Abstract
Huntington disease (HD) occurs only in humans. Thus, its natural pathogenesis takes place exclusively within the human brains expressing the causative, mutated protein huntingtin (mhtt). The techniques applicable to postmortem human HD brains are inadequate for investigating the cellular pathogenesis. The creation of genetically engineered animals represents a critical moment in neuroscience. Monitoring the actions of either normal, or abnormal proteins at subcellular levels, and at different time points is now possible thanks to these models. They are the necessary substitutes to investigate the wild type (whtt), or mhtt. The postmortem neuropathologic phenotype of the human HD is well documented. Its pattern and spectrum are highly predictable. From this point of view, the existent models do not exhibit the phenotypic constellation of changes seen in the human HD brains. On one hand, this deficit reflects the limitations of the methods of evaluation used in a clinical setting. On the other hand, it highlights the limitations of the animals. The validity of the models probably should be measured by their capacity of reproducing the cellular dysfunctions of HD rather than the phenotype of the postmortem human brains. Although not perfect, these models are essential for modeling the human disease in cells, which is not feasible with postmortem human HD brains. Nonetheless, their relevance to the patient population remains to be determined. Ultimately needed are means preventing the disease to occur, the discovery of which probably depends on these models.
Keywords: Huntington disease, Transgenic mouse, Knockin mouse, Mice gene carriers of the HD mutation
Introduction
Genetically engineered animals carrying part or the full length of the mutated gene causing Huntington disease (HD), which naturally occurs exclusively in humans, are the most powerful tools available for investigating its referent, neurotoxic mechanisms [81]. Genetically engineered animal models may offer the opportunity to explore the pathogenic cascade at the subcellular level at different time points during the degenerative process. The advent of these models induced original hypotheses, brought about new tools of investigations, and attracted brilliant neuroscientists in the field. The ongoing research is yielding essential data on neurodegenerative mechanisms. The animal models gained acceptance as novel, powerful devices of investigation. The research on HD, beyond what is currently feasible methodologically when using postmortem human brain samples, depends mostly on transgenic animals. Indeed, animal models will be essential for monitoring candidate agents aimed at preventing or delaying the toxicity of the mutant huntingtin (mhtt), and to ascertain their side effects. However, from the human neuropathology point of view of HD, the existent models do not exhibit the phenotypic constellation of changes characteristically seen in the human HD brain. This phenotypic deficit is in part due to the limitation of the usual methods that are conventionally applied in a clinical setting to evaluate neuropathologically human brains. Nonetheless, that the dedicated scientific publications often include statements such as “we believe…”, “our findings suggest…” reflects the intuitive awareness of the shortcomings of the models used thus far to probe the cellular pathogenesis of HD.
Comparing the neuropathologic changes in the HD human brains with those occurring in the brains of the engineered animals is challenging. First, a thorough knowledge of the type, distribution, and extent of the changes characteristically occurring in either group is required. Second, the validity of the comparison depends in part on the nature of the changes at different stages of the disease, and on the methods used to evaluate them. Third, the difference in the life expectancy of the species involved is up to 60 years, which greatly complicates the assessment of the analogy. Fourth, using mice, rats, or flies with distinctive genetic constructs complicates the discernment of the differences of significance. Finally, the nature, novelty, and complexity of the subject, and the ongoing yield of multifaceted data obtained from these models, impose an arbitrary selective approach of the topic addressed in this review.
The aim of this report is to evaluate the extent to which engineered animal models, notably HD-IT15 CAG repeat expansion mice (polyQ mice), phenotypically mimic the neuropathologic changes seen in postmortem brains from individuals diagnosed with HD. Thus, this assessment is made from the point of view of a neuropathologist dealing mainly with human neurodegenerative diseases, including the thorough, standardized evaluations of 1,250 human HD brains, using conventional methods of neuropathologic evaluation. This critique is not a verdict but an appraisal of what may be learnable, the potential ramifications, and limits of the animal models as per a contemporary context. The benefits and costs of the animal models are manifold. The new knowledge these animals provide outwardly justify the costs and efforts of the investigations involving them.
Mouse models of HD or mouse carriers of the HD mutation?
The first engineered mice carrying a fragment of the abnormal human gene causing HD became available in 1996 [60]. Since then, additional and distinctive models have been developed, triggering an extraordinary increase in the number of investigations, observations, and reports. Although many seductive pathogenic hypotheses have been tested, so far no affiliated therapy has been developed [4]. Indeed, despite the availability of these models and the abundance of cellular abnormalities allegedly due to the presence of mhtt they helped identify, the neuropathological phenotype of human HD remains the same. The exception is the identification of the ubiquitinated nuclear and cytoplasmic aggregates of mhtt involving nuclei of 1–7% of cortical, striatal, or thalamic neurons and neuropil of patients with adult onset of symptoms [5, 19, 25, 32, 46], and up to 21% of cortical neurons in patients with juvenile onset of symptoms [58]. The observation of these aggregates in human brains was made following the occurrence of widespread nuclear inclusions seen in neurons, glial, and ependymal cells of the first transgenic mice harboring exon 1 of the human gene [17] (Fig. 1). This genetic insertion encodes mhtt with expanded CAG-repeats, which translate into a series of consecutive glutamine residues or polyglutamine tract (polyQ).
