Abstract
Introduction
Quantitative proteomics using tandem mass spectrometry is an attractive approach for identification of potential cancer biomarkers. Fractionation of complex tissue samples into subproteomes prior to mass spectrometric analyses increases the likelihood of identifying cancer-specific proteins that might be present in low abundance. In this regard, glycosylated proteins are an interesting class of proteins that are already established as biomarkers for several cancers.
Materials and Methods
In this study, we carried out proteomic profiling of tumor and adjacent non-cancer liver tissues from hepatocellular carcinoma (HCC) patients. Glycoprotein enrichment from liver samples using lectin affinity chromatography and subsequent 18O/16O labeling of peptides allowed us to obtain relative abundance levels of lectin-bound proteins. As a complementary approach, we also examined the relative expression of proteins in HCC without glycoprotein enrichment. Lectin affinity enrichment was found to be advantageous to quantitate several interesting proteins, which were not detected in the whole proteome screening approach. We identified and quantitated over 200 proteins from the lectin-based approach. Interesting among these were fetuin, cysteine-rich protein 1, serpin peptidase inhibitor, leucine-rich alpha-2-glycoprotein 1, melanoma cell adhesion molecule, and heparan sulfate proteoglycan-2. Using lectin affinity followed by PNGase F digestion coupled to 18O labeling, we identified 34 glycosylation sites with consensus sequence N-X-T/S. Western blotting and immunohistochemical staining were carried out for several proteins to confirm mass spectrometry results.
Conclusion
This study indicates that quantitative proteomic profiling of tumor tissue versus non-cancerous tissue is a promising approach for the identification of potential biomarkers for HCC.
Keywords: Hepatocellular carcinoma, Lectin affinity enrichment, Quantitative proteomics, Mass spectrometry
Introduction
The incidence of hepatocellular carcinoma is rising, making it the fifth most common malignancy worldwide [1]. This increased incidence is attributed to viral hepatitis and carcinogenic toxins such as aflatoxin [2]. Alpha fetoprotein (AFP), one of the tumor markers used for diagnostic purposes in hepatocellular carcinoma (HCC), is limited in its diagnostic utility because 50% (35% in Johns Hopkins Hospital [3]) of the patients with HCC do not show high level of AFP [4–6]. Serum level of des-gamma-carboxy prothrombin (DCP) is also shown to be useful for the diagnosis of HCC in conjunction with AFP [7]. Several studies have reported potential diagnostic markers for HCC, which are associated with chromosomal alterations, mRNA expression alterations, protein-level alterations and post-translational modifications. Lately, proteomic strategies have been extensively used for biomarker discovery because of their ability for multiplex analysis using quantitative approaches. Quantitative proteomics using isotope-labeling techniques provide opportunities to analyze changes in protein levels in multiple samples in an unbiased fashion [8]. However, a limitation of these techniques is that, due to the complexity of samples, least abundant proteins are not detected.
O- and N-glycosylation of proteins have been associated with cancers including HCC. For example, an overall increase in fucosylation has been reported in HCC serum proteins [9]. Increased or altered glycosylation is also reported to be associated with secreted alpha fetoprotein [10] and transferrin [11] in HCC. Glycomic profiling is a complementary strategy used to identify cancer-specific glycans especially in ovarian cancer [12], HCC [13], and pancreatic cancer [14]. There are several reports on the application of lectin-based enrichment method to study the glycoproteome in cancers [15, 16]. Lectin reactive form of AFP is reported to be a more sensitive marker for HCC [17]. The profile of carbohydrate moieties of glycoproteins has also been used for differentiating normal from cancerous tissue in lung [16] and pancreatic cancers [18]. Since individual lectins have their own specificity while the nature of the carbohydrates present in samples may vary dramatically, it is advantageous to use a combination of lectins for enrichment purposes. Such a multiple lectin affinity approach has been used for breast cancer biomarker analysis [19]. A mixture of different lectins has also been used for lectin affinity enrichment method to identify biomarkers in bile secretome from cancer patients [20].
Stable isotope can be incorporated into proteins and peptides by different methods in vitro such as ICAT [21], iTRAQ [22], 18O-labeling methods [23], and in vivo labeling, using heavy-isotope-labeled amino acids in cell culture (SILAC) [24]. Labeling of peptides in the presence of 18O-labeled water is a simple method without chemical tagging and can be readily used to analyze the proteins separated by SDS-PAGE [25]. The present study is designed to analyze subproteome in HCC using affinity enrichment and relative quantitation based on 18O labeling. Further, analysis of glycopeptides treated with PNGase F provides definitive evidence of N-linked glycosylation [20]. Western blotting was used to confirm the differential expression results obtained by quantitative mass spectrometry. The candidate biomarkers were screened by immunohistochemical labeling using tissue microarrays containing hepatocellular carcinoma and non-cancerous liver tissue. We validated fetuin, a glycoprotein involved in endocytosis using immunohistochemical labeling. We also identified N-glycosylated peptides with sites from lectin-enriched protein. These results demonstrate advantages of lectin affinity enrichment method before quantitative proteomics profiling for identification of potential biomarkers for cancers.
Materials and Methods
Tissue Samples
Liver tissue samples were obtained after obtaining appropriate Institutional Review Board approval. One pair of tumor tissue and patient-matched non-cancerous tissue was collected at the time of surgery from patients and snap-frozen in liquid nitrogen. Serum samples were collected from HCC patients and stored at −80°C until analysis. A total of 114 HCC samples and 79 non-cancer liver sections from two tissue microarrays were used in this study. Formalin fixed and paraffin embedded tissue microarrays (Creative Biolabs, Cat No. CBL-TMA-070 and CBL-TMA-076), which consisted of 55 tissue sections from hepatocellular carcinoma with I, II, and III stages and 20 non-cancer tissues were used for immunohistochemical labeling. Tissue microarrays obtained from Imgenex consisted of 59 tumors (13 metastatic tissues, 46 poor to well-differentiated HCC, Cat. No. IMH-318) and 59 non-cancer tissues (Cat. No. IMH-342). Frozen tissues were used for mass spectrometric analysis and validation by Western blotting.
Lectin Affinity Enrichment
Tissue lysates were prepared from fresh frozen tissues from the tumor and adjacent non-cancerous region from a patient diagnosed with HCC in the presence of cocktail protease inhibitor and 10 mM phosphate buffer pH 7.8 (wash buffer). Lysates (4 mg protein) were subsequently incubated with 100 μl of each of ConA sepharose (Amersham BioSciences), wheat germ agglutinin and jacalin agarose for 12 h at 4°C. The beads were then washed three times using wash buffer and the bound proteins eluted using a mixture of carbohydrates (100 mM each of N-acetylglucosamine, melibiose, and galactose). The eluate was dialyzed against wash buffer (three volumes) to remove free sugars, concentrated and resolved by SDS-PAGE and visualized using Colloidal Coomassie staining.
Serum samples were obtained from patients diagnosed with hepatocellular carcinoma. Pooled normal serum was obtained from Invitrogen. Serum samples were prepared by incubating 100 μl (7 mg) of serum with 0.5 ml of mixture of lectins (ConA sepharose, jacalin, wheat germ lectins) overnight at 4°C in Tris-buffered saline (0.05 M Tris–HCL, pH 7.1, 0.15 M NaCl). Beads were washed three times in Tris buffer saline and boiled in Laemmli buffer for 10 min at 95°C to elute the proteins, and eluted proteins were run on 10% SDS-PAGE for further analysis by Western blot.
18O Labeling and In-gel Trypsin Digestion
Liver tissue samples from non-cancer and HCC tissues were run in parallel lanes on an SDS-PAGE gel for comparative analysis. Two gels were used for analysis, one from whole proteome and another from lectin-enriched proteins from HCC and normal tissues. Exact size gel slices corresponding to individual protein bands from HCC and normal tissues were excised and used for in-gel trypsin digestion as explained earlier [11] using Stratagene prolytica 18O labeling kit. In brief, sliced colloidal Coomassie-stained gel bands were destained and completely dehydrated gel slices were incubated with trypsin in the presence of heavy or normal water overnight. After completion of digestion, peptides were extracted from each sample, dried and subjected to post-trypsin digestion. Samples were incubated with immobilized trypsin (5 μl beads per sample) in the presence of heavy or normal water for 3 h. Finally, peptides derived from adjacent bands corresponding to normal and HCC were acidified, mixed, and analyzed on a quadrupole time-of-fight (QToF) mass spectrometer.
PNGase F Treatment for Identification of N-glycosylation Sites
Lectin affinity enriched proteins from HCC tissue were separated on 1D SDS-PAGE and stained with colloidal Coomassie. Protein bands were destained, reduced, and alkylated. Gel pieces were then subjected to in-gel trypsin (Promega) digestion and extracted peptides were dried completely. For the confirmation of N-glycosylation sites, peptides were incubated with PNGase F in the presence of 18O-water at 37°C for 3 h. After incubation, peptides were analyzed by LC-MS/MS (liquid chromatography–tandem mass spectrometry).
LC-MS/MS Analysis and Quantitation
In a separate study HCC and normal whole tissue lysates were analyzed on a SDS-PAGE gel and 18O labeling was done during in-gel trypsin digestion. For the common proteins found in this experiment, we were able to compare the changes in ratios to study glycoyslation status of proteins in normal and HCC. Bands were destained using ammonium bicarbonate in 50% ACN, dehydrated by acetonitrile and dried completely. Digestion was carried out in sequence grade trypsin (Promega, Southampton, UK), 20 μg/ml in 16O-water or 18O-water. Peptides were extracted using 16O-water or 18O-water three times and dried in SpeedVac. Subsequently, postdigest-end labeling was carried using prewashed immobilized trypsin (Stratagene; 2 μl/sample). Immobilized trypsin was added to the dried peptide samples and subsequently reconstituted in 8 μl of 18O-water or 16O-water, 2 μl of acetonitrile and incubated for 4 h at room temperature. Samples were centrifuged 10,000 ×g to remove the immobilized trypsin. Supernatant was acidified with 1 μl formic acid and stored at −80°C until LC-MS/MS analysis.
For LC-MS/MS analysis, peptides from paired samples were mixed and analyzed using Micromass quadrupole time-of-flight mass spectrometer (QToF) connected to reversed-phase nano LC system (Agilent 1100). The trap column consisted of (150 μM×3 cm, C18, 15 μm, 300A, YDAC, flow rate 1.5 μl/min) and analytical column (75 μM×10 cm, C18, 5 μm, 300A, YDAC, flow rate 200 nl/min), solvent system included 0.4% acetic acid, 0.004% heptafluorobutyric acid). The peptides were eluted using a gradient of acetonitrile up to 45% containing 0.4% acetic acid, 0.004% heptafluorobutyric acid. LC-MS/MS analysis was based on data dependent analysis (DDA) manner, performed using a 1-s MS survey scan m/z, 300-1,800 followed by four MS/MS scans (m/z, 50–1,800) for most intense ions and precursor ions were excluded for 60 s for the next MS/MS scans. A total of 4.4 s time was allowed for MS/MS spectrum acquisition.
