). For example, upon re-assigning grade based on WHO 2000 criteria versus WHO 1993 criteria, 35% of previously diagnosed grade III meningiomas were re-assigned to grade II whereas few original grade II meningiomas were re-assigned (13). Hence, understanding the clinical significance of the diagnosis of atypical meningioma is assuming increasing importance.
Primary treatment for meningiomas consists of maximal safe surgical resection. Although outcomes for benign meningiomas after surgery are favorable and those for the rarer anaplastic (malignant; grade III) subtype are uniformly poor, outcomes for atypical meningiomas are variable (1, 2, 10). Progression-free survival (PFS) at 5 years after definitive treatment has been reported to be 20%–50% when atypical and anaplastic meningiomas are combined and 38%–62% when atypical meningioma is evaluated alone (1, 2, 14). In addition, atypical meningioma is associated with 57% cause-specific survival at 15 years, compared to 86% for benign meningioma (15). But, in many cases, the significant individual variability in clinical behavior of atypical meningiomas cannot be accounted for by known clinical or pathological variables. Given the broad range of reported outcomes after radical surgery (1, 16) it remains unclear which atypical meningiomas will recur and which patients will benefit from additional treatment options such as adjuvant postoperative radiation therapy. We reasoned that detailed molecular-genetic characterization of these neoplasms could provide a better understanding of their pathogenesis and potentially reveal prognostic and therapeutic markers. In turn, such knowledge could further help guide clinical management and decrease inappropriate treatment, thereby minimizing treatment failure and reducing treatment-related toxicity. We therefore undertook a comprehensive genomic analysis of a series of atypical meningiomas.
Meningiomas are among the most studied human solid tumors by karyotype analysis. Among characterized genetic alterations, loss of an entire chromosome 22 is commonly reported in meningiomas and was among the first recurring cytogenetic alterations ever described in human solid tumors (17). Alteration of the neurofibromatosis type 2 gene (NF2) on 22q12 is associated with 30–60% of all sporadic meningiomas (18). Notably, however, relatively few studies have focused specifically on the genetic characteristics of atypical meningiomas. In addition to loss on 22q, loss of 1p is a second common chromosomal abnormality in atypical meningioma (19–22). Two main target regions on 1p36 and 1p32-p34 have been identified based on high resolution microsatellite markers (23). The putative 8.2 megabase minimal region of deletion on 1p36 includes TP73, CDKN2C, RAD54L and ALPL as potential candidate genes (24–28). The region of deletion on 1p33-34 has been further narrowed to 2.8 megabases using in silico sequence analysis and deletion mapping with microsatellite markers (29, 30). Other regions of chromosomal alteration reported in atypical meningioma include losses of 6q, 10, 14q, 18q and gains of 1q, 9q, 12q, 15q, 17q and 20q (19, 22, 31, 32).
Array-based comparative genomic hybridization (aCGH) detects DNA copy number changes and provides a global assessment of molecular events in the genome (33). Multiple studies using chromosomal CGH have been reported in the meningioma literature (19, 22, 32, 34). However, these studies used fewer samples, lack the improved resolution of aCGH, have not elucidated specific genes or loci associated with chromosomal changes and have not specifically studied large, carefully annotated series of atypical meningioma. Array CGH data can be integrated with underlying genome annotations, allowing identification of associations between clinical parameters, such as progression and death, and candidate tumor suppressor or oncogene loci. The potential clinical utility of aCGH-based studies is maximized with inclusion of tumor samples from patients with substantial clinical follow-up. To improve our understanding of meningioma genetics and to identify potentially useful prognostic markers for use in the setting of atypical meningioma, we studied a large series of atypical meningiomas using a comprehensive aCGH approach.
Materials and Methods
Tumor Samples and Clinical Data
The inclusion criteria for the study were: 1) diagnosis of atypical meningioma on primary resection; 2) frozen tissue in the brain tumor repository, and 3) at least 6 months of clinical follow-up. Exclusion criteria included history of prior brain irradiation and age < 18 years. The Massachusetts General Hospital Brain Tumor Repository contained fresh frozen tumor specimens from 85 cases of atypical meningioma treated surgically between August 1987 and September 2006. Histopathologic diagnosis of atypical meningioma was made by neuropathologists on original paraffin-embedded surgical specimens using WHO criteria: four or more mitotic figures per ten high-power fields; or at least three of five other histologic features (architectural sheeting, necrosis, prominent nucleoli, hypercellularity and high nuclear:cytoplasmic ratio) (35). Presence of atypical meningioma within the banked tissue was confirmed by an independent pathologist using hematoxylin-and-eosin stains of the frozen material. Informed consent for use of tissue was obtained from each patient at the time of resection. Medical records of each patient were reviewed for demographic information, tumor characteristics, treatment details, tumor progression and death under a protocol approved by the Institutional Review Board. Thirty-eight cases were excluded for the following reasons: recurrent tumor (23 cases), history of prior brain irradiation (8 cases), lack of follow-up (6 cases), age < 18 years (1 case). The study included the remaining 47 cases of primary atypical meningioma from the tumor bank (with at least 6 months clinical follow-up). Of the 47 cases, 25 (53%) were men and 22 (47%) were women with a median age at diagnosis of 59 years (range 31–90). The diagnosis of atypical meningioma was confirmed by review of hematoxylin and eosin-stained sections from formalin-fixed, paraffin-embedded tissue sections from each case. Only samples with 80–90% tumor cells were used for DNA extraction. All samples were anonymized and a database with detailed clinical follow-up information was created using File Maker Pro. Radiographic PFS was measured from the date of primary surgery until the date of first documented radiographic recurrence of tumor after gross total resection or growth of residual disease after subtotal resection or death, whichever occurred first. The median radiographic follow-up was 29 months (95% CI 24–55 months). Radiographic progression was seen in 13 patients. The estimated median PFS period for all patients was 56 months (95% CI 35 months-not estimable).
aCGH
Genomic DNA was isolated from 47 primary atypical meningioma samples and from normal whole blood from 10 anonymous donors using routine protocol. Array CGH was performed to determine DNA copy number changes using Agilent Human 105K oligonucleotide microarrays following the manufacturer’s instructions (http://www.home.agilent.com/agilent/home.jspx). Genomic coordinates for this array are based on the NCBI build 36, March 2006 freeze of the assembled human genome (UCSC hg18), available through the UCSC Genome Browser. This array includes a comprehensive probe coverage spanning both coding and non-coding regions, with emphasis on well known genes, promoters, micro RNAs, and telomeric regions and provides an average spatial resolution of 21.7 kb. For array hybridizations, 5 μg each of tumor and normal DNA were digested with Dpn II for 3 hours at 37°C and purified with PureLink PCR purification columns (Invitrogen). Purified tumor and normal DNA, 1 μg each, were labeled with Cy3-dCTP and Cy5-dCTP, respectively, using bioprime labeling kit (Invitrogen) following the manufacturer’s instructions. Unincorporated nucleotides were removed using Sephadex G-50 columns. Labeled tumor and reference samples were precipitated with 100 μg of human Cot-1 DNA and resuspended in 250 μl of hybridization buffer provided in the Agilent oligonucleotide array CGH kit. Prior to hybridization the probe mixtures were denatured for 5 minutes at 95°C and incubated at 37°C for 30 minutes. The samples were then hybridized onto the oligonucleotide array in the Agilent SureHyb microarray hybridization chamber and the hybridization was carried out for 42 hours at 65°C. Arrays were then disassembled and washed as recommended by the manufacturer. Array slides were scanned in Axon 4000B microarray scanner using GenePix Pro 4.0 software. Microarray images were analyzed and data points were generated using the Feature Extraction software (version 9.1, Agilent Technologies) with linear normalization (protocol-v4_91). Data were subsequently imported into CGH Analytics software (version 3.4.40, Agilent Technologies) for visualization.