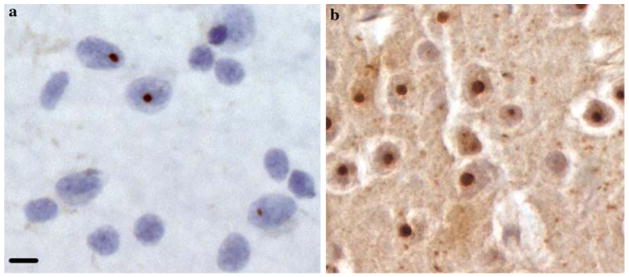
Fig. 1.
Neurons displaying ubiquitinated nuclear inclusions. a Smear obtained from Brodmann area (BA) 8 of a 54-year-old patient with HD grade 4/4. Three nuclei of neurons harbor an ubiquitinated inclusion. b Microphotograph from the frontal cortex of an R6/2 mouse. R6/2 mice were the first generated that are transgenic for the 5′ end of the human HD gene carrying (CAG)115–(CAG)150 repeat expansions. The R6/2 mouse was instrumental in the discovery of the occurrence of these aggregates in the human HD brain. a, b Immunohistochemistry, same magnification. Scale bar 6 μm
The disparate models employed and the profusion of data generated appear to obscure rather than reveal the relevance of the subcellular dysfunctions recorded in these models. A flood of reports documenting alterations of the many hitherto relatively ignored, interrelated tiers of RNA-based genetic regulation is likely to inundate the scientific literature in the near future.
Hence the question: how pertinent are these findings to human HD? Whatever the objective of the answer to this question, it would be premature. Unfortunately, no matter how genuine transgenic models are or will be, the fact is that the ultimate, global pathogenic cascade of HD occurs only within the human HD brain. The shortstop mouse, which remains asymptomatic despite expressing the HD polyQ and the referent nuclear and neuropil aggregates, supports this claim. Thus, the research on either humans or animal models must be much more convergent than it is now. The birth of the transgenic polyQ mouse, which was accepted from the start as a model of HD, suddenly shifted the focus of basic research away from postmortem human HD brains. This occurred on the premise that the resulting engineered mice were indeed a reliable model of HD, and they were referred to as such, albeit their neuropathological phenotype was not analogous to that of the HD human.
Indeed, criteria for defining the degree of validity of the artificial imitation of HD are required. The assessment of the relevance of data obtained from the models to the natural form of the disease depends on the degree of validity of the models. Lack of such criteria fosters soft relativism, biased conclusions, and questionable claims when assessing results of investigations made with these models. The labor-intensive endeavor bestowed on investigations involving animal models, their novelty, and the commitment they require may condition the processing and analyses of the data they yield. Therefore, the relevance of the models to the human phenotype of HD may be intuitively amplified. The modus operandi for equating the findings made in animal models with those occurring in the human HD brains is as diverse as are the number of investigators, thus of debatable cogency.
A consensual language is missing between investigators using engineered animal models and neuropathologists or neuroscientists involved mainly with human HD brains. The lack of working definitions of key terms of the field results in broad meanings and ambiguities. Possibly, our current understanding of the data gathered from the artificially induced huntingtin related abnormal mice, rats, or flies is insufficient. Hence the claim that these animals are models for HD may be, and probably is, an overstatement. Thus, the significance of the findings they provide remains relative. The relevance of these findings should be carefully tested against the human HD brains as much as possible, and by developing new methods of investigations. This would validate the extrapolations of the observations made in the HD-IT15 CAG repeat expansion mice to human HD.
In fact, what are, at present, the accepted standards for an engineered mouse to qualify as a model of HD?
Currently, a mouse qualifies as being a transgenic model of HD if it carries either a fragment or a full length of the human, pathologic huntingtin gene inserted randomly within its genome, thus driven by variable promoters expressing the mhtt usually in addition to its own wild type.
A mouse qualifies as being a knockin model of HD if it carries a fragment or a full length of the human pathologic huntingtin gene inserted within its own huntingtin gene (Hdh). Thus, in this model, the natural mouse promoter drives the foreign gene expressing the expanded polyglutamine tract.
In short, a mouse referred to as a model of HD is a mouse the genome of which includes merely a fragment or the full length of the human gene encoding for the mhtt triggering or causing the disease. None of these models exhibit the constellation of changes in their brains that would meet the criteria used to establish the neuropathologic diagnosis of HD, as applied in the setting of clinical neuropathology.
Hence the question: is this artificial genetic modification sufficient to qualify the engineered product as a “model for HD,” which naturally does not occur in non-human animals? A “no” answer makes sense. Indeed, from a clinical neuropathological point of view these engineered animals are gene carriers of the HD mutation, rather than models for HD.