The identification and quantitation of the peptide was done after formatting the mass spectrometry data. LC-MS/MS data acquired using Masslynx (Micromass) were searched using Mascot (Matrixscience, Manchester, UK). While searching the database, 18O at C-terminal carboxyl group (+4 Da) and oxidation of methionine were allowed as variable modification. Relative abundance of proteins was quantitated using MSQuant software downloaded from http://msquant.sourceforge.net [26] and expressed as fold changes (±S.D.). Essentially, Mascot search results were parsed with LC-MS/MS instrument data file using MSQuant. The new_MSQ_quantitationModes.xml files were modified to read the mascot output files and identify the correct light and heavy isotopic MS peaks. The quantitation data was also verified by manual inspection of heavy- and light-peptide-derived MS and MS/MS spectra in MSQuant.
Western Blot
Validation of the mass spectrometry results was carried out for a subset of proteins by Western blotting analysis using specific antibodies. These proteins were selected based on their functional relevance and differential expression between the cancerous and normal liver tissue. Eighty micrograms of protein was resolved on SDS-PAGE and transferred electrophoretically onto a nitrocellulose membrane. The membrane was blocked using 5% bovine serum albumin prepared in phosphate-buffered saline containing 0.1% Tween 20 (PBS-T) for 1 h at room temperature. The membrane was probed with specific primary antibody for 3 h and washed thrice with PBS-T and incubated in the HRP-conjugated secondary antibody for 1 h. After the incubation, the membrane was washed thrice in PBS-T. The signal was visualized by an enhanced chemiluminescence solution and exposed to Hyperfilm. Following antibodies were used for Western blot analysis. Fetuin (AF 1184, 1:1000, R&D systems) and CSRP1 (SC-33331, 1:1000, Santa Cruz).
Immunohistochemical Labeling
Immunohistochemical labeling was performed on liver tissue sections and liver cancer tissue microarrays for a subset of proteins based on their biological relevance. Tissue microarrays were obtained from Creative Biolabs, NY (tumor TMA, CBL-TMA-070 and normal TMA, CBL-TMA-076) and Imgenex, San Diego, CA (tumor TMA, IMH 318, lot CS3 and normal TMA, IMH-342 lot CSN3). The Envision kit (DAKO) was used according to the manufacturer's specifications. Briefly, the slides were first deparaffinized by xylene and rehydrated with ethanol. Antigen retrieval was done by heating the slides in 0.01 mol/L of sodium citrate buffer for 20 min on a steamer. After blocking by peroxidase, the sections were incubated with primary antibody anti-Fetuin A (dilution 1:400). After rinsing with wash buffer, the slides were incubated with HRP-conjugated appropriate secondary antibody. The signal was developed using chromogen supplied for peroxidase. Tissue sections were observed using Nikon DS-Fi1, microscope-operated using NIS-Elements F package. The immunostaining was assessed by an experienced liver pathologist and intensity was scored as negative, weak (1+), moderate (2+), and strong (3+). The distribution of staining of cancer cells was scored as 3+ for maximum distribution and, accordingly, 2+ and 1+ score was given for lower distribution. The following antibodies were used for immunohistochemical analysis: Fetuin (1:100) and CSRP1 (1:100).
Results and Discussion
The strategy used for quantitative profiling of lectin binding proteins from HCC liver tissue is shown in scheme Fig. 1. Recently, mass-spectrometry-based relative quantitation approach has been used to quantitate peptides containing N-linked glycosylation and also to identify the site of glycosylation [27]. Our approach of lectin affinity enrichment followed by 18O-labeling strategy was aimed at identifying proteins undergoing both N- and O-linked modifications in which the mass-spectrometry-based quantitation is based on any peptide derived from lectin-bound proteins and not glycopeptides alone. A mixture of lectins was used for enrichment of a broader group of glycoproteins in samples from normal individual and HCC patients. 16O/18O labeling allows quantitative profiling of a pair of samples and it relies on 18O labeling of peptides at C-terminal carboxyl groups during trypsin digestion in the presence of heavy water, which allows relative quantitation using mass spectrometry. Interesting candidate proteins found in this study were validated using tissue microarrays. Peptides containing N-linked glycosylation sites were identified by enzymatic incorporation 18O at the sites of glycosylation.
Fig. 1.
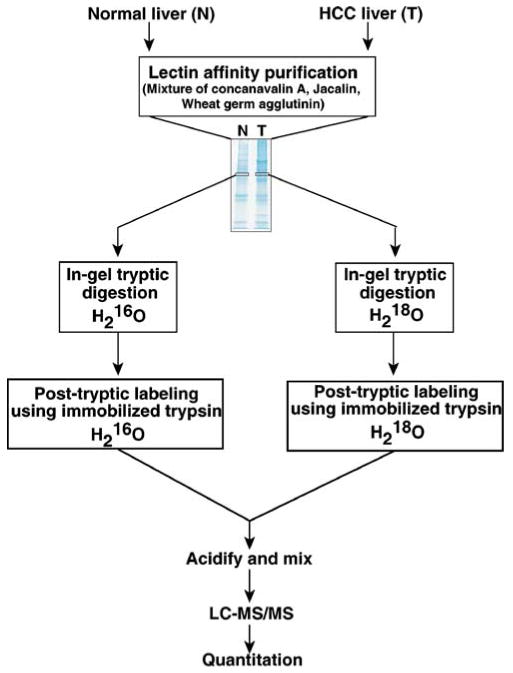
Strategy used for the enrichment of glycosylated proteins for 16O/18O labeling based quantitative mass spectrometric analysis of liver proteins in HCC. Tissue lysates (4 mg) were prepared from tumor and non-tumor liver tissues from a diagnosed case of HCC and incubated with mixture of lectins. The lectin-bound proteins were eluted using mixture of sugars and bound proteins derived from tumor and non-tumor samples were resolved separated on SDS-PAGE. In-gel trypsin digestion was carried out in the presence of 16O/18O-water differential labeling of peptides derived from non-tumor/tumor samples. After mixing the labeled and unlabeled peptides, the LC-MS/MS analysis was done using quadrupole time-of-flight mass spectrometer
Lectin Affinity Chromatography for Enrichment of Glycoproteins
It has been established that aberrant glycosylation serves as a marker of tumor progression in cancers. Several reagents that block glycosylation have been shown to inhibit tumor metastasis. N- or O-glycosylation of membrane components play an important role in altering tumor cell adhesion or motility [28]. In general, glycosylation modification is involved in cell adhesion, enhanced matrix destructive properties and cell motility in tumors.
Lectins are proteins that bind carbohydrates and can be used to enrich glycoproteins. Lectins can be distinguished based on their carbohydrate specificity. Different lectins can be used for isolation of specific glycosylated proteins from a complex sample. Concanavalin A preferentially binds alpha-mannosyl groups, wheat germ agglutinin has a higher specificity for N-acetylglucosamine-containing glycoproteins and jacalin interacts readily with mannose, glucose, and N-acetylneuraminic acid. Since these lectins show affinity for a broad spectrum of glycoproteins, we used the mixture of the above lectins and glycoproteins eluted using corresponding mixture of sugars. In our study, we were able to capture several proteins by lectin affinity purification which were not detected in unfractionated samples, which include transforming growth factor, beta-induced, 68 kDa, apolipoprotein A-1, destrin, and transgelin (Table 1). A complete list of overexpressed proteins is given in Supplementary Table 1.
Table 1.
A partial list of identified proteins previously reported to be overexpressed in HCC
| RefSeq accession number | Gene symbol | Protein name | 18O/16O ratio (tumor/non-tumor) | References |
|---|---|---|---|---|
| NP_002151.1 | TNC | Tenascin C | 8.5 | [33] |
| NP_997639.1 | FN1 | Fibronectin 1 | 3.5 | [36] |
| NP_000087.1 | CP | Ceruloplasmin | 2.4 | [40] |
| NP_001017963.1 | HSP90A1 | Heat shock protein 90 kDa alpha | 3.0 | [39] |
| NP_005134.1 | HP | Haptoglobin | 15 | [31] |
| NP_976084.1 | CLU | Clusterin isoform 2 | 5.1 | [45, 46] |
| NP_001002029.1 | C4B | Complement component 4B | 5.6 | [68] |
| NP_003093.1 | SOD3 | Superoxide dismutase 3, extracellular | 6.6 | [69] |
| NP_000030.1 | APOA1 | Apolipoprotein A-I | 7.1 | [70] |
| NP_001744.2 | CAV1 | Caveolin 1 | 4.2 | [37, 38] |
| NP_001653.1 | ARF5 | ADP-ribosylation factor 5 | 4.3 | [71] |
| NP_005498.1 | CFL1 | Cofilin 1 (non-muscle) | 6.7 | [72] |
| NP_002296.1 | LGALS1 | Galectin 1 | 2.8 | [41] |
18O Labeling and Mass Spectrometry
Differential labeling of peptides by 18O labeling using trypsin has been shown to be a simple and efficient method of relative quantitation of proteins [20, 29]. We used 18O-labeling method of quantitation for proteins obtained from tissue lysates using lectin-bound beads. Peptides derived from HCC tissue and non-cancerous tissue were labeled with heavy and light oxygen, respectively. After mixing, the peptides were analyzed using LC-MS/MS on a quadrupole time-of-flight mass spectrometer. 18O/16O-labeled peptides appear as doublets in MS, and ratio of peak intensities indicates relative differences in level of protein expression. Labeling at both oxygen atoms from the C-terminal carboxyl group is confirmed by visual inspection of spectrum. MSQuant assigned monoisotopic peaks from 16O-labeled and two 18O-labeled peptides. Corrections to the fold changes applied when there is a mixed labeling [30]. Peptide ratios were grouped and the average value is taken as protein-level fold changes. With a cutoff of threefold, we found 30 proteins to be overexpressed in HCC (Table 2).
Table 2.