aCGH data analysis methods
DNA copy number alteration (CNA) was identified through dynamic thresholding of segmented aCGH data. First, circular binary segmentation (CBS) (36) was used to segment each hybridization into regions of common mean. Second, for each hybridization, the scaled median absolute deviation (MAD) across all segments was obtained. A default scaling factor of 1.48 was utilized. Finally, probes assigned to segments with mean value greater than 0.75 MADs were identified as gain. Likewise, probes corresponding to segments with mean value less than 0.75 MADs were identified as loss. Segment classification and choice of thresholds was based on the approach of Korkola et al. (2008) (37).
Statistical analysis
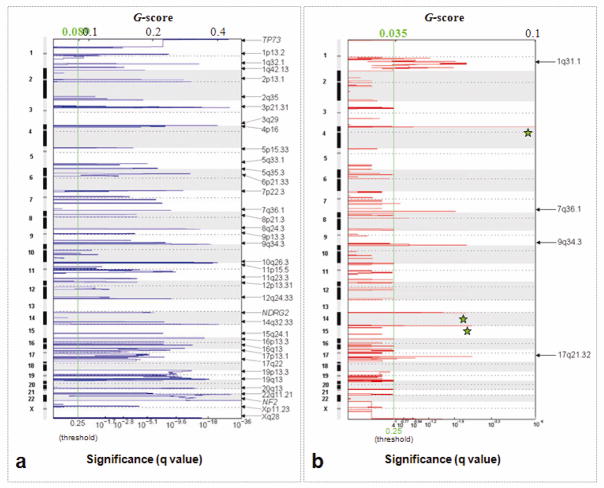
To identify minimal regions of common alteration across all hybridizations, the Genomic Identification of Significant Targets in Cancer (GISTIC) approach was utilized (38). Under this approach, regions from among the CBS identified segments that are aberrant more often than would be expected by chance are identified, with greater weight given to high amplitude events. GISTIC assigns each region of the genome two G-scores (Figure 2), each representing the combined frequency and amplitude of either losses or gains. It then compares these scores with similar scores generated from random permutations of the data to determine false discovery rate q-values (39), representing the likelihood of obtaining the observed G-scores from chance events alone. This approach has been shown to be more likely to identify regions highly associated with cancer pathogenesis (38). Threshold selection for the GISTIC procedure was based, conservatively, on the maximum threshold for alteration (across all hybridizations) identified under the MAD approach described above; 0.4 was selected as the gain and loss threshold and 0.25 was selected as the significance threshold. Each analyzed CBS segment consisted of at least four markers. Segments that contained fewer than four markers were combined with the adjacent segment closest in segment value. A q-value was then obtained for each region. Each peak (i.e., region associated with a low q-value) was tested to determine whether the signal was primarily due to broad events, focal events or overlapping events of both types.
Figure 2.
GISTIC analysis of copy number alterations in atypical meningiomas. The statistical significance of the aberrations identified by GISTIC are displayed as false discovery rate q values to account for multiple hypothesis testing (q values; green line is 0.25 cut-off for significance). Scores for each alteration are plotted along x axis and the genomic positions are plotted along y axis; dotted lines indicate the centromeres. a) Loss of both broad and focal regions are identified by GISTIC (copy number threshold = log2 ratio ≤ 0.4 for broad and ≤ 0.1 for focal events). Sixty five broad regions of losses were identified. Three focal events indicated by the gene names were identified in the background of broad regions. b) GISTIC reveals five broad regions of gain (copy number threshold = log2 ratio ≥ 0.4). Green stars indicate known or presumed copy number polymorphisms.
In an additional attempt to assess common patterns across hybridizations, unsupervised hierarchical clustering of samples was also conducted. Following segmentation and classification, data were further reduced, with out compromising the continuity and breakpoints, to facilitate downstream analyses (40). The log2 ratios from this reduced dataset were then clustered using the Cluster and Treeview program (41). The distance matrix was computed using the Pearson distance and clustered using gplots module within Bioconductor (http://www.r-project.org/).
To assess association of DNA copy number alteration with radiographic progression free survival in primary tumors, univariate marker-by-marker analyses were conducted under the Cox proportional hazards model. The effects of gain and loss were assessed separately. That is, for the analysis evaluating the effect of gains (losses), each marker was classified as either gain or no-gain (loss or no-loss). Assignment of each marker was made according to the CBS-MAD classification (described above) of the CBS segment to which the given marker belonged. Only markers identified as potentially informative were utilized in each analysis. Potentially informative markers were defined as those with some variation in gain or loss across all hybridizations; it was assumed that markers with nearly uniform gain or loss (or absence thereof) across all hybridizations were not potentially informative with regard to progression. Hence, markers whose percentage of gain/loss across all hybridizations exceeded 90% or was less than 10% were omitted from the analysis. To adjust for multiple testing via the false discovery rate, the q-value (distinct from the q-value obtained under the GISTIC analysis described above) for each test was obtained (42).
Results
Patient and tumor characteristics
Between 1987 and 2006, atypical meningiomas comprised 20.9% (171/817) of all surgically resected meningiomas collected in the MGH brain tumor repository. The percentage of atypical meningiomas increased over time, corresponding with refinements in meningioma grading, including WHO classifications in 1993 and 2000: 1987–1993: 17.6% (56/318), 1994–2000: 22.1% (64/290), 2001–2006: 24.4% (51/209). Fifty percent of the atypical meningiomas collected between 1987–2006 had frozen tissue available for evaluation (n=85). The 47 atypical meningioma cases included in this study consisted of 25 males (53%) and 22 females (47%) (Table 1). Ninety percent of patients (42/47) were Caucasian. Median age at diagnosis was 59 years (range, 31–90 years). Seventy-four percent (34/46) of patients had no or only minor comorbidities (comorbidity score 0, 1). Twenty-six percent (12/46) of patients had multiple/major comorbidities (comorbidity scores >1). Information about tumor size was available in the neuroradiology or operative reports for 41/47 cases. Median tumor diameter was 5.5 cm (range, 2–11.6 cm) based on these reports. The majority of tumors were located in the convexity (28%) and parasagittal (28%) regions. Other regions of involvement included the falx (17%), sphenoid (15%), skull base (6%), olfactory groove (2%), orbit (2%) and cerebellopontine angle (CPA; 2%). Gross total resection (GTR) was achieved in 74% of cases (35/47), compared with 26% (12/47) subtotal resection (STR). Bone involvement was documented in 32% of cases (15/47). Radiographic evidence of bone involvement included hyperostosis, bone sclerosis and/or osteolytic lesions. Intraoperative evidence of bone involvement included hyperostosis, extension of tumor mass into bone, and/or bony destruction. Bone samples were sent for histopathologic evaluation in 8/15 cases of bone involvement, with 100% concordance. Brain invasion was noted in only 4% (2/47).