Neuropathology of human Huntington disease
Overview
Huntington disease is an autosomal dominant illness usually with mid-life onset of psychiatric, cognitive, and motor symptoms. Death occurs 12–15 years from the time of symptomatic onset [21, 35]. An unstable expansion of CAG (trinucleotide) repeats within the coding region of the gene “IT15” (for “interesting transcript,” referred to as HD-IT15 CAG repeats) causes the disease. This gene, on chromosome 4 (4p16.3), encodes the 350 kDa protein huntingtin, which is endowed with important functions in the healthy brain [13, 30, 86]. An expanded polyglutamine residue (polyQ) distinguishes the mhtt (with about 37–250 polyQ) from the wild type [with 8 to about 34–36 polyQ (whtt)] [70, 87]. The disease occurs when the critical threshold of about 37 polyQ is exceeded [45]. The expansion confers on the mutant protein a novel attribute that may initiate disease by changing an activity of huntingtin or an interacting protein, assuming that these are critical to the targeted neurons [37, 40, 93]. This phenomenon is observed in a group of nine inherited, neurodegenerative diseases caused by polyQ extension, referred to as polyglutaminopathies [8, 61, 73, 74]. The actual causative pathway from the HD gene mutation to neuronal dysfunction and loss, which is under intense scrutiny, has not yet been established. Proteolytic processing and aggregation of polyQ fragments apparently participate to the pathogenesis of polyglutaminopathies. Neuronal, nuclear inclusions and neuropil aggregates in postmortem brains of HD patients and in several mouse models were shown to contain short N-terminal fragments of htt [19, 111]. Therefore, proteolysis (e.g., at the caspase-6 cleavage site) seems to play a critical role in the degenerative process by the production and accumulation of putatively toxic fragments of mhtt [27, 113]. Transgenic mice expressing only N-terminal htt fragments display pathologic changes earlier than mice expressing longer or full-length transgenes indicating that the cleavage fragments, rather than the full-length proteins that are toxic.
Patients homozygous for the HD gene produce only mhtt, but most of them are clinically like heterozygous, which reveals that mhtt retains some functions of the whtt [67, 99, 110]. The mhtt is expressed in all organs, yet the brunt of the changes of HD identified so far occurs in the brain [92]. The degeneration initially involves the striatum (neuronal loss, gliosis), then the cerebral cortex, and eventually may appear throughout the brain as a constellation of the toxic effect of the mutation and the ensuing secondary changes. Features that HD shares with the other eight known polyglutaminopathies are ubiquitinated, neuronal, nuclear inclusions involving a subset of neurons, and dystrophic neurites; and neuronal loss in regions more or less distinctive for each disease of this group [19, 73, 82, 84, 103].
Among the theories for the selective cellular damage in HD, the most compelling involve impaired energy metabolism, excitotoxicity, and relative, selective endotoxicity [56]. Perhaps endotoxicity results from misfolding of mhtt [3, 74]. Wild type huntingtin is soluble. In contrast, mhtt is insoluble and forms aggregates that are small enough to enter the nucleus either actively or passively [19, 43, 93, 111]. Within the nucleus, mhtt may act as a transcription factor or alter transcription [14, 29, 71, 72]. Indeed, data indicate that mhtt impairs gene transcription following the sequestration of key transcription factors within the nucleus [43, 100]. The engineered mice provide opportunities to evaluate the aggregation and interaction of mhtt with proteins, and to identify factors responsible for the neuronal loss.
Neuropathology: human HD brains
Distinct topographic and cellular alterations, notably in the striatum and cerebral cortex, are characteristic of HD. A grading system (see below) that stages the extent of striatal degeneration has been developed and widely used as a research tool.
The landmarks of special interest in assessing the neuropathological changes of HD are the basal ganglia, which consist of the corpus striatum and the amygdaloid nucleus [12]. Because of their connections, the subthalamic nucleus and substantia nigra are often included among the basal ganglia. The corpus striatum consists of the neostriatum [caudate nucleus (CN) and putamen] and paleostriatum (globus pallidus). The globus pallidus (GP) is divided into external (GPe) and internal (GPi) segments. The neostriatum is commonly referred to as the striatum. The substantia nigra (SN) has two main zones: the pars reticulata (SNr), and the pars compacta (SNc), the gradual pigmentation of which becomes visible on gross examination at puberty.
On external examination, 80% of the human HD brains show atrophy of the frontal lobes, and 20% are apparently normal. Halliday et al. [34] measured the total brain volumes of seven HD brains and recorded a 19% loss compared to controls (n = 12).
Examination of coronal slices reveals bilateral atrophy of the striatum in 95% of the HD brains. The striatal atrophy is prominent in 80%, mild in 15%, and subtle, if at all, in 5% of the brains [109].
Non-striatal regions show atrophy of variable severity or have normal appearance.