Partial list of proteins overexpressed in HCC
| RefSeq accession number | Gene symbol | Protein name | Lectin affinity (HCC:normal) | Whole tissue analysis (HCC:normal) |
|---|---|---|---|---|
| NP_005498.1 | CFL1 | Cofilin 1 (non-muscle) | 13* | 3.2 |
| NP_000604.1 | HPX | Hemopexin | 12* | 3.7 |
| NP_066953.1 | PPIA | Cyclophilin A | 12 | 1.4 |
| NP_003509.1 | HIST1H2BG | H2B histone family, member A | 9.3* | 0.9 |
| NP_006477.2 | FBLN1 | Fibulin 1 isoform D | 7.8* | NF |
| NP_000349.1 | TGFBI | Transforming growth factor, beta-induced, 68 kDa | 7.5* | 5.1 |
| NP_000030.1 | APOA1 | Apolipoprotein A-I | 7.1 | 3.8 |
| NP_003093.1 | SOD3 | Superoxide dismutase 3, extracellular | 6.6 | NF |
| NP_000005.2 | A2M | Alpha-2-macroglobulin | 6.5 | 2.7 |
| NP_006861.1 | DSTN | Destrin isoform A | 6.5* | NF |
| NP_003177.2 | TAGLN | Transgelin | 6.0* | 4 |
| NP_008949.4 | CEP110 | Centrosomal protein 110 kDa | 6.0 | NF |
| NP_001076.2 | SERPINA3 | Serpin peptidase inhibitor, clade A, | 5.6* | 5.1 |
| NP_003364.1 | VCL | Vinculin | 5.6* | 2.8 |
| NP_001054.1 | TF | Transferrin | 5.6 | 3.5 |
| NP_001002029.1 | C4B | Complement component 4B | 5.6 | 2.5 |
| NP_005727.1 | ACTR1A | ARP1 actin-related protein 1 homolog A, centractin | 5.6 | NF |
| NP_006491.2 | MCAM | Melanoma cell adhesion | 5.3 | NF |
| NP_997639.1 | FN1 | Fibronectin 1 isoform 6 | 5.2 | 5.8 |
| NP_976084.1 | CLU | Clusterin isoform 2 | 5.1 | NF |
| NP_001701.2 | CFB | Complement factor B | 5.0 | NF |
| NP_001613.1 | AHSG | Fetuin | 5.0* | NF |
| NP_443204.1 | LRG1 | Leucine-rich alpha-2-glycoprotein 1 | 4.7* | NF |
| NP_001176.1 | AZGP1 | Alpha-2-glycoprotein 1, zinc | 4.6* | NF |
| NP_000497.1 | F2 | Coagulation factor II | 4.5 | NF |
| NP_005520.3 | HSPG2 | Heparan sulfate proteoglycan 2 | 4.3* | NF |
| NP_001653.1 | ARF5 | ADP-ribosylation factor 5 | 4.3 | 0.8 |
| NP_001744.2 | CAV1 | Caveolin 1 | 4.2 | 6.7 |
| NP_005338.1 | HSPA5 | Heat shock 70 kDa protein 5 | 4.0 | 1.5 |
| NP_001210.1 | CALU | Calumenin | 3.8 | NF |
| NP_001649.1 | ARF1 | ADP-ribosylation factor 1 | 3.8 | 0.7 |
| NP_937895.1 | GSN | Gelsolin isoform b | 3.5 | 4.4 |
| NP_031381.2 | HSP90AB1 | Heat shock 90 kDa protein 1, beta | 3.3 | 2.2 |
| NP_001124.1 | AFM | Afamin | 3.2* | NF |
| NP_002281.1 | LAMA4 | Laminin, alpha 4 | 3.2 | 6.9 |
| NP_000087.1 | CP | Ceruloplasmin | 3.2 | NF |
| NP_570602.2 | A1BG | Alpha 1B-glycoprotein | 3.1 | 4.9 |
| NP_002209.2 | ITIH4 | Inter-alpha (globulin) inhibitor H4 | 3.0 | NF |
| NP_001035166.1 | IGHG1 | Anti-RhD monoclonal T125 gamma1 heavy chain | 2.9 | 1.7 |
| NP_002296.1 | LGALS1 | Beta-galactoside-binding lectin (Galectin 1) | 2.8 | NF |
| NP_008977.1 | EMILIN1 | Elastin microfibril interfacer 1 | 2.7 | NF |
| NP_004069.1 | CSRP1 | Cysteine-rich protein 1 | 1.1* | 2.9 |
Fold change in the proteins detected in lectin enrichment method is compared with total protein level wherever available. Novel HCC-associated proteins detected in this study are highlighted with an asterisk.
NF Not found
Overexpressed Proteins in HCC
We used the 18O-labeling method for the relative quantitation of proteins from HCC and adjacent normal tissue. Both unfractionated sample and eluates of lectin-bound proteins from HCC and adjacent normal were separated on 1D SDS-PAGE. Nearly 50% of the proteins identified from lectin affinity methods were also found in whole proteome analysis allowing the comparison of glycosylation status of these proteins in cancer. Both quantitation approaches revealed many known and novel upregulated proteins in cancer. Table 1 shows a partial list of overexpressed proteins detected in lectin affinity methods. Among them haptoglobin [31], alpha-2-microglobulin [32], tenascin C [33], transferrin [34, 35], fibronectin [36], caveolin 1 [37, 38], heat shock 70 kDa protein 5 [39], and ceruloplasmin [40] have been reported to be upregulated in HCC. We also found several interesting proteins, which are highly overexpressed and they have not been reported in HCC. These include serpin peptidase inhibitor (clade A, member 3), alpha-2-HS-glycoprotein, calumenin, cysteine and glycine-rich protein 1, beta-galactoside-binding lectin, peroxiredoxin 2, and clusterin isoform 2. Similarly, many of the proteins enriched by lectin affinity have been shown to be associated with HCC, demonstrating effectiveness of our approach. Lectin affinity method enriched several known glycosylated proteins. Galectin 1 is one such protein overexpressed (3.0-fold) in HCC, which has been reported to be overexpressed at mRNA level in HCC [41]. Alpha-2-glycoprotein 1, zinc and alpha 1B-glycoprotein were found to be 5.0 and 3.1-fold overexpressed in HCC, respectively. Many studies on cancer-associated glycosylation reported upregulated proteins such as haptoglobin related protein and hemopexin which are known to undergo glycosylation in HCC [42, 43]. Clusterin isoform 2 (apolipoprotein J), a sulfated glycoprotein involved in apoptosis, cell death and complement activation, was found to be five-fold upregulated in the present study. Clusterin is a known potential biomarker involved in HCC metastasis [44–46]. Similarly, a glycated form of haptoglobin (>15-fold in HCC) has been shown to be elevated in sera from an HCC patient [31], indicating that quantitative proteomics coupled to lectin affinity method is a promising strategy for cancer biomarker study.
Figure 2 (panels A, B, and C) shows some of the MS and MS/MS spectra of proteins respectively, fetuin, destrin and SERPINA1, which were detected only in lectin affinity enrichment method. Fetuin is a serum glycoprotein, which has earlier been shown to contain both N-glycans and O-glycans [47, 48]. SERPINA1 has been shown to contain N-glycans and mass-spectrometry-based quantitation was successfully used for the measurements of extent of glycosylation [49]. Figure 3 shows the MS and MS/MS spectra of peptides derived proteins, which were also found in whole proteome analysis. MS spectra of peptides from hemopexin (panel A) and peptidylprolyl isomerase B (PPIB; panel B) show increase in glycosylated form when compared to MS spectra from whole proteome analysis. Hemopexin has been found to contain many N-glycan and O-glycan sites [50]. Interestingly, PPIB is not known to be glycosylated; however, in the present study, we observe an elevated glycosylated form in tumor tissue. Similarly, transgelin and cofilin 1 (Fig. 4, panels A and B) indicate different levels of glycosylation in cancer and normal tissues. Although, further studies are necessary to validate our approach, results indicate it is a promising method to study cancer-specific glycosylation versus normal glycosylation events. All proteins identified in lectin affinity method need not represent the cancer-associated hyperglycosylation. In fact, in this study, glycosylation of CSRP1 and peroxiredoxin 6 (Fig. 5, panels A and B) shows no change or decreased level in tumor tissue compared to normal, respectively. In case of tenascin C and HSP 27 kDa, the known upregulated proteins associated with HCC show an HCC-associated increase in both protein level and glycosylated form as shown in Fig. 6 panels A and B, respectively. In general, increased fold changes seen in lectin affinity enrichment method indicate that transgelin, cofilin 1, tenascin C and HSP 27 are upregulated compared to normal tissue, whereas glycated form of CSRP1 did not alter in tumor, glycated form of peroxiredoxin 6 seems to downregulated in tumor compared to normal. Complete lists of proteins identified with their relative quantitation from the two large-scale experiments involving whole lysates and lectin-affinity-enriched proteins from tumor and non-tumor are given in Supplementary Tables 1 and 2, respectively. We further analyzed the overexpressed proteins identified in lectin affinity method for functional annotations [51]. Among the 120 overexpressed proteins, 40 proteins (33%) localized to the extracellular region, 49 (40%) of proteins contain predicted or assigned signal peptide and 25 (20%) were plasma proteins. Our data indicate marked enrichment of glycoproteins (40%) and membrane-related proteins (20%) which would otherwise be under represented in the whole proteome analysis.
Fig. 2.
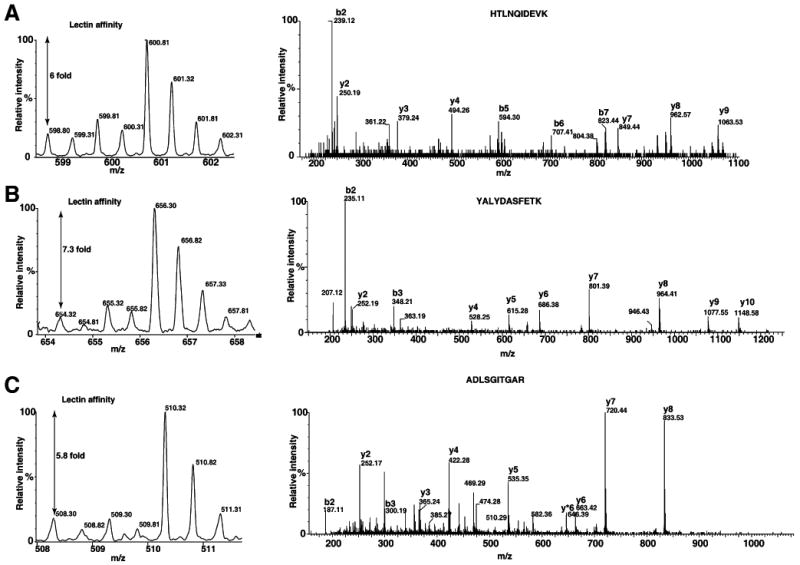
Differential proteomic analysis by 18O labeling: Proteins identified only in lectin affinity enrichment. Panel of MS spectra and MS/MS spectra showing fold changes and peptide sequence identification from overexpressed proteins in HCC. Panels A, B, and C show the MS and MS/MS spectra of HTLNQIDEVK (fetuin), YALYDAS-FETK (destrin) and ADLSGITGAR (SERPINA1), respectively. The fold changes after lectin affinity enrichment is indicated in MS spectra
Fig. 3.