Table 1.
Patient and tumor characteristics.
| Characteristic | Number of patients |
|---|---|
| All | 47 |
| Gender | |
| Male | 25 (53%) |
| Female | 22 (47%) |
| Race | |
| Caucasion | 42 (90%) |
| African American | 1 (2%) |
| Asian | 2 (4%) |
| Hispanic | 2 (4%) |
| Median age | 59 yrs (range, 31–90) |
| Comorbidity Score | |
| 0, 1 | 34 (74%) |
| > 1 | 12 (26%) |
| Median tumor size | 5.5 cm (range, 2 cm – 11.6 cm) |
| Tumor location | |
| Convexity | 13 (28%) |
| Falx | 8 (17%) |
| Sphenoid | 7 (15%) |
| Skull base | 3 (6%) |
| Parasagittal | 13 (28%) |
| Olfactory groove | 1 (2%) |
| Orbit | 1 (2%) |
| CPA | 1 (2%) |
| Resection | |
| GTR | 35 (74%) |
| STR | 12 (26%) |
| Brain invasion | |
| Yes | 2 (4%) |
| No | 45 (96%) |
| Bone involvement | |
| Yes | 15 (32%) |
| No | 32 (68%) |
GTR = Simpson grade I-III. STR = Simpson grade IV.
Global DNA copy number alterations
Genomic copy number for each of over 99,000 probes was determined by calculating the log2 ratio of median signal intensities of the tumor and normal reference DNAs. A genome-wide view of the DNA copy-number alterations (CNAs) in 47 primary atypical meningiomas is shown in Figure 1. Most chromosomal arms undergo either loss or gain across a large proportion of the samples, suggesting high degree aneuploidy in the atypical meningioma genomes (Figure 1). Extensive erosion of telomeric regions was frequently observed for many chromosomes. There were no high level amplification or homozygous deletion events detected in this set of tumors. In order to identify consistent regions of copy-number alterations associated with gains and losses, and to identify minimal regions of loss and gain, we applied the statistical method GISTIC to the data set (figure 2). GISTIC identified 65 regions of losses along 35 chromosome arms (Figure 2a) and 5 regions of gains along 4 chromosome arms (Figure 2b) distributed throughout the genome. Several chromosome arms had more than one minimal region of loss or gain. For each alteration we selected the peak region (ie, the highest frequency and amplitude of events) as the region most likely to contain a cancer gene. Several tumor suppressor genes previously shown to have copy number changes in meningiomas, such as TP73, NDRG2, and NF2, were readily identified by GISTIC. Chromosomal locations, frequencies, genomic intervals, gene contents and candidate cancer genes of these changes are listed in Table 2. The size of deletions ranged from 400kb to 3 Mb and the number of genes mapping to these regions ranged from 1–137 respectively. There were only 5 regions of gains identified with the number of genes ranging from 2–42. Genes with known or possible function in cancer are listed in Table 2.
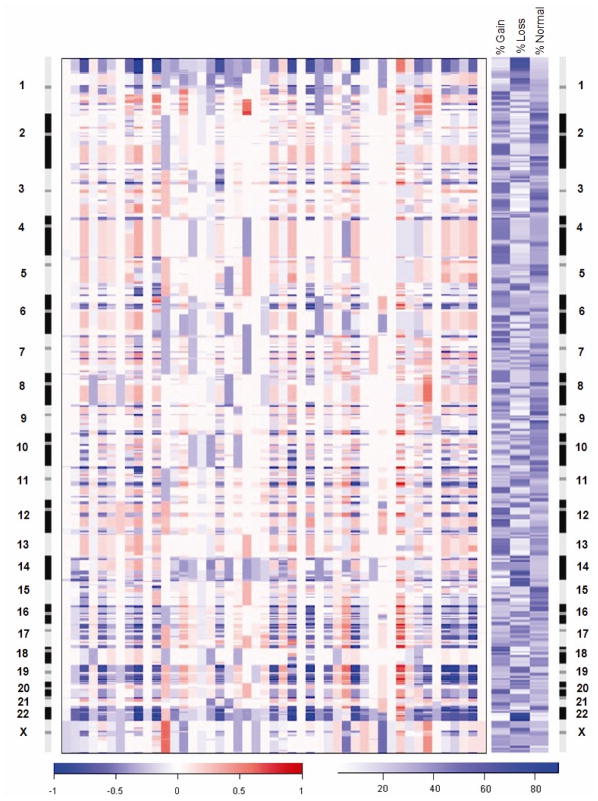
Figure 1.
Genomic profiles of 47 primary atypical meningiomas generated by oligonucleotide array CGH. Each column in the left panel represents a tumor sample and rows represent losses and gains of DNA sequences along the length of chromosomes 1 through X as determined by the segmentation analysis of normalized log2 ratios. The color scale ranges from blue (loss) through white (two copies) to red (gain). The right panel indicates the frequencies of gain and loss of oligonucleotide probes on a probe-by-probe basis for all autosomes and the X chromosome. The color scale ranges from white (no changes) to blue (frequent changes). Loss of 22q12.2 including the NF2 gene was the most frequent alteration observed in 85% of the atypical meningiomas.
Table 2.
Broad and focal regions of losses and gains in atypical meningioma identified by GISTIC analysis.