As a rule, the postmortem HD brain is diffusely smaller than normal in the late stage of disease. Morphometric analyses revealed a 21–29% area loss in the cerebral cortex, a 28% loss in the thalamus, a 57% loss in the CN, a 64% loss in the putamen, and a 29–34% loss in the telencephalic white matter [18]. With volumetric analyses from six HD brains, Lange et al. [49] found a 20% loss of the “hemisphere and cortex”, 58% loss of the “striatum”, 57% loss of the GPe, 50% loss of the GPi, and a 24% loss of the subthalamic nucleus. Hence their suggestion that HD is a polytopic disorder with primary and secondary involvements of the brain. Increased atrophy may occur in non-striatal regions of the HD brain with comorbidity. For example, enhanced atrophy of the limbic system may occur when HD coexists with Alzheimer disease.
In HD, the striatum is probably the only site where neuronal loss and “active” reactive, fibrillary astrocytosis coexist. Indeed, by conventional methods of evaluation, reactive, astrocytic gliosis is not detected in the non-striatal parts of the HD brain. Yet an increased density of oligodendrocytes, up to twice that of controls, is observed in the neostriatum (Fig. 2) [22, 33, 63, 68]. Data gathered in the tail of the CN of asymptomatic gene carriers revealed an increased oligodendrocytic density, which may precede the onset of symptoms by many years [24]. More reactive microgliocytes occur within the HD striatum, neocortex, and white matter than in controls [89]. Lymphoplasmocytic infiltration is not a feature of HD.
Fig. 2.
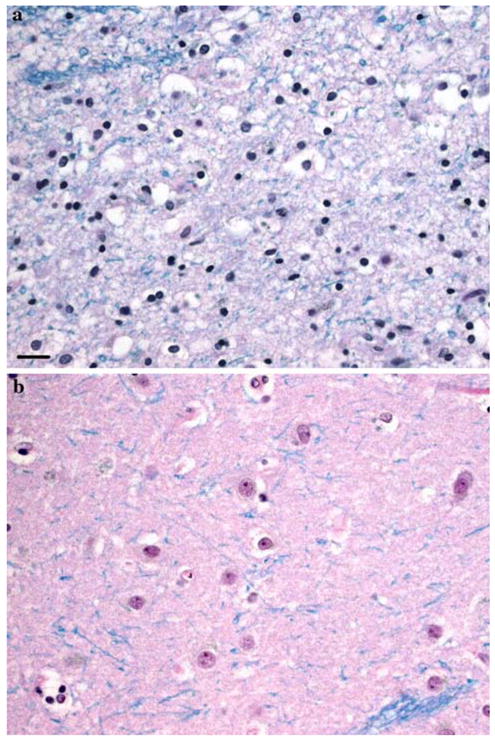
Tail of the caudate nucleus. The density of oligodendrocytes is increased and that of the neuron is decreased in the HD neostriatum (a) compared to the control (b). a A 54-year-old man with HD, grade 4/4; b 76-year-old woman without history of neurologic or psychiatric disorder. This change in the density of oligodendrocytes is not observed in the animal models currently available. a, b LHE, same magnification. Scale bar 50 μm
As mentioned, the occurrence of prominent, nuclear inclusions in neurons and glial cells in HD transgenic mice (R6/2) contributed to the identification of nuclear inclusions in scarce neurons in human HD brains [17, 19]. These inclusions are not visible in tissue sections stained with hematoxylin and eosin (HE) or with Luxol-fast-blue counterstained with HE (LHE), or in sections processed according the Bielschowsky method. However, they are labeled with antibodies directed against ubiquitin or against mhtt, e.g., EM48, 1C2 [19, 26, 103]. These inclusions can be detected long before the onset of symptoms in otherwise apparently normal brains of asymptomatic gene carriers [24]. Ubiquitinated, neuronal, nuclear inclusions are a common feature of the now-known nine polyQ diseases or polyglutaminopathies.
Grading of severity of striatal neuropathologic changes
The gradual atrophy of the striatum, which sequentially involves the neostriatum, GPe, then the GPi, typifies HD. In turn, neostriatal loss has an ordered, topographic distribution [33, 63, 83, 108]. The tail of the caudate nucleus (TCN) shows more degeneration than the body (BCN), which is more involved than the head (HCN). Similarly, the caudal portion of the putamen is more degenerated than the rostral portion. Along the coronal (or dorsoventral) axis of the neostriatum, the dorsal neostriatal regions are more involved than the ventral ones (Fig. 3a). Along the medio-lateral axis, the paraventricular half of the CN is more involved than the paracapsular half. With the evolution of the disease, neostriatal degeneration appears to move simultaneously in a caudo-rostral direction, a dorso-ventral direction, and a medio-lateral direction. Fibrillary astrogliosis parallels the loss of neurons along the caudo-rostral and dorso-ventral gradients of decreasing severity. Most remaining neostriatal neurons in the postmortem brains have normal morphology but contain more lipofuscin and may be smaller than normally expected. In addition, scattered, atrophic neurons stain darker with LHE, HE, or Cresyl violet than the apparently healthy neurons. Thus, they are referred to as neostriatal dark neurons (NDN) [109]. These neurons have a scalloped cellular membrane, a granular dark cytoplasm, and a nucleus with condensed chromatin. They are scarce in both the atrophic and the relatively preserved zones. However, their density increases in the intermediary zone, which lies between the two other zones. They have a tendency to form small groups.