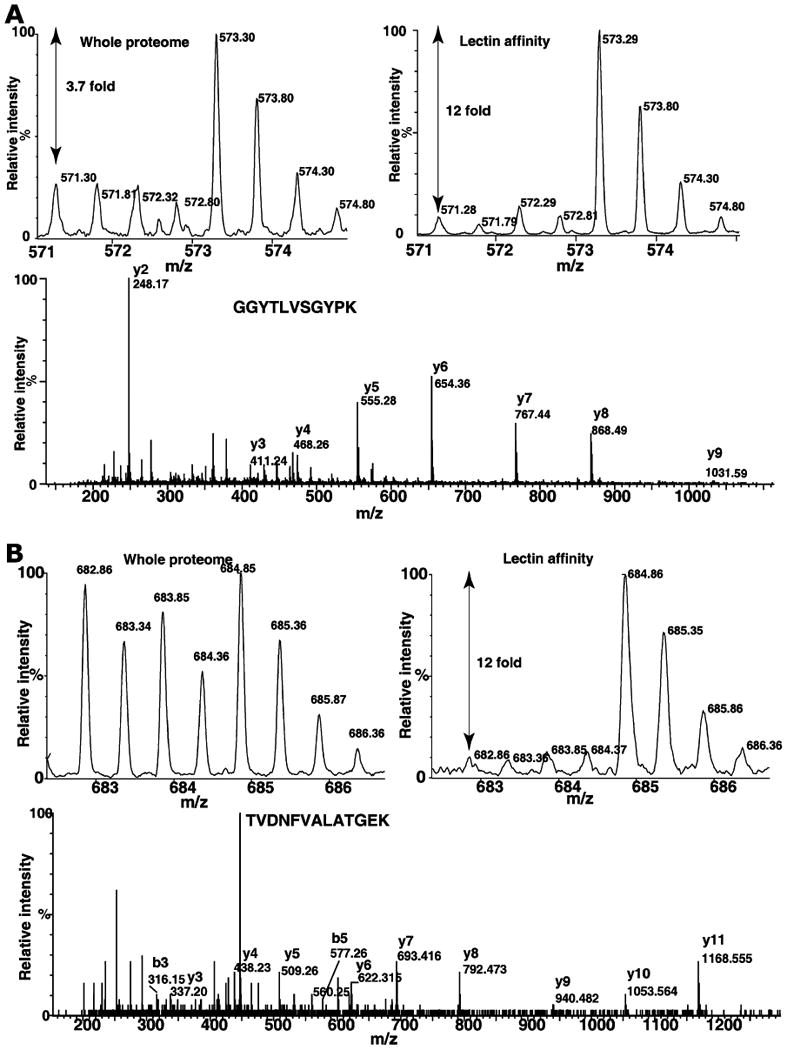
Quantitative analysis of protein changes by whole proteome and lectin enrichment analysis. Panels A and B show the MS and MS/MS spectra of GGYTLVSGYPK (hemopexin) and TVDNFVALATGEK (peptidylprolyl isomerase B), respectively. The fold changes at the whole proteome level and after lectin affinity enrichment are indicated in MS spectra
Fig. 4.
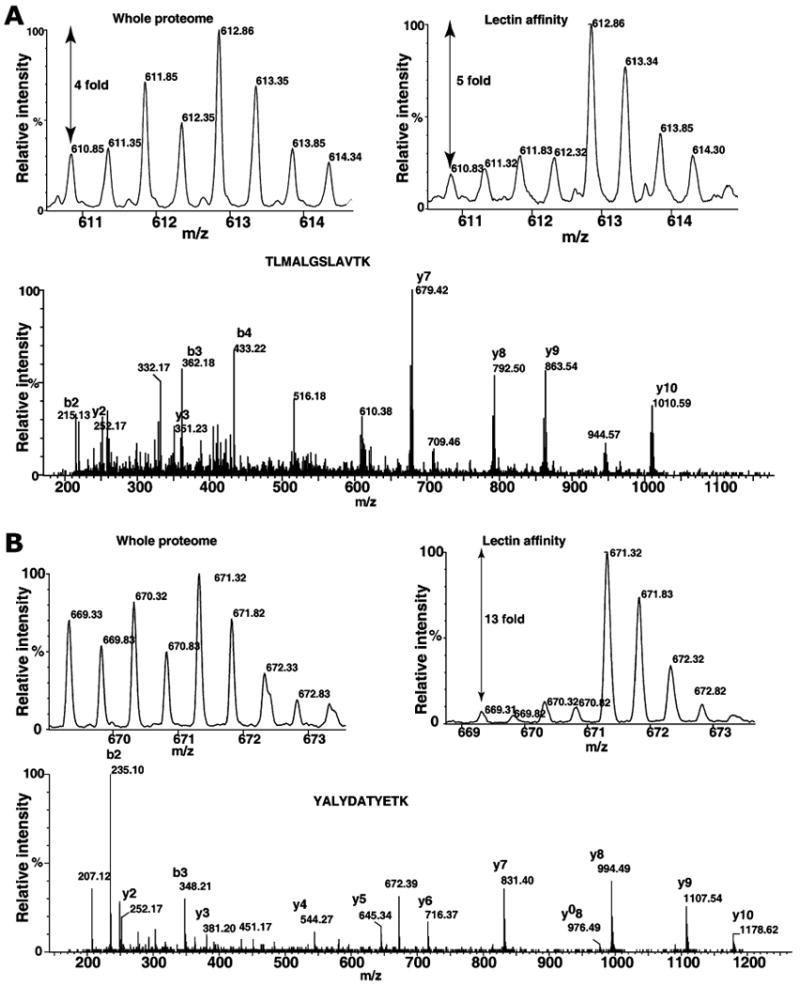
Quantitative analysis of protein changes by whole proteome and lectin enrichment analysis. Panel A and B show the MS and MS/MS spectra of TLMALGSLAVTK (transgelin) and YALYDATYETK (cofilin 1), respectively. The fold changes at the whole proteome level and after lectin affinity enrichment are indicated in MS spectra
Fig. 5.
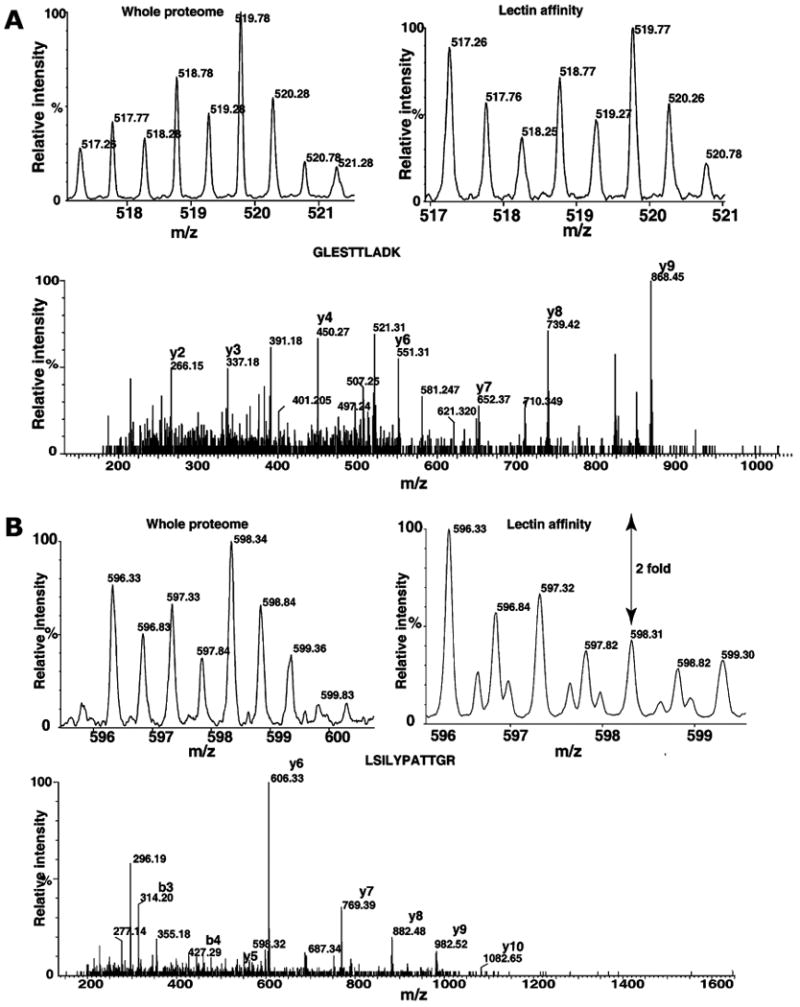
Quantitative analysis of protein changes by whole proteome and lectin enrichment analysis. Panels A and B show the MS and MS/MS spectra of GLESTTLADK (CSRP 1) and LSILYPATTGR (perox-iredoxin 6), respectively. The fold changes at the whole proteome level and after lectin affinity enrichment are indicated in MS spectra
Fig. 6.