| Cytoband | Broad or focal | Peak region (Mb) | q value | Frequency (%) | Number of genes in peak | Known cancer genes |
|---|---|---|---|---|---|---|
| Copy Number Losses | ||||||
| 1p36.32 | both | chr1:1810386-2847377(probes 67:111) | 1.8236E-32 | 70 | 18 | TP73 |
| 1p13.2 | focal | chr1:111326775-111897939(probes 3987:4016) | 1.55E-10 | 64 | 11 | |
| 1q32.1 | focal | chr1:200252797-200463129(probes 6396:6404) | 7.53E-16 | 40 | 5 | |
| 1q42.13 | both | chr1:225643390-226726883(probes 7266:7322) | 0.000485 | 40 | 19 | |
| 1q44 | both | chr1:246911252-247249719(probes 8035:8044) | 6.70E-07 | 49 | 6 | |
| 2q35 | broad | chr2:216778698-220212213(probes 14809:14987) | 0.00819 | 47 | 65 | WNT6, CDK5R2 |
| 2q37.3 | broad | chr2:238972240-242951149(probes 15638:15805) | 1.63E-06 | 47 | 40 | |
| 2p13.3 | both | chr2:71595156-72012567(probes 10400:10409) | 0.0004362 | 21 | 1 | |
| 2p13.1 | both | chr2:74536424-74606525(probes 10532:10542) | 2.33E-10 | 32 | 5 | |
| 2q11.2 | broad | chr2:96035700-96341316(probes 11018:11032) | 2.32E-13 | 45 | 9 | |
| 3q29 | broad | chr3:196924341-197049555(probes 22343:22348) | 4.53E-22 | 40 | 2 | |
| 3p21.31 | broad | chr3:50156214-50688743(probes 17620:17660) | 4.13E-17 | 49 | 23 | TUSC2&4, RASSF1 |
| 3p21.1 | broad | chr3:51948378-52059380(probes 17723:17733) | 1.04E-26 | 49 | 9 | PARP3, PCBP4, DUSP7 |
| 4p16.3 | broad | chr4:146101-2164782(probes 22466:22566) | 4.16E-08 | 51 | 35 | |
| 4p16.1 | broad | chr4:5508457-9468999(probes 22705:22864) | 1.60E-05 | 51 | 35 | |
| 5p15.33 | both | chr5:1034536-1570159(probes 28100:28126) | 0.0011558 | 40 | 7 | TERT |
| 5q31.3 | both | chr5:140995024-141299875(probes 32197:32210) | 9.42E-05 | 33 | 5 | PCDH1 |
| 5q33.1 | both | chr5:148181565-150843781(probes 32452:32597) | 0.0043584 | 34 | 35 | PDGFRB, IL17B |
| 5q35.3 | broad | chr5:175886313-177743535(probes 33368:33458) | 3.32E-06 | 40 | 39 | FGFR4 |
| 6p21.33 | broad | chr6:31654033-32398882(probes 34782:34856) | 1.25E-13 | 55 | 55 | |
| 7q36.1 | broad | chr7:150277900-150468342(probes 44309:44322) | 2.27E-09 | 47 | 10 | |
| 7p13 | broad | chr7:43911979-45251561(probes 40723:40789) | 2.39E-05 | 45 | 26 | |
| 7q11.23 | broad | chr7:71945710-76475359(probes 41411:41566) | 4.47E-09 | 50 | 58 | |
| 7p22.3 | broad | chr7:962086-1559228(probes 39157:39190) | 6.43E-12 | 54 | 12 | |
| 7q22.1 | broad | chr7:99327158-102122525(probes 42354:42510) | 3.84E-09 | 52 | 65 | |
| 8q24.3 | broad | chr8:145534920-145756763(probes 49163:49181) | 1.01E-16 | 55 | 17 | VPS28, FOXH1 |
| 8p21.3 | broad | chr8:22009601-22164198(probes 45357:45369) | 0.0001077 | 43 | 9 | |
| 9q32 | both | chr9:115818205-116113463(probes 52044:52060) | 0.0003482 | 21 | 4 | KIF12 |
| 9q34.11 | broad | chr9:129185484-131549479(probes 52562:52702) | 2.34E-08 | 47 | 60 | CDK9, ENG |
| 9q34.3 | broad | chr9:138978944-139324935(probes 53067:53093) | 1.29E-20 | 51 | 28 | |
| 9p13.3 | broad | chr9:34231161-35777161(probes 50268:50363) | 0.0001502 | 42 | 40 | FANCG |
| 10q26.3 | broad | chr10:133846906-135145826(probes 57675:57746) | 1.08E-14 | 55 | 23 | INPP5A |
| 11p15.5 | broad | chr11:1-1515184(probes 57758:57844) | 2.21E-23 | 49 | 49 | HRAS |
| 11q23.3 | broad | chr11:118043425-119502992(probes 61928:62001) | 9.65E-05 | 44 | 29 | |
| 11q12.1 | broad | chr11:56850038-57193055(probes 59612:59634) | 5.27E-05 | 36 | 12 | |
| 11q13.1 | broad | chr11:66780417-67594725(probes 60196:60243) | 2.66E-10 | 49 | 29 | |
| 12q24.33 | broad | chr12:130850076-131736884(probes 67143:67185) | 1.67E-09 | 47 | 9 | |
| 12p13.31 | broad | chr12:6616267-6903061(probes 62836:62865) | 8.56E-05 | 49 | 18 | |
| 14q32.33 | broad | chr14:104589610-105050674(probes 73764:73792) | 4.85E-24 | 65 | 10 | JAG2, CRIP1 &2 |
| 14q11.2 | both | chr14:23956846-24348799(probes 70746:70760) | 9.88E-07 | 68 | 8 | NDRG2 |
| 14q32.12 | both | chr14:90665049-90858909(probes 73154:73162) | 3.26E-08 | 68 | 3 | |
| 15q24.1 | broad | chr15:72051988-73168030(probes 76042:76114) | 5.47E-08 | 40 | 26 | PML |
| 16p13.3 | broad | chr16:1-3105058(probes 77207:77422) | 4.80E-18 | 51 | 137 | TSC2, SOX8 |
| 16p11.2 | broad | chr16:27341930-31408441(probes 78244:78442) | 3.71E-08 | 47 | 117 | MAPK3 |
| 16q13 | broad | chr16:56068807-56658226(probes 78870:78898) | 4.33E-08 | 49 | 13 | DOK4 |
| 16q22.1 | broad | chr16:65749906-66612926(probes 79108:79177) | 2.29E-14 | 49 | 41 | |
| 16q24.3 | broad | chr16:86172468-88827254(probes 79912:80047) | 2.70E-12 | 49 | 45 | IL17C, FANCA, CDK10 |
| 17p11.2 | broad | chr17:20618475-20845678(probes 80907:80912) | 0.0006169 | 45 | 2 | |
| 17q21.2 | broad | chr17:34989301-39828444(probes 81445:81756) | 4.14E-06 | 45 | 47 | STAT3 |
| 17q22 | broad | chr17:53603427-53812876(probes 82242:82252) | 3.26E-08 | 34 | 7 | SUPT4H1 |
| 17q23.3 | broad | chr17:58901630-60399623(probes 82454:82525) | 0.0006169 | 32 | 30 | |
| 17q25.1 | broad | chr17:70825746-70934827(probes 82864:82868) | 4.05E-05 | 32 | 1 | |
| 17p13.1 | broad | chr17:7217430-8017270(probes 80410:80478) | 3.13E-09 | 45 | 42 | TP53 |
| 19p13.3 | broad | chr19:1-3747215(probes 85650:85832) | 4.74E-14 | 52 | 109 | |
| 19q13.12 | broad | chr19:40168317-41398102(probes 86948:87029) | 1.50E-08 | 51 | 56 | |
| 19q13.33 | broad | chr19:54489423-56393197(probes 87670:87779) | 5.53E-14 | 51 | 60 | |
| 19q13.42 | broad | chr19:59337767-60853685(probes 87917:87997) | 7.33E-20 | 51 | 42 | |
| 19q13.43 | both | chr19:63654029-63811651(probes 88142:88157) | 2.00E-18 | 51 | 9 | |
| 20p13 | both | chr20:3576278-3741605(probes 88329:88344) | 5.47E-08 | 45 | 9 | |
| 20q13.33 | broad | chr20:61334277-61867592(probes 90310:90342) | 2.15E-15 | 45 | 21 | |
| 21q22.3 | broad | chr21:44375425-46717428(probes 91623:91770) | 1.52E-06 | 51 | 29 | |
| 22q11.21 | broad | chr22:18070267-18691703(probes 91909:91947) | 2.71E-30 | 78 | 15 | SEPT5 |
| 22q12.2 | both | chr22:27,901,842-28,852,291(probes 92365:92430) | 3.16E-37 | 85 | 17 | NF2 |
| Xq28 | focal | chrX:152278550-152830969(probes 97921:97955) | 2.97E-17 | 57 | 19 | TREX2 |
| Xp11.23 | focal | chrX:47780775-49008093(probes 94924:95005) | 5.97E-05 | 55 | 43 | GATA1 |
| Copy Number Gains | ||||||
| 1q25.1 | broad | chr1:172161698-172889501(probes 5543:5570) | 0.0098287 | 55 | 2 | |
| 1q31.1 | broad | chr1:182779475-197636068(probes 5944:6019) | 0.004383 | 59 | 42 | |
| 7q36.1 | broad | chr7:150277900-150468342(probes 44309:44322) | 0.0098287 | 26 | 10 | CDK5 |
| 9q34.3 | broad | chr9:138940849-139254452(probes 53064:53086) | 0.004383 | 30 | 34 | COBRA1 |
| 17q21.32 | broad | chr17:41604766-42212584(probes 81839:81848) | 0.0027607 | 44 | 7 | WNT3 |
The most common genomic alteration was loss of 22q including the NF2 gene (85% of samples). The second most frequent regions of loss were confined to the short arm of chromosome 1. Most cases displayed copy number alteration patterns consistent with complex structural rearrangements involving both arms. Two distinct regions of loss along 1p included 1p33-p36.2 (70%) and 1p13.2 (64%). Both regions spanned several megabases of DNA sequences and included 134 genes (Table 2). Other frequent regions of loss, detected in more than 50% of samples, included 14q (68%), 10q (55%), 8q (55%), 7p (54%), 21q (51%), 19 (51%), 9q34 (51%), and 4p16 (51%). Frequent gains were detected along 1q (59%), 17q (44%), 9q34 (30%) and 7q36 (26%).