Fig. 3.
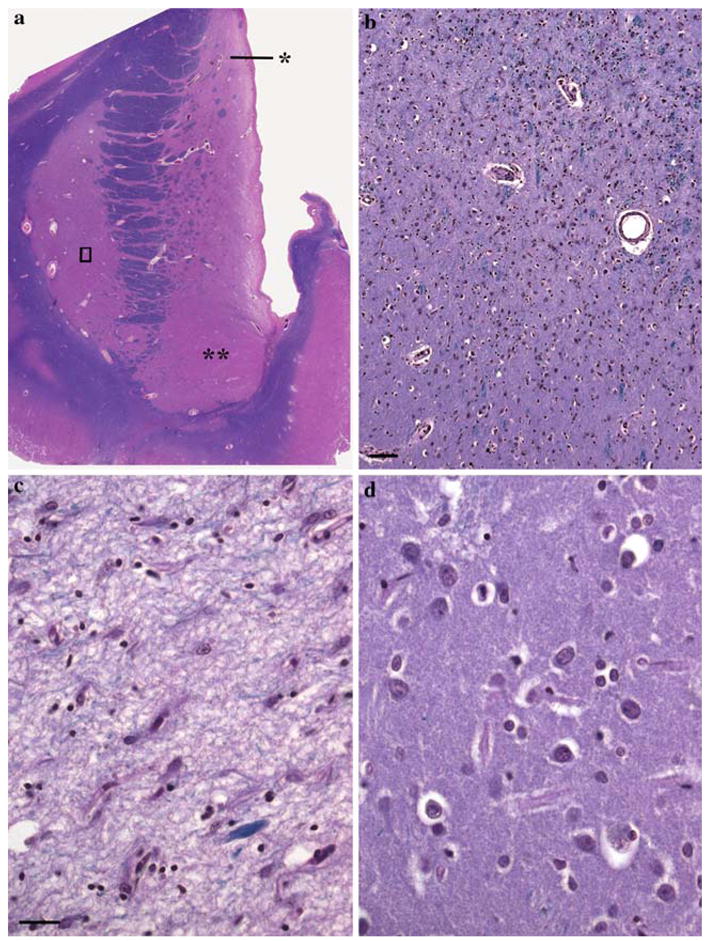
Rostral neostriatum (a—level CAP) including the head of the caudate nucleus (single asterisk), nucleus accumbens (double asterisks), and putamen. A 46-year-old man with HD, grade 3/4. The medial outline of the head of the caudate nucleus is flattened. b Is the magnified area of the putamen included in the rectangle in “a”, which depicts the transition zone between the severely involved dorsal and the relatively preserved ventral part. The demarcation is below and more or less parallel to the diagonal joining the upper left angle with the lower right angle of the picture. Scale bar 100 μm. c Is a magnified field of the dorsal third of the head of the caudate nucleus (single asterisk in a) showing subtotal neuronal loss, severe astrogliosis, and a loose-textured neuropil. d Is a magnified field of the nucleus accumbens (double asterisks in a), which is relatively preserved. This gradient is a hallmark or human HD, and does not occur in models. c and d same magnification, scale bar 65 μm. LHE
In essence, the dorsal third of the rostral neostriatum is especially prone to degenerate in contrast to the relatively preserved ventral third, including the nucleus accumbens (Fig. 3). That the transition third exhibits the microscopic features of its two flanked zones supports this deduction (Fig. 3b). Within the degenerated part, reactive astrocytes are the predominant cells, while neurons are virtually absent (Fig. 3c). In contrast, the relatively preserved area displays almost the normally expected cellular population (Fig. 3d), and is distinguishable from the intercalated zone, in which a mixture of apparently normal or atrophic neurons and reactive astrocytes is identifiable.
Five percent of the HD brains show unusual microscopical changes, especially in the anterior neostriatum. They consist of one to five discrete (0.5–1.0 mm in diameter) islets of relatively intact parenchyma. The density of neurons in these islets is the same as, or slightly lower than, that of the normal neostriatum, but the density of astrocytes is increased [107]. Islets are found more frequently in patients with juvenile than adult onset of clinical symptoms. These islets may reflect a degenerative mosaicism.
The framework of the neuropathologic grading system of HD is the distinctive, temporospatial pattern of degeneration in the striatum [108]. The assignment of a grade of neuropathologic severity is based on gross and microscopic findings using conventional methods of examination obtained from three standardized, coronal sections that include the striatum [(1) at the level of the nucleus accumbens; (2) just caudal to the edge of the anterior commissure; and (3) at the level of the lateral geniculate body]. This system has five grades (0–4) of severity of striatal involvement.