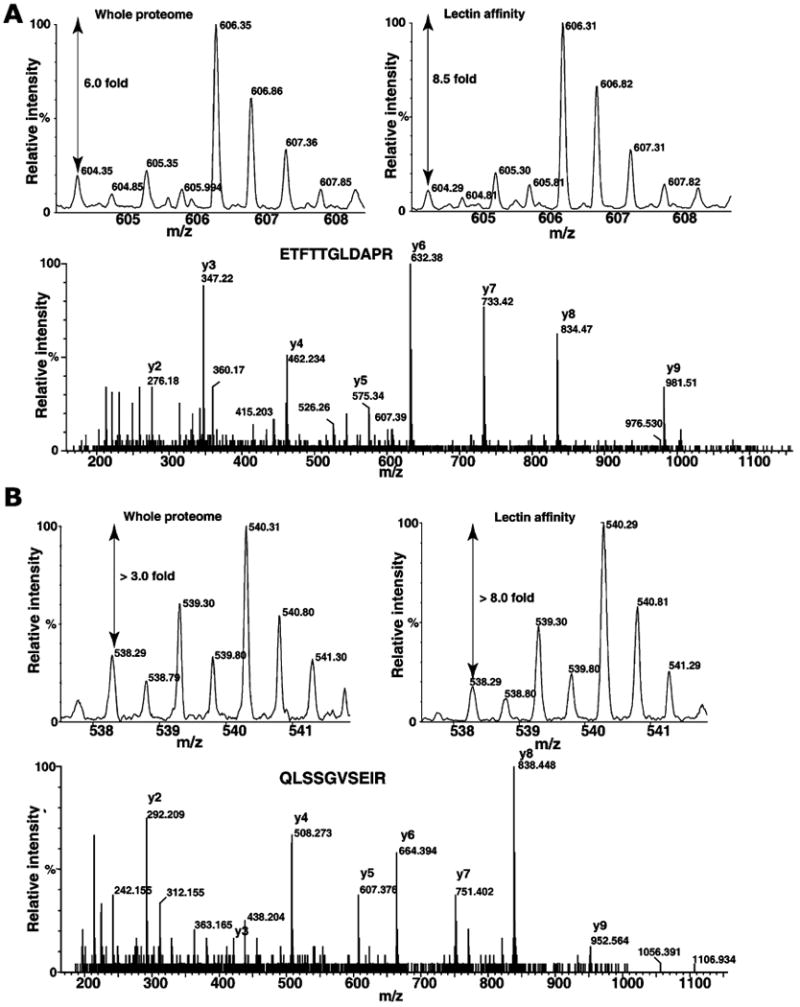
Quantitative analysis of protein changes by whole proteome and lectin enrichment analysis. Panels A and B show the MS and MS/MS spectra of ETFTTGLDAPR (tenascin C) and QLSSGVSEIR (HSP 27-kDa), respectively. The fold changes at the whole proteome level and after lectin affinity enrichment are indicated in MS spectra
Novel HCC-associated Proteins Identified by Lectin Affinity Method
Fetuin (also called as alpha-2-HS-glycoprotein) is a glycoprotein present in serum, which is synthesized by hepatocytes and forms dimers. Although its exact function is unknown, it has been postulated to play a role in tissue growth and brain development. It is found to be a circulating inhibitor of vascular calcification [52]. Fetuin has been shown to be associated with head and neck cancers [53]. In our study, fetuin was overexpressed 3.8-fold in HCC. Alpha-2-glycoprotein 1, zinc-binding (ZAG) is a glycoprotein which has 18.2% carbohydrate and binds zinc ions. It is a secreted protein which stimulates lipid degradation in adipocytes [54]. ZAG is not only expressed in normal liver and breast but also reported to be expressed in breast cancer [55, 56], prostate cancer [57, 58], and bladder cancer [58]. In our study, alpha-2 glycoprotein 1 zinc was overexpressed 4.6-fold in HCC. Transgelin is an actin cross-linking protein found in fibroblasts and smooth muscles. Downregulation of transgelin expression in many cell lines may be a sensitive marker for the onset of transformation. It is reported to be overexpressed in breast cancer, cervical cancer, colorectal cancer, endometrial cancer, head and neck cancer, malignant lymphoma, ovarian cancer, pancreatic cancer, prostate cancer, skin cancer, stomach cancer, and urothelial cancer [59]. Transgelin is reported to be an inhibitor of ARA54-enhanced androgen receptor transactivation which gives an insight into the suppressor role of transgelin in prostate cancer cell growth [60]. In our study, transgelin was overexpressed 6.0-fold in HCC when compared to 2.5 in whole liver proteome without lectin affinity purification.
Destrin belongs to F-actin polymerizing factor family. It is ubiquitously expressed and reported to be associated with colon cancer [61] and Alzheimer's disease [62]. Destrin is overexpressed 6.5-fold in HCC with lectin affinity purification. Leucine-rich alpha-2 glycoprotein 1 belongs to a leucine-rich family of proteins which are involved in protein–protein interactions and signal transduction. This protein consists of a single polypeptide chain with one galactosamine and four glucosamine oligosaccharides attached. Leucine-rich alpha-2 glycoprotein 1 is reported to be a marker involved in neutrophilic granulocyte differentiation [63]. It is also shown to be overexpressed in several cancers similar to transgelin [59]. In our study, leucine-rich alpha-2 glycoprotein 1 was overexpressed 4.7-fold with lectin affinity purification. Heparan sulfate proteoglycan 2 or perlecan is a major component of basement membrane and has angiogenic and growth-promoting attributes. It acts as a co-receptor for basic fibroblasts growth factor. It is reported that antisense targeting of perlecan subsides tumor growth and angiogenesis, and could be a potential target for therapeutics [64]; it has been shown to be overexpressed in prostate cancer [65]. In our study, perlecan was overexpressed 4.3-fold in HCC with lectin affinity enrichment. Hemopexin is a heme-binding plasma glycoprotein which is synthesized in liver and forms about 1.4% of total serum proteins. Each molecule of hemopexin binds to one heme and transports it to hepatocytes for salvage of the iron; it plays an important in receptor mediated cellular uptake of heme [66]. It has been reported that expression of hemopexin is low in fetal liver and its expression increases in adult liver [67]. In our study, hemopexin was overexpressed threefold in HCC.
Validation of CSRP1 and Fetuin Western Blot and IHC
Validation of proteomic data is crucial for eliminating the artifacts of sample heterogeneity. Western blotting and immunohistochemical labeling methods were used for confirming and validating the expression level in tissue microarrays. Based on the availability of antibodies, we checked the expression levels of cysteine and glycine-rich protein (CSRP 1) and fetuin in HCC and adjacent normal tissue and found that these two proteins were overexpressed in HCC when compared to adjacent normal in correlation to our mass spectrometry data. Western blot results shown in Fig. 7a indicates upregulation of CSRP 1 and fetuin from whole proteome. However, as observed in 18O-labeling analysis after lectin enrichment, Western blot shows no change in CSRP1 and increase in level of fetuin in HCC. Fig. 7b shows the increased staining CSRP 1 and fetuin in tumor tissue compared to non-tumor tissue used for proteomic analysis.
Fig. 7.
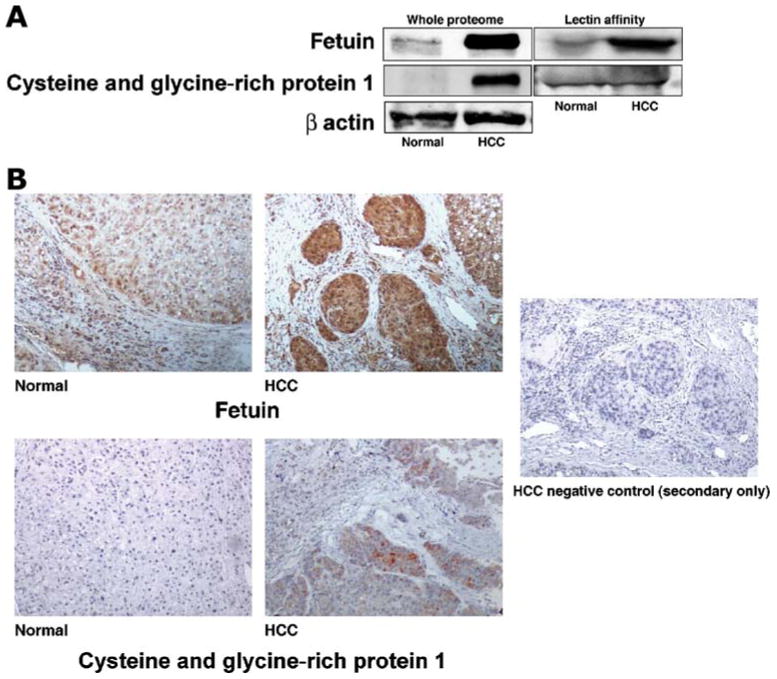
Validation of fetuin and cysteine-rich protein by Western blotting and immunohistochemical labeling in tissues used for proteomic analysis. A The homogenates from the tissue samples were resolved by SDS-PAGE and subsequently electroblotted onto nitrocellulose membrane and probed with specific antibodies as indicated. B Shows immunohistochemical labeling for fetuin and CSRP1, respectively
Validation of Fetuin as an Overexpressed Protein in HCC Using Tissue Microarrays
We used a total of 114 tissue samples from liver cancer patients and 79 non-cancerous liver tissues for the validation of fetuin expression using immunohistochemical labeling. Tumor tissues were grouped into grade I (21 cases), grade II (47 cases), grade III (28 cases), grade IV (five cases), and 13 metastasis tissues. The scoring of the TMAs was done by an expert pathologist, in which the intensity and distribution were each scored from 0 to 3 points. Although fetuin was detectable in both non-cancer and HCC sections, the overall intensity and distribution of staining pattern indicates that fetuin overexpression in HCC compared non-cancer tissues (Fig. 8). No significant difference in expression levels of fetuin among different tissue grades was found. TMAs obtained from a different commercial source contained 32 non-neoplastic matching tissues among them 28 samples showed increased distribution of staining for fetuin compared to matching non-cancer tissues (p<0.05). In all, fetuin was overexpressed in 60/114 (52%) of HCC cases with the total score ranging from 2 to 5 whereas in non-HCC cases, 27/79 (34%) showed expression of fetuin with total score less than 2.
Fig. 8.