Cluster analysis
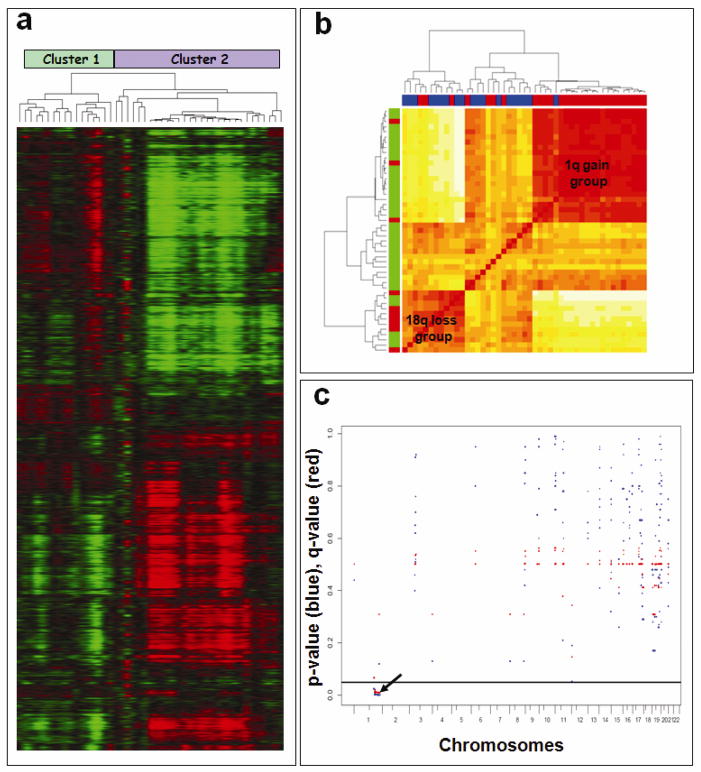
Figure 3a illustrates the two groups that resulted from the unsupervised hierarchical clustering of unfiltered aCGH data described above. Results of the hierarchical clustering are presented in Figures 3a and 3b. In Figure 3a, rows correspond to probes and columns correspond to subjects. Gain of 1q was much more common in cluster 2, shown on the right in Figure 3a (25 of 30 cases), than in cluster 1 (3 of 17 cases). Loss of 18q12.3-q21.31 in 7 of 17 in cluster 1, compared with 3 of 30 cases in cluster 2. The known tumor suppressor gene DCC (deleted in colorectal carcinoma) maps to the minimal region of 18q deletion. This distinction between cluster 1 and cluster 2 is further displayed in Figure 3b which consists of a heatmap based on the distance matrix underlying the hierarchical clustering. Along the left vertical axis, red indicates 18q loss. Along the upper horizontal axis, red indicates 1q gain. The heatmap shows two large clusters associated with gain of 1q and loss of 18q (Fig 3b). This finding confirms the mutual exclusivity of these two alterations in atypical meningiomas.
Figure 3.
Cluster analysis of copy number alterations in atypical meningiomas. a) Unsupervised hierarchical clustering of CGHsmooth transformed dataset derived from 47 primary atypical meningiomas. Copy number values are color coded as follows: green (loss), black (normal) and red (gain). The pattern of dendrogram suggests two major genomic subgroups of atypical meningiomas. b) The distance matrix of all copy number alterations included in the heatmap (Pearson distance). Majority of the samples with 1q gain (indicated in Red on the horizontal side bar) and 18q loss (indicated in Red on the vertical side bar) cluster together. c) Marker-by-marker assessment of DNA copy number gains reveals association between 1q gain and shorter progression-free survival. P-values (Cox model) are denoted in red. Q-values are denoted in blue. The horizontal black line at 0.05 denotes the chosen significance level. Long tick marks along the bottom axis denote chromosome ends whereas short tick marks denote the centromere.
Correlation with clinical outcome
To evaluate relationships between DNA copy number alterations and clinical end points, such as PFS, we chose to perform univariate marker-by-marker analysis for all probes showing gain or loss throughout the genome. On univariate marker-by-marker analysis, multiple markers with DNA copy number gains on 1q were found to be significantly associated with shorter PFS (p < 0.05). Figure 3c shows marker-by-marker p-value (red) and q-value (blue) results for copy number gains across all chromosome arms for potentially informative markers. Correlation between loss on 18q and longer PFS was also investigated. However, due to the small number of observed PFS events, no statistically significant association was identified for 18q loss.
Minimal region of 1q gain
GISTIC approach identified two regions of frequent CNAs along 1q. The first one maps to 1q25.1 and the peak region (chr1:172161698–172889501) spans 727 kilobases of DNA sequence, includes 30 probes and 2 genes (Table 2). The second peak region (chr1:182,779,475–197,636,068) of gain along 1q25.3-q32.1 spans 1.48 megabases of DNA sequence, includes 75 markers and 42 genes (Table 2).
Discussion
In this study, we profiled a cohort of 47 clinically annotated primary atypical meningiomas using high resolution oligonucleotide aCGH. The goals of this study were to generate a high resolution genomic map that would allow identification of genes targeted by frequent loss and gain of DNA sequences, and to develop prognostic markers for atypical meningioma risk stratification. We analyzed data obtained by aCGH to explore the relationship between CNAs and PFS. As a result of this analysis, we found a significant correlation between the gain of 1q and PFS.