Grade 0 comprises <1% of all HD brains (n = 1,250). Grade 0 is assigned to brains from individuals with clinical signs consistent with HD, but whose brains are apparently normal on gross and on microscopical examinations when conventional methods of evaluation are used [69]. Obviously, the diagnostic threshold to assign grade 0 depends on the methods used to evaluate postmortem brains from individual clinically diagnosed as having HD. For example, although neuronal density was assessed as having been within normal limits on general survey, cell counts of grade 0 HD brains revealed a 30–40% loss of neurons in the HCN and no visible reactive astrocytosis [108]. Furthermore, as alluded to before, a study using sections including the tail of the CN of three asymptomatic gene carriers revealed nuclear inclusions in all three brains, including one individual, with 37 polyQ, who died three decades before the expected age for onset of symptoms. In addition, cell counts of the TCN revealed an increased density of oligodendrocytes among the asymptomatic gene carriers (Fig. 2) [24]. Thus, although these brains appeared normal when conventional methods of evaluation were applied, further evaluations using morphometric or immunohisto-chemical techniques revealed abnormalities.
The discrepancy between the paucity of detectable changes in this group of brains despite the occurrence of symptoms consistent with HD may result from a subset of neostriatal neurons that are gradually becoming dysfunctional. The presence of these dysfunctional neurons probably contributes to the misleading impression that there is no abnormality because the density of neurons within the cortex or neostriatum is apparently normal on general survey of the microscopic slides. Furthermore, the deficit of diagnostic sensitivity of the standard methods of assessment used in the clinical setting is highlighted by cell counts, which show that the neuronal density of the neostriatum is indeed decreased in grade 0 compared to controls [108].
Interestingly, in relation to grade 0 human HD brains, are the recurring observations made in many mouse carriers of the HD mutations showing compelling motor and behavioral changes before any neuronal loss is detected [52, 57, 60, 64–66, 85]. That these symptoms precede the loss of striatal or cortical neurons, reflect a dysfunction of a subset of vulnerable neurons with subsequent premature death later during the disease. The potential of the mice models is enormous in the attempts to prevent, or delay, or reverse this degenerative phase especially of the neostriatal spiny neurons [4, 86].
Grade 1 comprises 4% of all HD brains. The TCN is smaller than normal and atrophy of the BCN may also be detectable. Neuronal loss and astrogliosis are evident in the TCN, and less so in the BCN, dorsal portion of both the head and nearby dorsal putamen. Cell counts show a 50% or greater loss of neurons in the HCN. A careful examination of the entire length of the TCN is necessary for assignment of grade 1, since the body and head of the caudate nucleus and putamen may appear normal on gross examination. The TCN of neurologically normal subjects may show variations, including periodic constrictions or segmentations. In contrast to HD, the normal variations are focal and, therefore, likely to be apparent in only one or two coronal slices of the cerebral hemisphere.
Brains assigned grade 2 comprise 16%, those assigned grade 3 comprise 52%, and those assigned grade 4 comprise 28% of all HD brains. Gross striatal atrophy is mild to moderate in grade 2 (the medial outline of the HCN is only slightly convex but still bulges into the lateral ventricle), and severe in grade 3 (the medial outline of the HCN forms a straight line or is slightly concave medially (Fig. 3a). Thus, the microscopical changes in grades 2 and 3 are more severe than in grade 1, and less than in grade 4 brains.
In grade 4, the striatum is severely atrophic (the medial contour of the HCN is concave, as is the anterior limb of the internal capsule. The neostriatum loses 95% or more neurons. In at least 50% of grade 4 brains, the underlying nucleus accumbens remains relatively preserved, although smaller than normal.
Relationship of neostriatal degeneration to changes in other brain regions
In general, the striatal atrophy correlates with that of other brain regions. In grades 1 and 2, non-striatal structures of the brain show no or mild atrophy, unless there is age-related volumetric loss or a coexistent disease. However, in grades 3 and 4, non-neostriatal structures, including GP, neocortex, thalamus, subthalamic nucleus, SN, white matter, and cerebellum, are slightly to markedly smaller than normally expected. As further detailed below, these structures may show minimal or marked neuronal loss, usually without reactive astrocytosis. Similarly, the white matter atrophy may be severe, yet without any microscopical abnormality recognized by conventional methods, the exception being an increased microglial reaction, as mentioned.
Globus pallidus
The globus pallidus shows atrophy in grades 3 and 4, with the external segment more involved than the internal segment. In grade 4, there is a 50% volume reduction of the GP. The neurons are more densely packed than normal in grade 3, and even more so in grade 4, suggesting that, although tissue bulk decreases, neurons are relatively preserved. The absolute number of pallidal neurons decreases up to 40%, but the neuronal density is up to 42% higher than normal in the GPe and 27% higher in the GPi [49]. Thus, the pallidal atrophy is apparently chiefly due to loss of neuropil, and hence of striatal fiber connections and fiber passage, and to a lesser extent to loss of neurons [11, 108]. Reactive gliosis is usually confined to the external segment and is visible in grade 4, and to a lesser extent in grade 3. The ansa lenticularis is thinner than normal in grades 3 and 4.