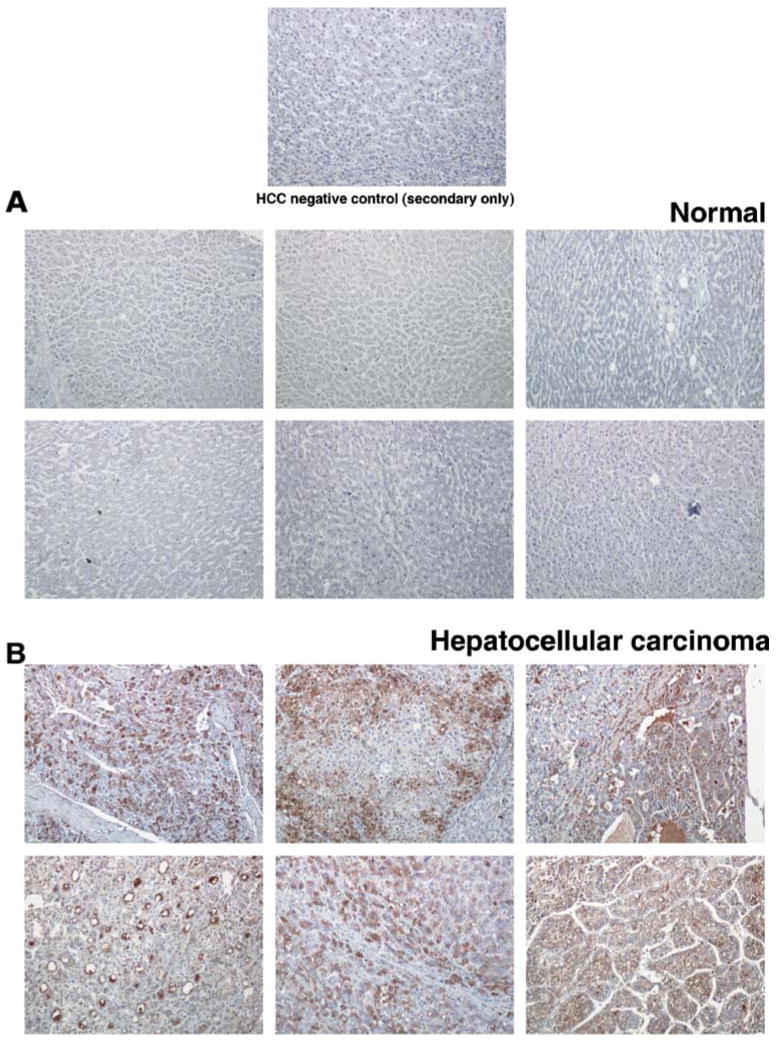
Immunohistochemical labeling for alpha-2HS-glycoprotein proteins in tissue microarrays. Six representative sections from tissue microarrays (TMAs) containing 114 tumor and 79 normal tissues are shown
Identification of N-glycosylation Sites
Using PNGase F and 18O-water, we identified 34 N-glycosylation sites from 28 HCC-associated proteins. The peptides containing the N-glycosylation sites are presented in Table 3, which also shows the consensus sequences identified for each peptide, Mascot score and expect values. *N-X-T/S motif indicates cleavage of glycosylated group at aspargine, incorporation of an 18O atom, and conversion of Asn to Asp 18O (+3 Da). PNGase F cleaves innermost GlcNAc residue linked to Asn resulting in deamidation of Asn to Asp. Earlier, Kristiansen et al. used PNGase F and 18O-water successfully for the identification of N-glycosylated proteins in human bile [20]. Sites identified using PNGaseF treatment contain N-X-T/S motif. Comparison of these sites with the well-annotated human protein database (www.hprd. org) showed majority of known glycosylation sites which indicate glycopeptide enrichment using lectin affinity method, indeed an efficient approach for the enrichment of subproteome. We also identified 14 novel N-glycosylation sites from 12 HCC-associated proteins, two of the novel N-linked glycosylation sites are annotated in MS/MS spectrum as shown in Fig. 9.
Table 3.
N-glycosylated peptides identified from lectin affinity enriched proteins of HCC tissue
| RefSeq accession number | Protein name | Expect value | Mascot score | Peptide sequence | Known? | Consensus motif |
|---|---|---|---|---|---|---|
| NP_000005.2 | Alpha-2-macroglobulin | 1.0E–01 | 42 | SLGNV*NFTVSAEALESQELCGTEVPSVPEHGRK | YES | NFT |
| 2.1E–03 | 57 | VS*NQTLSLFFTVLQDVPVR | YES | NQT | ||
| NP_000375 | Apolipoprotein B | 8.3E–05 | 62 | F*NSSYLQGTNQITGR | YES | NSS |
| NP_001638.1 | Apolipoprotein D | 7.2E–02 | 42 | ADGTVNQIEGEATPV*NLTEPAKLEVK | YES | NLT |
| P00450 | Ceruloplasmin | 3.4E–06 | 85 | E*NLTAPGSDSAVFFEQGTTR | YES | NLT |
| NP_001822.2 | Clusterin isoform 1 | 2.1E–05 | 76 | LA*NLTQGEDQYYLR | YES | NLT |
| NP_009224.2 | Complement component 4A | 1.3E–02 | 47 | GL*NVTLSSTGR | NO | NVT |
| NP_001747.2 | Corticosteroid binding globulin | 3.4E–04 | 54 | AQLLQGLGF*NLTER | YES | NLT |
| NP_005132.2 | Fibrinogen, beta chain | 9.5E–05 | 59 | GTAGNALMDGASQLMGE*NR(T) | NO | NRT |
| NP_000500.2 | Fibrinogen, gamma chain isoform gamma-A | 1.5E–06 | 78 | DLQSLEDILHQVE*NK(T) | YES | NKT |
| NP_006673.1 | Fibrinogen-like 2 | 9.9E–02 | 38 | LDGST*NFTR | NO | NFT |
| 7.1E–05 | 62 | LHVGNY*NGTAGDALR | NO | NGT | ||
| NP_997640.1 | Fibronectin 1 isoform 2 | 4.3E–02 | 44 | DQCIVDDITYNV*NDTFHK | YES | NDT |
| NP_997640 | Fibronectin 1 isoform 2 | 8.9E–07 | 82 | LDAPTNLQFV*NETDSTVLVR | NO | NET |
| NP_009201 | FK506 binding protein 9 | 1.1E–03 | 51 | YHYNASLLDGTLLDSTWNLGK | NO | NAS |
| NP_005657.1 | H factor (complement)-like 3 | 6.5E–05 | 61 | LQNNEN*NISCVER | NO | NIS |
| NP_005134 | Haptoglobin | 1.5E–05 | 78 | MVSHH*NLTTGATLINEQWLLTTAK | YES | NLT |
| 3.5E–07 | 85 | NLFL*NHSENATAK | YES | NHS | ||
| 2.9E–06 | 71 | VVLHP*NYSQVDIGLIK | YES | NYS | ||
| NP_000604 | Hemopexin | 2.1E–05 | 76 | ALPQPQ*NVTSLLGCTH | YES | NVT |
| 3.5E–08 | 94 | SWPAVG*NCSSALR | YES | NCS | ||
| NP_000403.1 | Histidine-rich glycoprotein | 5.6E–03 | 52 | VIDF*NCTTSSVSSALANTK | YES | NCT |
| NP_002207 | Inter-alpha globulin inhibitor H2 | 2.3E–02 | 37 | GAFISNFSMTVDGK | YES | NFS |
| NP_000884 | Kininogen 1 isoform 2 | 5.4E–06 | 73 | ITYSIVQTNCSK | NO | NCS |
| NP_071751 | Leucine proline-enriched proteoglycan 1 | 2.7E–03 | 47 | VPLQSAHLYYNVTEK | NO | NVT |
| NP_006491 | Melanoma cell adhesion molecule | 7.2E–03 | 42 | CVASVPSIPGLNR(T) | NO | NRT |
| NP_002395 | Microfibrillar-associated protein 4 | 2.3E–04 | 57 | FNGSVSFFR | NO | NGS |
| 1.1E–04 | 69 | VDLEDFE*NNTAYAK | NO | NNT | ||
| NP_006380.1 | Oxygen regulated protein | 3.2E–09 | 104 | LSALDNLL*NHSSMFLK | YES | NHS |
| CAJ15161 | Alpha-antitrypsin | 4.0E–08 | 102 | YLG*NATAIFFLPDEGK | YES | NAT |
| NP_000479.1 | Serine (or cysteine) proteinase inhibitor, clade C | 8.1E–06 | 70 | LGAC*NDTLQQLMEVFK | YES | NDT |
| NP_001076.2 | serpin peptidase inhibitor, clade A | 2.3E–04 | 65 | F*NLTETSEAEIHQSFQHLLR | NO | NLT |
| NP_001031.2 | Sex hormone-binding globulin | 9.5E–03 | 41 | LDVDQAL*NR(S) | NO | NRS |
| NP_001054 | Transferrin | 7.4E–06 | 72 | CGLVPVLAENY*NK(S) | YES | NKS |
| 3.0E–02 | 46 | QQQHLFGS*NVTDCSGNFCLFR | YES | NVT |
Accession number and protein names were from NCBI RefSeq database, and mascot score and expect values were listed for N-glycosylated peptides. Human protein reference database (www.hprd.org) was used for the identification of previously reported N-glycosylation sites. An asterisk in *N-X-T/S implies 18 O labeling of Asn after PNGase F mediated cleavage of glycan.
Fig. 9.
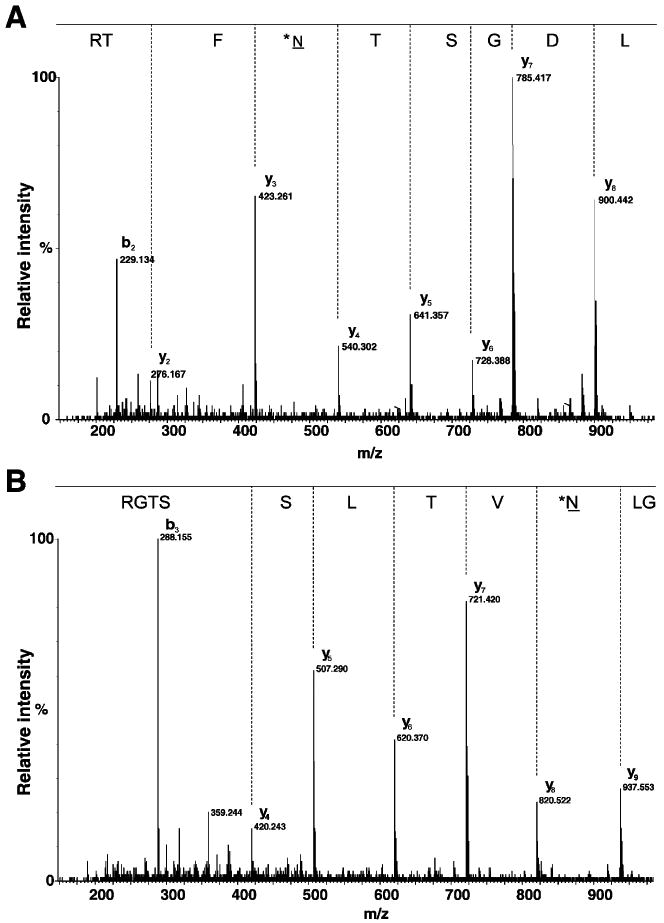
MS/MS spectra of peptides from fibrinogen-like 2 and complement component 4A analyzed using QToF. The spectra A and B show the N-glycosylation sites of the peptide LDGST*NFTR from fibrinogen-like 2 and GL*NVTLSSTGR from complement component 4A respectively (where *N indicates the N-glycosylation site modified by 18O). Spectrum shows the conversion of Asn to Asp after PNGase F treatment
Levels of Transgelin and Destrin in Lectin-enriched Serum
To evaluate the expression of some of the interesting candidate proteins found in our study, we used lectin affinity enrichment of serum for Western blot analysis to see whether tissue-level fold change is reflected in the serum from HCC patients. In this experiment, sera from eight HCC patients and pooled normal sera (Invitrogen) were chosen for lectin affinity enrichment followed by Western blot analysis. Figure 10 shows an average increase level of transgelin and destrin in serum of HCC compared to normal.
Fig. 10.