Eighty-five percent of atypical meningiomas exhibited loss of 22q, including the NF2 gene. The second most frequent regions of loss were confined to the short arm of chromosome 1, particularly 1p33-p36.2 (70%) and 1p13.2 (64%). Other common regions of loss, detected in more than 50% of samples, included 14q, 10q, 8q, 7p, 21q, 19, 9q34, and 4p16. Frequent regions of gain were detected along 1q, 17q, 9q34 and 7q36. Unsupervised clustering of aCGH data revealed two groups of atypical meningioma: one group was composed of the majority of cases with 1q gain and the other group was composed of the majority of the cases with 18q loss (Fig 3a, b). Loss of genetic information from 1p may represent early progression-associated changes in the transition from benign to atypical meningioma (39). Analysis of the data in the current study revealed focal regions of frequent loss on 1p36.32 (70%) and 1p13.2 (64%). The TP73 gene, encoding a p53-related protein, is located at 1p36.2 and may represent a candidate gene warranting further investigation for its potential role in atypical meningioma tumorigenesis. Other studies of atypical meningioma using loss of heterozygosity and/or comparative genomic hybridization analysis have reported loss on 1p in 40%–86% (21, 22, 32, 43) of cases.
Evaluation of chromosome 1 copy number imbalances using a genomic tiling path method of aCGH on 82 meningiomas revealed frequent, aberrations of chromosome 1 of varying type (deletions and amplifications), size (monosomy and homozygous deletion) and distribution (44). Deletion on 1p was the most frequent alteration, ranging from whole p-arm deletion (120 MB) to homozygous loss (3.5 Mb). Three distinct candidate loci on 1p and one locus on 1q exhibited tumor-specific aberrations of potential importance for tumorigenesis. Aberrations of 1q were less common and were always accompanied by full or partial loss of 1p. Elsewhere in the literature, gains on 1q have been shown to occur in 30%–50% of meningioma cases (19, 22). Our data revealed chromosomal gains on 1q in 59% of atypical meningioma cases. The 1q gain centered on two regions: 1q25.1 (chr1:172,161,698-172,889,501) and 1q25.3-q32.1 (chr1:182,779,475-197,636,068). The 1q25.1 interval includes two genes: RC3H1 (roquin) encodes a highly conserved member of the RING type ubiquitin ligase protein family and RABGAP1L (RAB GTPase activating protein 1-like isoform A) encodes a 298-amino acid GTPase-activating protein, containing a putative phosphotyrosine binding domain (45). The 1q25.3-q32.1 interval includes 42 genes, some of which may have relevance to tumorigenesis: EDEM3 (ER degradation enhancer, mannosidase alpha like), which ensures that only properly folded proteins are retained in the cell; RNF2 (ring finger protein 2), a polycomb protein, part of the multiprotein complexes involved in transcriptional control; PRG4 (proteoglycan 4), a large proteoglycan synthesized by chondrocytes; TPR (nuclear pore complex associated protein), which encodes a large coiled-coil protein that is required for the nuclear export of mRNAs and some proteins, with oncogenic fusions of the 5′ end of this gene with several different kinase genes occuring in some tumors; PTGS2 (prostaglandin endoperoxidase synthase 2 precursor), a key enzyme in prostaglandin biosynthesis, the expression of which is deregulated in epithelial tumors; CDC73 (parafibromin), a member of the PAF protein complex, which associates with the RNA polymerase II subunit POLR2A and with a histone methyltransferase complex (46); ASPM (abnormal spindle)-like, microcephaly), a human ortholog of the Drosophila ‘abnormal spindle’ gene, which is essential for normal mitotic spindle function in embryonic neuroblasts (47); LHX9 (Homo sapiens LIM homeobox 9), which encodes a member of the LIM homeobox gene family of developmentally expressed transcription factors; NEK7 (Homo sapiens NIMA (never in mitosis gene a)-related kinase 7), which shares high amino acid sequence identity with the gene product of the Aspergillus nidulans ‘never in mitosis A’ gene and which controls initiation of mitosis. The potential functional relevance of these genes in the progression of atypical meningioma is not known and will require further investigation.
There is a paucity of data evaluating the clinical significance of genomic abnormalities in atypical meningioma. To our knowledge, this is the first comprehensive genomic analysis of a large clinically-annotated cohort of atypical meningiomas. Previous studies have focused on genetic progression of meningiomas and relatively small numbers of atypical meningiomas have been included (22, 32). The overall pattern of losses and gains detected in our study is consistent with what has been reported in the general meningioma literature. However, the increased resolution and the relatively large sample of atypical meningiomas allowed us to identify novel large-scale and focal chromosomal alterations. Most importantly, each case evaluated in the current study was fully annotated in a clinical database with information regarding treatment course and clinical endpoints. This provided the opportunity to evaluate for potential clinically relevant genomic markers.
Analysis of all frequently identified CNAs showed a significant correlation between gain of 1q and shorter PFS (p<0.05). Gain of 1q has previously been associated with increased recurrence in neuroblastoma, Wilms’ tumor and ependymoma (48–50) and reduced overall survival in multiple myeloma, medulloblastoma and Ewing sarcoma (51–53). To our knowledge this is the first study to correlate chromosome 1q gain with clinical endpoints in atypical meningioma.
Although an association of 1q gain with shorter PFS was evident in our series of tumors and although the sample size was large relative to other atypical meningioma investigations, the study remained under-powered to reveal other clinical associations of identified CNAs. Clarification of molecular-genetic signatures useful in atypical meningioma diagnosis and treatment will therefore require investigational efforts on even larger clinically annotated series. Given the increasing diagnosis and heterogeneous treatment outcomes of atypical meningioma, such efforts are warranted and should become a priority to guide clinical management, decrease inappropriate treatment, minimize treatment failure and reduce undue treatment-related toxicity in this disease.
Acknowledgments
DG was supported by NIH T32CA09216.
The authors thank Shumin Dong, Anna Levitz, Andrew Potts, Candice Romany and Jennifer Roy for histopathology expertise, technical support and software management. DG was supported by NIH T32CA09216.
Footnotes
Authors’ contributions. DG participated in the study design, reviewed patient charts, collected clinical information, set up the clinical data base, collected samples and isolated DNA. DE and RAB participated in statistical analyses, SG carried out bioinformatic analyses, GAS participated in DNA isolation, probe preparations and microarray hybridizations, FGB and JSL participated in study design and provided clinical input, DNL conceived of the study, participated in study design, provided clinical and histopathological input. GM participated in study design, performed microarray hybridizations, carried out data analysis and drafted the manuscript. All authors contributed to the edits and approved the final manuscript.