Cerebral cortex
Atrophy of the cerebral cortex may or may not be pronounced in grades 3 and 4. Even when atrophy is marked, neuronal loss in the HD cerebral cortex is hard to appreciate on general survey of histological sections, resulting in contradictory statements in the literature. Little or no neuronal loss, normal astrocytes, and a relatively normal content of glial fibrillary acidic protein (GFAP) were recorded in the cortex of 14 HD brains [114]. Sotrel et al. [98] documented a loss of large, pyramidal neurons in layers III, V, and VI in grades 2–4, with the greatest loss in grade 4. There was no astrogliosis; however, cortical oligodendrocytic density was increased. Hedreen et al. found a 57% neuronal loss in layer VI, and a 71% loss in layer V in Brodmann area (BA) ten of five grade 4 brains [36]. Macdonald et al. [59] recorded up to a 55% loss of pyramidal neurons in the angular gyrus without correlation with the grade of striatal atrophy in six brains.
Braak and Braak [7] detected “a layer-specific” loss of neurons without increase of astrocytes in the entorhinal cortex and subiculum of seven patients with grade 3 or 4 striatal atrophy.
The extent of the cerebral cortical degeneration in HD depends in part on the stage of the disease at the time of the evaluation. It apparently varies from area to area; it is difficult to assess on general survey. Probably a reliable assessment of the cerebral cortical degeneration in HD requires morphometric evaluations.
Thalamus, substantia nigra, and subthalamic nucleus
In the thalamus, neuronal loss with or without astrocytosis involving the centrum medianum is regularly observed in grade 4, and to a lesser extent in grade 3 brains; otherwise, the thalamus is apparently normal in lower grades. There is a loss of neurons in the SNr [11, 33, 79]. The SNc is thinner than normal, yet its number of neurons is apparently normal in all grades, giving the impression of an increased density of pigmented neurons. In the subthalamic nucleus, there is a discrepancy between the marked atrophy present in grades 3 and 4 (up to 25% volumetric loss) and the scarcity of reactive astrocytes [49].
Cerebellum
Among our 1,250 HD brains, we found that the cerebellum is smaller than normally expected in grades 3 and 4. Although the volume loss of the cerebellum may be discrete, neuronal density frequently appears within normal limits. Segmental loss of Purkinje cells with or without Bergmann gliosis may occur; however, these changes are inconsistent and seem not to be specific for HD.
Cerebellar atrophy is often reported in patients with juvenile onset [10, 42, 62]. Juvenile HD patients are prone to seizures, which may account for some neuronal loss in the cerebellum or hippocampus, two sites notably vulnerable to hypoxic-ischemic events. Extensive loss of neurons in the cerebellar cortex was rarely encountered in our series, and then concomitant neuronal changes as those caused by ischemia were found. These changes were found either in the Sommer sector of the hippocampal formation, which is particularly vulnerable to hypoxia, or in neocortical watershed territories especially at the junction between the middle and anterior cerebral arteries, where ischemia tends to be worse in the event of hypotensive episodes.
Relative vulnerability of neostriatal neurons
Medium, spiny, projection neurons are especially vulnerable in HD. Evidence of relative, selective degeneration of the spiny neurons includes curved dendrites, and decreased or increased density of dendrites [28]. Variable rates of degeneration occur among different types of spiny neurons. Enkephalin-containing spiny neurons projecting to the GPe are more affected than substance P-containing neurons projecting to the GPi [78, 80, 88].
The NADPH-d aspiny interneurons are relatively resistant, perhaps because of the types of glutamate receptors they express [20]. In short, striatal projection neurons degenerate in a specific temporal sequence: striato-SNc followed by striato-GPe and striato-SNr, then by striato-GPi [1]. The extreme striatal atrophy and the loss of neurons in grade 4 indicate that both spiny and aspiny neurons are vulnerable at the end stage of the disease.
Characteristics of wild type and mutant huntingtin
The human HD gene includes 67 exons spanning more than 200 kb, and encoding 3,144 amino acids. The translated whtt contains a polymorphic stretch of up to 36 polyQ [6]. It is expressed in neurons throughout the brain regardless of their relative vulnerability [31, 48]. Huntingtin is found in the cytoplasm and nuclei of neurons, dendrites, and axon terminals [6, 19, 32].
Mutant huntingtin, with its elongated polyQ (>36), co-distributes with wild type in all regions, and in both gray and white matter. In the HD human brain, huntingtin immunoreactivity is increased in neurons compared to control brains. Labeling appears diffusely elevated in the nucleus and cytoplasm, and in multivesicular organelles of affected neurons [91]. The truncated, N-terminal region of mhtt accumulates in the nucleus and cytoplasm of the neocortex, entorhinal cortex, subiculum, pyramidal neurons of the hippocampus, and neostriatum, but apparently not in the GP, cerebellum, or pars compacta of the substantia nigra [19,32, 58]. Cortical neurons with nuclear or cytoplasmic inclusions, or both, are more widespread in the brains of patients with juvenile rather than adult onset of symptoms [19, 90]. Cortical projections to the striatum may be among the earliest affected by mhtt. The labeling of neurons with antibodies directed against mhtt apparently does not correlate with their relative vulnerability [46].