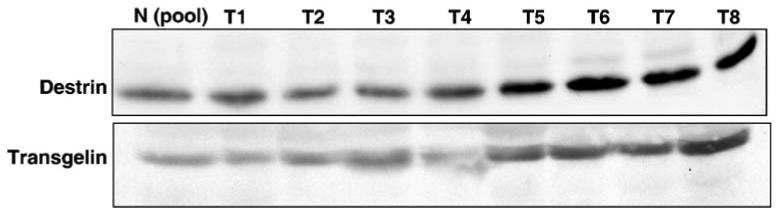
Validation of candidate proteins by Western blotting in serum. Serum samples (7 mg) from HCC patients and normal pooled sample was enriched for glycoproteins using mixture of lectins as explained in methods. Eluted proteins were resolved by SDS-PAGE and subsequently electroblotted onto nitrocellulose membrane and probed with antibodies for destrin and transgelin
Conclusions
Quantitative profiling of proteins in HCC revealed many overexpressed proteins relevant to HCC. Many proteins that have been previously associated with HCC were detected in this study. Important among those were complement component H, tenascin C, ceruloplasmin, haptoglobin, and alpha-2 macroglobin. More importantly, a number of novel HCC-associated proteins discovered in this study are candidates for validation in HCC using alternate platforms such as immunohistochemical labeling or ELISA. One of the novel candidates, fetuin was validated as a potential biomarker using tissue microarrays containing 114 tumor and 79 normal liver tissue sections. Although many lectin-bound proteins were found to be overexpressed in HCC, whole protein levels were also high in both normal and tumor cells. We also compared the extent of fold changes in proteins before and after lectin affinity enrichment to evaluate the status of glycosylation in HCC. Many of the proteins identified in this study showed differences in fold changes before and after lectin affinity enrichment indicating that this subset of proteins may represent hyper-glycosylation associated with cancer. This approach is useful in identifying cancer-associated proteins which are otherwise undetectable in a complex mixture. For example, fetuin, SERPINA1, and destrin were identified only by lectin affinity method because of reduced complexity. Hemopexin, peptidyl prolyl isomerase B, cofilin 1, and HSP 27-kDa protein showed more fold changes in lectin affinity method indicating glycation specific changes in these proteins in HCC. Our study showed many N-linked glycosylation sites from HCC including novel sites, which may be of interest for further study of glycoprotein marker. Nearly 30% of the protein-bound lectins contain confirmed N-glycosylation sites indicating this technical approach of quantitative proteomics using multiple lectin affinity method can be used for simple screening of biomarkers in cancer. Majority of the confirmed N-glycosylation sites were found in proteins upregulated in HCC such as haptoglobin, hemopexin, clusterin, and ceruloplasmin. While many potential biomarker studies in HCC are based on identification and validation of single upregulated proteins which involve unfractionated serum or tissues, lectin affinity allows enrichment of subproteomes containing aberrantly glycated proteins and help in identification of multiple markers in a single analysis. A significant number of upregulated proteins (40%) identified in this study were also predicted to be N-glycosylated proteins [51]. In our study, PNGase F treatment in the presence of 18O-water found to be useful for the identification of N-glycosylation sites in addition to the presence of consensus motif N-X-T/S. In conclusion, quantitative proteomic approach can be successfully used for the analysis of subproteomes, such as glycosylated proteins, to specifically identify and quantitate N-glycosylated proteins and peptides.
Supplementary Material
Acknowledgments
This work is supported by the family of Margaret Lee. A. P. is supported by National Institutes of Health Grant CA62924 and the Alexander and Margaret Stewart Trust and a grant from the Cancer Center (GI SPORE).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s12014-008-9013-0) contains supplementary material, which is available to authorized users.
Contributor Information
Raghothama Chaerkady, Institute of Bioinformatics, International Technology Park, Bangalore, 560066, India; McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Biological Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Paul J. Thuluvath, Departments of Hepatology and Liver Transplantation, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
Min-Sik Kim, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Biological Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Anuradha Nalli, Institute of Bioinformatics, International Technology Park, Bangalore, 560066, India; McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Biological Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Perumal Vivekanandan, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Jessica Simmers, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Biological Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Michael Torbenson, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Akhilesh Pandey, Email: pandey@jhmi.edu, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Biological Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA; Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–6. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Reuben A. The crab, the turkey and a malignant tale from the year of the rooster. Hepatology. 2005;41:944–50. doi: 10.1002/hep.20674. [DOI] [PubMed] [Google Scholar]
- 3.Torbenson M, Kannangai R, Abraham S, Sahin F, et al. Concurrent evaluation of p53, beta-catenin, and alpha-fetoprotein expression in human hepatocellular carcinoma. Am J Clin Pathol. 2004;122:377–82. doi: 10.1309/YH0H-3FKY-M4RM-U1JF. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura S, Nouso K, Sakaguchi K, Ito YM, et al. Sensitivity and specificity of des-gamma-carboxy prothrombin for diagnosis of patients with hepatocellular carcinomas varies according to tumor size. Am J Gastroenterol. 2006;101:2038–43. doi: 10.1111/j.1572-0241.2006.00681.x. [DOI] [PubMed] [Google Scholar]
- 5.Toyoda H, Kumada T, Kiriyama S, Sone Y, et al. Prognostic significance of simultaneous measurement of three tumor markers in patients with hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2006;4:111–7. doi: 10.1016/s1542-3565(05)00855-4. [DOI] [PubMed] [Google Scholar]
- 6.Ohhira M, Ohtake T, Saito H, Ikuta K, et al. Increase of serum des-gamma-carboxy prothrombin in alcoholic liver disease without hepatocellular carcinoma. Alcohol Clin Exp Res. 1999;23:67–70. doi: 10.1111/j.1530-0277.1999.tb04537.x. [DOI] [PubMed] [Google Scholar]
- 7.Sassa T, Kumada T, Nakano S, Uematsu T. Clinical utility of simultaneous measurement of serum high-sensitivity des-gamma-carboxy prothrombin and Lens culinaris agglutinin A-reactive alpha-fetoprotein in patients with small hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 1999;11:1387–92. doi: 10.1097/00042737-199912000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Raghothama C, Pandey A. Absolute systems biology—measuring dynamics of protein modifications. Trends Biotechnol. 2003;21:467–70. doi: 10.1016/j.tibtech.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Comunale MA, Lowman M, Long RE, Krakover J, et al. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J Proteome Res. 2006;5:308–15. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- 10.Aoyagi Y. Carbohydrate-based measurements on alpha-fetoprotein in the early diagnosis of hepatocellular carcinoma. Glycoconj J. 1995;12:194–9. doi: 10.1007/BF00731319. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita K, Koide N, Endo T, Iwaki Y, et al. Altered glycosylation of serum transferrin of patients with hepatocellular carcinoma. J Biol Chem. 1989;264:2415–23. [PubMed] [Google Scholar]
- 12.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, et al. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J Proteome Res. 2006;5:1626–35. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 13.Dai Z, Liu YK, Cui JF, Shen HL, et al. Identification and analysis of altered alpha1,6-fucosylated glycoproteins associated with hepatocellular carcinoma metastasis. Proteomics. 2006;6:5857–67. doi: 10.1002/pmic.200500707. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Simeone DM, Heidt D, Anderson MA, et al. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: application to pancreatic cancer serum. J Proteome Res. 2006;5:1792–802. doi: 10.1021/pr060034r. [DOI] [PubMed] [Google Scholar]
- 15.Xu Z, Zhou X, Lu H, Wu N, et al. Comparative glycoproteomics based on lectins affinity capture of N-linked glycoproteins from human Chang liver cells and MHCC97-H cells. Proteomics. 2007;7:2358–70. doi: 10.1002/pmic.200600041. [DOI] [PubMed] [Google Scholar]
- 16.Ueda K, Katagiri T, Shimada T, Irie S, et al. Comparative profiling of serum glycoproteome by sequential purification of glycoproteins and 2-nitrobenzensulfenyl (NBS) stable isotope labeling: a new approach for the novel biomarker discovery for cancer. J Proteome Res. 2007;6:3475–83. doi: 10.1021/pr070103h. [DOI] [PubMed] [Google Scholar]
- 17.Li D, Mallory T, Satomura S. AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin Chim Acta. 2001;313:15–9. doi: 10.1016/s0009-8981(01)00644-1. [DOI] [PubMed] [Google Scholar]
- 18.Patwa TH, Zhao J, Anderson MA, Simeone DM, et al. Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem. 2006;78:6411–21. doi: 10.1021/ac060726z. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem. 2006;52:1897–905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- 20.Kristiansen TZ, Bunkenborg J, Gronborg M, Molina H, et al. A proteomic analysis of human bile. Mol Cell Proteomics. 2004;3:715–28. doi: 10.1074/mcp.M400015-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Gygi SP, Rist B, Gerber SA, Turecek F, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 22.Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–69. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Yao X, Freas A, Ramirez J, Demirev PA, et al. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem. 2001;73:2836–42. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 24.Amanchy R, Kalume DE, Pandey A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci STKE. 2005;2005:pl2. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 25.Bantscheff M, Dumpelfeld B, Kuster B. Femtomol sensitivity post-digest (18)O labeling for relative quantification of differential protein complex composition. Rapid Commun Mass Spectrom. 2004;18:869–76. doi: 10.1002/rcm.1418. [DOI] [PubMed] [Google Scholar]