References
- 1.Goyal LK, Suh JH, Mohan DS, Prayson RA, Lee J, Barnett GH. Local control and overall survival in atypical meningioma: a retrospective study. Int J Radiat Oncol Biol Phys. 2000;46:57–61. doi: 10.1016/s0360-3016(99)00349-1. [DOI] [PubMed] [Google Scholar]
- 2.Dziuk TW, Woo S, Butler EB, Thornby J, Grossman R, Dennis WS, Lu H, Carpenter LS, Chiu JK. Malignant meningioma: an indication for initial aggressive surgery and adjuvant radiotherapy. J Neurooncol. 1998;37:177–88. doi: 10.1023/a:1005853720926. [DOI] [PubMed] [Google Scholar]
- 3.Louis D, Ohgaki H, Wiestler O, Cavenee W, editors. WHO Classification of Tumours of the Central Nervous System. Lyon, France: IARC Press; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willis J, Smith C, Ironside JW, Erridge S, Whittle IR, Everington D. The accuracy of meningioma grading: a 10-year retrospective audit. Neuropathol Appl Neurobiol. 2005;31:141–9. doi: 10.1111/j.1365-2990.2004.00621.x. [DOI] [PubMed] [Google Scholar]
- 5.Palma L, Celli P, Franco C, Cervoni L, Cantore G. Long-term prognosis for atypical and malignant meningiomas: a study of 71 surgical cases. J Neurosurg. 1997;86:793–800. doi: 10.3171/jns.1997.86.5.0793. [DOI] [PubMed] [Google Scholar]
- 6.Milker-Zabel S, Zabel A, Schulz-Ertner D, Schlegel W, Wannenmacher M, Debus J. Fractionated stereotactic radiotherapy in patients with benign or atypical intracranial meningioma: long-term experience and prognostic factors. Int J Radiat Oncol Biol Phys. 2005;61:809–16. doi: 10.1016/j.ijrobp.2004.07.669. [DOI] [PubMed] [Google Scholar]
- 7.Kallio M, Sankila R, Hakulinen T, Jaaskelainen J. Factors affecting operative and excess long-term mortality in 935 patients with intracranial meningioma. Neurosurgery. 1992;31:2–12. doi: 10.1227/00006123-199207000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Ayerbe J, Lobato RD, de la Cruz J, Alday R, Rivas JJ, Gomez PA, Cabrera A. Risk factors predicting recurrence in patients operated on for intracranial meningioma. A multivariate analysis. Acta neurochirurgica. 1999;141:921–32. doi: 10.1007/s007010050398. [DOI] [PubMed] [Google Scholar]
- 9.Pearson BE, Markert JM, Fisher WS, Guthrie BL, Fiveash JB, Palmer CA, Riley K. Hitting a moving target: evolution of a treatment paradigm for atypical meningiomas amid changing diagnostic criteria. Neurosurg Focus. 2008;24:E3. doi: 10.3171/FOC/2008/24/5/E3. [DOI] [PubMed] [Google Scholar]
- 10.Perry A, Gutmann DH, Reifenberger G. Molecular pathogenesis of meningiomas. J Neurooncol. 2004;70:183–202. doi: 10.1007/s11060-004-2749-0. [DOI] [PubMed] [Google Scholar]
- 11.Perry A, Scheithauer BW, Stafford SL, Lohse CM, Wollan PC. “Malignancy” in meningiomas: a clinicopathologic study of 116 patients, with grading implications. Cancer. 1999;85:2046–56. doi: 10.1002/(sici)1097-0142(19990501)85:9<2046::aid-cncr23>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Lohmann CM, Brat DJ. A conceptual shift in the grading of meningiomas. Adv Anat Pathol. 2000;7:153–7. doi: 10.1097/00125480-200007030-00004. [DOI] [PubMed] [Google Scholar]
- 13.Yang SY, Park CK, Park SH, Kim DG, Chung YS, Jung HW. Atypical and anaplastic meningiomas: prognostic implications of clinicopathological features. Journal of neurology, neurosurgery, and psychiatry. 2008;79:574–80. doi: 10.1136/jnnp.2007.121582. [DOI] [PubMed] [Google Scholar]
- 14.Hug EB, Devries A, Thornton AF, Munzenride JE, Pardo FS, Hedley-Whyte ET, Bussiere MR, Ojemann R. Management of atypical and malignant meningiomas: role of high-dose, 3D-conformal radiation therapy. J Neurooncol. 2000;48:151–60. doi: 10.1023/a:1006434124794. [DOI] [PubMed] [Google Scholar]
- 15.Condra KS, Buatti JM, Mendenhall WM, Friedman WA, Marcus RB, Jr, Rhoton AL. Benign meningiomas: primary treatment selection affects survival. Int J Radiat Oncol Biol Phys. 1997;39:427–36. doi: 10.1016/s0360-3016(97)00317-9. [DOI] [PubMed] [Google Scholar]
- 16.Perry A. Unmasking the secrets of meningioma: a slow but rewarding journey. Surg Neurol. 2004;61:171–3. doi: 10.1016/s0090-3019(03)00488-9. [DOI] [PubMed] [Google Scholar]
- 17.Zang KD. Meningioma: a cytogenetic model of a complex benign human tumor, including data on 394 karyotyped cases. Cytogenet Cell Genet. 2001;93:207–20. doi: 10.1159/000056986. [DOI] [PubMed] [Google Scholar]
- 18.Ruttledge MH, Sarrazin J, Rangaratnam S, Phelan CM, Twist E, Merel P, Delattre O, Thomas G, Nordenskjold M, Collins VP, et al. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat Genet. 1994;6:180–4. doi: 10.1038/ng0294-180. [DOI] [PubMed] [Google Scholar]
- 19.Ozaki S, Nishizaki T, Ito H, Sasaki K. Comparative genomic hybridization analysis of genetic alterations associated with malignant progression of meningioma. J Neurooncol. 1999;41:167–74. doi: 10.1023/a:1006086723607. [DOI] [PubMed] [Google Scholar]
- 20.Simon M, Kokkino AJ, Warnick RE, Tew JM, Jr, von Deimling A, Menon AG. Role of genomic instability in meningioma progression. Genes Chromosomes Cancer. 1996;16:265–9. doi: 10.1002/(SICI)1098-2264(199608)16:4<265::AID-GCC7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 21.Simon M, von Deimling A, Larson JJ, Wellenreuther R, Kaskel P, Waha A, Warnick RE, Tew JM, Jr, Menon AG. Allelic losses on chromosomes 14, 10, and 1 in atypical and malignant meningiomas: a genetic model of meningioma progression. Cancer Res. 1995;55:4696–701. [PubMed] [Google Scholar]
- 22.Weber RG, Bostrom J, Wolter M, Baudis M, Collins VP, Reifenberger G, Lichter P. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci U S A. 1997;94:14719–24. doi: 10.1073/pnas.94.26.14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bello MJ, de Campos JM, Vaquero J, Kusak ME, Sarasa JL, Rey JA. High-resolution analysis of chromosome arm 1p alterations in meningioma. Cancer Genet Cytogenet. 2000;120:30–6. doi: 10.1016/s0165-4608(99)00249-6. [DOI] [PubMed] [Google Scholar]
- 24.Murakami M, Hashimoto N, Takahashi Y, Hosokawa Y, Inazawa J, Mineura K. A consistent region of deletion on 1p36 in meningiomas: identification and relation to malignant progression. Cancer Genet Cytogenet. 2003;140:99–106. doi: 10.1016/s0165-4608(02)00653-2. [DOI] [PubMed] [Google Scholar]
- 25.Lomas J, Bello MJ, Arjona D, Gonzalez-Gomez P, Alonso ME, de Campos JM, Vaquero J, Ruiz-Barnes P, Sarasa JL, Casartelli C, Rey JA. Analysis of p73 gene in meningiomas with deletion at 1p. Cancer Genet Cytogenet. 2001;129:88–91. doi: 10.1016/s0165-4608(01)00430-7. [DOI] [PubMed] [Google Scholar]