- 26.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–4. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 27.Tian Y, Zhou Y, Elliott S, Aebersold R, et al. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2:334–9. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakomori S. Tumor malignancy defined by aberrant glycosylation and sphingo(glyco)lipid metabolism. Cancer Res. 1996;56:5309–18. [PubMed] [Google Scholar]
- 29.Heller M, Mattou H, Menzel C, Yao X. Trypsin catalyzed 16O-to-18O exchange for comparative proteomics: tandem mass spectrometry comparison using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J Am Soc Mass Spectrom. 2003;14:704–18. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 30.Lane CS, Wang Y, Betts R, Griffiths WJ, et al. Comparative cytochrome P450 proteomics in the livers of immunodeficient mice using 18O stable isotope labeling. Mol Cell Proteomics. 2007;6:953–62. doi: 10.1074/mcp.M600296-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ang IL, Poon TC, Lai PB, Chan AT, et al. Study of serum haptoglobin and its glycoforms in the diagnosis of hepatocellular carcinoma: a glycoproteomic approach. J Proteome Res. 2006;5:2691–700. doi: 10.1021/pr060109r. [DOI] [PubMed] [Google Scholar]
- 32.Otegbayo JA, Arinola OG, Aje A, Oluwasola OA, et al. Usefulness of acute phase proteins for monitoring development of hepatocellular carcinoma in hepatitis B virus carriers. West Afr J Med. 2005;24:124–7. doi: 10.4314/wajm.v24i2.28181. [DOI] [PubMed] [Google Scholar]
- 33.Zhao M, Laissue JA, Zimmermann A. Tenascin and type IV collagen expression in liver cell dysplasia and in hepatocellular carcinoma. Histol Histopathol. 1996;11:323–3. [PubMed] [Google Scholar]
- 34.Moertel CG, Douglass HO, Hanley J, Carbone PP. Phase II study of methyl-CCNU in the treatment of advanced pancreatic carcinoma. Cancer Treat Rep. 1976;60:1659–61. [PubMed] [Google Scholar]
- 35.Suzuki Y, Aoyagi Y, Mori S, Suda T, et al. Microheterogeneity of serum transferrin in the diagnosis of hepatocellular carcinoma. J Gastroenterol Hepatol. 1996;11:358–65. doi: 10.1111/j.1440-1746.1996.tb01384.x. [DOI] [PubMed] [Google Scholar]
- 36.Gupta N, Kakkar N, Vasishta RK. Pattern of fibronectin in hepatocellular carcinoma and its significance. Indian J Pathol Microbiol. 2006;49:362–4. [PubMed] [Google Scholar]
- 37.Yerian LM, Anders RA, Tretiakova M, Hart J. Caveolin and thrombospondin expression during hepatocellular carcinogenesis. Am J Surg Pathol. 2004;28:357–64. doi: 10.1097/00000478-200403000-00008. [DOI] [PubMed] [Google Scholar]
- 38.Yokomori H, Oda M, Yoshimura K, Nomura M, et al. Elevated expression of caveolin-1 at protein and mRNA level in human cirrhotic liver: relation with nitric oxide. J Gastroenterol. 2003;38:854–60. doi: 10.1007/s00535-003-1161-4. [DOI] [PubMed] [Google Scholar]
- 39.Chignard N, Shang S, Wang H, Marrero J, et al. Cleavage of endoplasmic reticulum proteins in hepatocellular carcinoma: detection of generated fragments in patient sera. Gastroenterology. 2006;130:2010–22. doi: 10.1053/j.gastro.2006.02.058. [DOI] [PubMed] [Google Scholar]
- 40.Zhang YJ, Zhao DH, Huang CX. The changes in copper contents and its clinical significance in patients with liver cirrhosis and hepatocarcinoma. Zhonghua Nei Ke Za Zhi. 1994;33:113–6. [PubMed] [Google Scholar]
- 41.Kondoh N, Hada A, Ryo A, Shuda M, et al. Activation of Galectin-1 gene in human hepatocellular carcinoma involves methylation-sensitive complex formations at the transcriptional upstream and downstream elements. Int J Oncol. 2003;23:1575–83. [PubMed] [Google Scholar]
- 42.Kotaka M, Chen GG, Lai PB, Lau WY, et al. Analysis of differentially expressed genes in hepatocellular carcinoma with hepatitis C virus by suppression subtractive hybridization. Oncol Res. 2002;13:161–7. [PubMed] [Google Scholar]
- 43.Zhao J, Patwa TH, Qiu W, Shedden K, et al. Glycoprotein microarrays with multi-lectin detection: unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res. 2007;6:1864–74. doi: 10.1021/pr070062p. [DOI] [PubMed] [Google Scholar]
- 44.Feng JT, Liu YK, Song HY, Dai Z, et al. Heat-shock protein 27: a potential biomarker for hepatocellular carcinoma identified by serum proteome analysis. Proteomics. 2005;5:4581–8. doi: 10.1002/pmic.200401309. [DOI] [PubMed] [Google Scholar]
- 45.Kang YK, Hong SW, Lee H, Kim WH. Overexpression of clusterin in human hepatocellular carcinoma. Hum Pathol. 2004;35:1340–6. doi: 10.1016/j.humpath.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 46.Lau SH, Sham JS, Xie D, Tzang CH, et al. Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene. 2006;25:1242–50. doi: 10.1038/sj.onc.1209141. [DOI] [PubMed] [Google Scholar]
- 47.Bunkenborg J, Pilch BJ, Podtelejnikov AV, Wisniewski JR. Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics. 2004;4:454–65. doi: 10.1002/pmic.200300556. [DOI] [PubMed] [Google Scholar]
- 48.Yoshioka Y, Gejyo F, Marti T, Rickli EE, et al. The complete amino acid sequence of the A-chain of human plasma alpha 2HS-glycoprotein. J Biol Chem. 1986;261:1665–76. [PubMed] [Google Scholar]
- 49.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–6. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi N, Takahashi Y, Putnam FW. Structure of human hemopexin: O-glycosyl and N-glycosyl sites and unusual clustering of tryptophan residues. Proc Natl Acad Sci U S A. 1984;81:2021–5. doi: 10.1073/pnas.81.7.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 52.Ziolkowska H, Wojnar J, Panczyk-Tomaszewska M, Roszkowska-Blaim M. Fetuin A in children with renal diseases. Przegl Lek. 2006;63(Suppl 3):54–6. [PubMed] [Google Scholar]
- 53.Suarez Nieto C, Cuesta Garcia A, Fernandez Bustillo E, Mendez Colunga JC, et al. Serum glycoproteins and prognosis in cancer of the head and neck. Clin Otolaryngol Allied Sci. 1986;11:41–5. doi: 10.1111/j.1365-2273.1986.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 54.Bao Y, Bing C, Hunter L, Jenkins JR, et al. Zinc-alpha2-glycoprotein, a lipid mobilizing factor, is expressed and secreted by human (SGBS) adipocytes. FEBS Lett. 2005;579:41–7. doi: 10.1016/j.febslet.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 55.Diez-Itza I, Sanchez LM, Allende MT, Vizoso F, et al. Zn-alpha 2-glycoprotein levels in breast cancer cytosols and correlation with clinical, histological and biochemical parameters. Eur J Cancer. 1993;29A:1256–60. doi: 10.1016/0959-8049(93)90068-q. [DOI] [PubMed] [Google Scholar]
- 56.Freije JP, Fueyo A, Uria J, Lopez-Otin C. Human Zn-alpha 2-glycoprotein cDNA cloning and expression analysis in benign and malignant breast tissues. FEBS Lett. 1991;290:247–9. doi: 10.1016/0014-5793(91)81271-9. [DOI] [PubMed] [Google Scholar]
- 57.Descazeaud A, de la Taille A, Allory Y, Faucon H, et al. Characterization of ZAG protein expression in prostate cancer using a semi-automated microscope system. Prostate. 2006;66:1037–43. doi: 10.1002/pros.20405. [DOI] [PubMed] [Google Scholar]
- 58.Irmak S, Tilki D, Heukeshoven J, Oliveira-Ferrer L, et al. Stage-dependent increase of orosomucoid and zinc-alpha2-glycoprotein in urinary bladder cancer. Proteomics. 2005;5:4296–304. doi: 10.1002/pmic.200402005. [DOI] [PubMed] [Google Scholar]
- 59.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–32. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Yang Z, Chang YJ, Miyamoto H, Ni J, et al. Transgelin functions as a suppressor via inhibition of ARA54-enhanced androgen receptor transactivation and prostate cancer cell growth. Mol Endocrinol. 2007;21:343–58. doi: 10.1210/me.2006-0104. [DOI] [PubMed] [Google Scholar]
- 61.Estornes Y, Gay F, Gevrey JC, Navoizat S, et al. Differential involvement of destrin and cofilin-1 in the control of invasive properties of Isreco1 human colon cancer cells. Int J Cancer. 2007;121:2162–71. doi: 10.1002/ijc.22911. [DOI] [PubMed] [Google Scholar]
- 62.Heredia L, Helguera P, de Olmos S, Kedikian G, et al. Phosphorylation of actin-depolymerizing factor/cofilin by LIM-kinase mediates amyloid beta-induced degeneration: a potential mechanism of neuronal dystrophy in Alzheimer's disease. J Neurosci. 2006;26:6533–42. doi: 10.1523/JNEUROSCI.5567-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Donnell LC, Druhan LJ, Avalos BR. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J Leukoc Biol. 2002;72:478–85. [PubMed] [Google Scholar]
- 64.Sharma B, Handler M, Eichstetter I, Whitelock JM, et al. Antisense targeting of perlecan blocks tumor growth and angiogenesis in vivo. J Clin Invest. 1998;102:1599–608. doi: 10.1172/JCI3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Datta MW, Hernandez AM, Schlicht MJ, Kahler AJ, et al. Perlecan, a candidate gene for the CAPB locus, regulates prostate cancer cell growth via the Sonic Hedgehog pathway. Mol Cancer. 2006;5:9. doi: 10.1186/1476-4598-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu ML, Morgan WT. Conformational analysis of hemopexin by Fourier-transform infrared and circular dichroism spectroscopy. Proteins. 1994;20:185–90. doi: 10.1002/prot.340200208. [DOI] [PubMed] [Google Scholar]
- 67.Naylor SL, Altruda F, Marshall A, Silengo L, et al. Hemopexin is localized to human chromosome 11. Somat Cell Mol Genet. 1987;13:355–8. doi: 10.1007/BF01534930. [DOI] [PubMed] [Google Scholar]
- 68.Tsai JF, Tsai JH, Chang WY. Relationship of serum alpha-fetoprotein to circulating immune complexes and complements in patients with hepatitis B surface antigen-positive hepatocellular carcinoma. Gastroenterol Jpn. 1990;25:388–93. doi: 10.1007/BF02779456. [DOI] [PubMed] [Google Scholar]
- 69.Yang LY, Chen WL, Lin JW, Lee SF, et al. Differential expression of antioxidant enzymes in various hepatocellular carcinoma cell lines. J Cell Biochem. 2005;96:622–31. doi: 10.1002/jcb.20541. [DOI] [PubMed] [Google Scholar]
- 70.Steel LF, Shumpert D, Trotter M, Seeholzer SH, et al. A strategy for the comparative analysis of serum proteomes for the discovery of biomarkers for hepatocellular carcinoma. Proteomics. 2003;3:601–9. doi: 10.1002/pmic.200300399. [DOI] [PubMed] [Google Scholar]
- 71.Nomura F, Yaguchi M, Togawa A, Miyazaki M, et al. Enhancement of poly-adenosine diphosphate-ribosylation in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2000;15:529–35. doi: 10.1046/j.1440-1746.2000.02193.x. [DOI] [PubMed] [Google Scholar]
- 72.Ding SJ, Li Y, Shao XX, Zhou H, et al. Proteome analysis of hepatocellular carcinoma cell strains, MHCC97-H and MHCC97-L, with different metastasis potentials. Proteomics. 2004;4:982–94. doi: 10.1002/pmic.200300653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