- 26.Bostrom J, Meyer-Puttlitz B, Wolter M, Blaschke B, Weber RG, Lichter P, Ichimura K, Collins VP, Reifenberger G. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001;159:661–9. doi: 10.1016/S0002-9440(10)61737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendiola M, Bello MJ, Alonso J, Leone PE, Vaquero J, Sarasa JL, Kusak ME, De Campos JM, Pestana A, Rey JA. Search for mutations of the hRAD54 gene in sporadic meningiomas with deletion at 1p32. Molecular carcinogenesis. 1999;24:300–4. doi: 10.1002/(sici)1098-2744(199904)24:4<300::aid-mc8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 28.Niedermayer I, Feiden W, Henn W, Steilen-Gimbel H, Steudel WI, Zang KD. Loss of alkaline phosphatase activity in meningiomas: a rapid histochemical technique indicating progression-associated deletion of a putative tumor suppressor gene on the distal part of the short arm of chromosome 1. J Neuropathol Exp Neurol. 1997;56:879–86. [PubMed] [Google Scholar]
- 29.Sulman EP, Dumanski JP, White PS, Zhao H, Maris JM, Mathiesen T, Bruder C, Cnaan A, Brodeur GM. Identification of a consistent region of allelic loss on 1p32 in meningiomas: correlation with increased morbidity. Cancer Res. 1998;58:3226–30. [PubMed] [Google Scholar]
- 30.Sulman EP, White PS, Brodeur GM. Genomic annotation of the meningioma tumor suppressor locus on chromosome 1p34. Oncogene. 2004;23:1014–20. doi: 10.1038/sj.onc.1206623. [DOI] [PubMed] [Google Scholar]
- 31.Khan J, Parsa NZ, Harada T, Meltzer PS, Carter NP. Detection of gains and losses in 18 meningiomas by comparative genomic hybridization. Cancer Genet Cytogenet. 1998;103:95–100. doi: 10.1016/s0165-4608(97)00394-4. [DOI] [PubMed] [Google Scholar]
- 32.Arslantas A, Artan S, Oner U, Durmaz R, Muslumanoglu H, Atasoy MA, Basaran N, Tel E. Comparative genomic hybridization analysis of genomic alterations in benign, atypical and anaplastic meningiomas. Acta Neurol Belg. 2002;102:53–62. [PubMed] [Google Scholar]
- 33.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 34.Buschges R, Ichimura K, Weber RG, Reifenberger G, Collins VP. Allelic gain and amplification on the long arm of chromosome 17 in anaplastic meningiomas. Brain Pathol. 2002;12:145–53. doi: 10.1111/j.1750-3639.2002.tb00429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis DN, Scheithauer BW, Budka H, von Deimling A, Kepes JJ. Meningiomas. In: Kleihues P, Cavenee WK, editors. World Health Organization Classification of Tumors. Pathology and Genetics of Tumors of the Nervous System. IARC Press; Lyon: 2000. pp. 176–184. [Google Scholar]
- 36.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 37.Korkola JE, Heck S, Olshen AB, Reuter VE, Bosl GJ, Houldsworth J, Chaganti RS. In vivo differentiation and genomic evolution in adult male germ cell tumors. Genes Chromosomes Cancer. 2008;47:43–55. doi: 10.1002/gcc.20504. [DOI] [PubMed] [Google Scholar]
- 38.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behavioural brain research. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 40.van de Wiel MA, van Wieringen WN. CGHregions: dimension reduction for array CGH data with minimal informatics loss. Cancer Informatics. 2007;2:55–63. [PMC free article] [PubMed] [Google Scholar]
- 41.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruno M, Yoshimine T, Muhammad AK, Ninomiya H, Hayakawa T. Chromosomal losses and gains in meningiomas: comparative genomic hybridization (CGH) study of the whole genome. Neurological research. 1998;20:612–6. doi: 10.1080/01616412.1998.11740572. [DOI] [PubMed] [Google Scholar]
- 44.Buckley PG, Jarbo C, Menzel U, Mathiesen T, Scott C, Gregory SG, Langford CF, Dumanski JP. Comprehensive DNA copy number profiling of meningioma using a chromosome 1 tiling path microarray identifies novel candidate tumor suppressor loci. Cancer Res. 2005;65:2653–61. doi: 10.1158/0008-5472.CAN-04-3651. [DOI] [PubMed] [Google Scholar]
- 45.Seki N, Ohira M, Nagase T, Ishikawa K, Miyajima N, Nakajima D, Nomura N, Ohara O. Characterization of cDNA clones in size-fractionated cDNA libraries from human brain. DNA Res. 1997;4:345–9. doi: 10.1093/dnares/4.5.345. [DOI] [PubMed] [Google Scholar]
- 46.Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–20. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, Walsh CA, Woods CG. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–20. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 48.Pezzolo A, Rossi E, Gimelli S, Parodi F, Negri F, Conte M, Pistorio A, Sementa A, Pistoia V, Zuffardi O, Gambini C. Presence of 1q gain and absence of 7p gain are new predictors of local or metastatic relapse in localized resectable neuroblastoma. Neuro-oncology. 2009;11:192–200. doi: 10.1215/15228517-2008-086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hing S, Lu YJ, Summersgill B, King-Underwood L, Nicholson J, Grundy P, Grundy R, Gessler M, Shipley J, Pritchard-Jones K. Gain of 1q is associated with adverse outcome in favorable histology Wilms’ tumors. Am J Pathol. 2001;158:393–8. doi: 10.1016/S0002-9440(10)63982-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendrzyk F, Korshunov A, Benner A, Toedt G, Pfister S, Radlwimmer B, Lichter P. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12:2070–9. doi: 10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 51.Fonseca R, Van Wier SA, Chng WJ, Ketterling R, Lacy MQ, Dispenzieri A, Bergsagel PL, Rajkumar SV, Greipp PR, Litzow MR, Price-Troska T, Henderson KJ, Ahmann GJ, Gertz MA. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–40. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 52.Lo KC, Ma C, Bundy BN, Pomeroy SL, Eberhart CG, Cowell JK. Gain of 1q is a potential univariate negative prognostic marker for survival in medulloblastoma. Clin Cancer Res. 2007;13:7022–8. doi: 10.1158/1078-0432.CCR-07-1420. [DOI] [PubMed] [Google Scholar]
- 53.Hattinger CM, Potschger U, Tarkkanen M, Squire J, Zielenska M, Kiuru-Kuhlefelt S, Kager L, Thorner P, Knuutila S, Niggli FK, Ambros PF, Gadner H, Betts DR. Prognostic impact of chromosomal aberrations in Ewing tumours. British journal of cancer. 2002;86:1763–9. doi: 10.1038/sj.bjc.6600332. [DOI] [PMC free article] [PubMed] [Google Scholar]