Neuropathology of engineered mice gene carriers of the HD mutation
The creation of transgenic mouse models provides unprecedented opportunities to gain insights into disease pathogenesis. A mouse model developing distinct, quantifiable symptoms with discrete onset, and selective loss of striatal projection neurons would be ideal. The mouse models currently available can be categorized in three groups: (1) The transgenic HD mouse models express exon-1 fragments of the human htt gene containing polyQ in addition to both alleles of murine whtt. (2) The knockin HD mouse models are mouse with CAG repeats inserted into the existing CAG expansion in murine Hdh gene. (3) One group consists of mice that express the full-length human HD gene and the murine Hdh.
As stated, HD occurs only in humans. Thus, the singular, causal, molecular, cellular, and cerebral pathogenic process of HD is a natural phenomenon occurring exclusively within the human brains expressing the mutation. Consequently, only an affected human brain provides the actual biologic environment in which the chronic, long term, deleterious process evolves. However, the claim that only the human brain provides the intrinsic plasticity favorable to the harmful mutation is challenged notably by the recent thorough comparative investigations of the engineered rodents [47]. Nonetheless, theoretically, the human brain that expresses the mutation should be the most suitable substrate to elucidate the pathogenesis of HD. Unfortunately, the techniques currently available for investigations relying on postmortem human HD brains are inadequate, therefore, these brains are not fully exploited. These limitations impede progress in understanding the causal chains and their ramifications that are toxic to neurons, and occur naturally in HD brains. Because of these constraints combined with the trend of the time, research on the pathogenesis of HD rests mainly on mice carriers of the HD mutation. Determining which data obtained from these models are relevant to human HD is problematic, in part because the mouse’s short life span precludes comparison of glutamine thresholds and may limit the ultimate toxic consequences [111]. The quantitative, qualitative, relational, and modal aspects of the data obtained with the models must be carefully assessed before assuming that they reflect what actually happens in the HD human brain.
Macroscopically, the differences between healthy humans and healthy mice are obvious. In contrast, microscopically, many aspects and functions of normal cells are similar whether of human or murine origin [9, 102]. The criteria used for comparing the metabolism of normal human cells with that of normal murine cells may not be accurate for comparing the metabolism of human cells that naturally express the HD mutation with that of murine cells engineered to express it. Despite these reservations, the engineered models are useful and probably the best tools now available to clarify the pathogenesis of HD.
The models available so far do not reproduce the constellation of changes that compose the characteristic, neuropathologic phenotype seen in the postmortem human HD brains. A cross-sectional review of the neuropathological findings displayed by the animal models reveals a range of changes. The variance includes features that are either shared by the animals despite the different genetic constructs, including the HD humans, or that are confined to the models excluding the HD humans, or features that are exclusive to a specific model. This heterogeneity accounts for the various opinions toward models reported up to now.
Major features occurring in a subgroup of models, which are likewise detectable in the postmortem human HD brains, are the ubiquitinated, nuclear inclusions, dystrophic neurites, and dark neurons [60, 104]. As mentioned, the R6/2 model was instrumental in the detection of the ubiquitinated, nuclear inclusions in neurons of human HD brains [17, 19] (Fig. 1).
Transgenic mouse carriers of human HD mutation
The first reported transgenic mouse carriers of HD mutation were the R6 lines [60]. The mice express exon 1 of the human huntingtin gene, with an expanded CAG repeat under control of the human huntingtin promoter. The R6/1 line have about 120 CAG repeats. The transgene is expressed at 31% of endogenous levels. The R6/2 line have about 150 or more CAG repeats that are expressed at 75% of endogenous levels [60]. Because the life expectancy of the mouse models is short compared to humans, an excessive polyQ repeat is used to accelerate the neurodegenerative process. Furthermore, because of its long polyQ, the R6/2 mouse is considered more representative of the juvenile than the adult phenotype of the human HD.
The R6/2 mouse has been the most widely investigated, thus the most informative model whose characteristics are well documented [4]. Definite symptoms occur at age 6 weeks and death at about 13–15 weeks. Atrophy of the cortex and striatum is detectable at age 12 weeks, but without or with mild cortical, striatal, cerebellar neuronal loss, which occurs at the late stage. The symptoms happen later and are milder, and the peristriatal cerebral volume loss is less prominent if the transgenic mice are living in an enriched environment than in non-enriched living conditions [38, 105]. In regard to this observation, analysis of the 83 kindreds of the Venezuelan HD cohort demonstrates that residual variability in age of onset has both genetic and environmental components [76].
Occasional dark neurons confined to limited areas of the cortex and striatum (the equivalent of the neostriatal dark neurons described in the human HD neostriatum—see above) occur after age 14 weeks [104]. Ubiquitinated, nuclear inclusions are found in many cortical and subcortical neurons, in occasional glial cells, and ependymal cells (Fig. 1). In contrast, neurons with ubiquitinated, nuclear inclusions are much less frequent in humans: Up to 21% of cortical neurons were found to harbor such inclusions in one patient with juvenile HD and 86 CAG repeats; and up to 16.4% in one with 84 CAG repeats; or between 0.8 and 3.8% in five patients with the number of CAG repeats of ≥40 but ≤47 [58].
In summary, the R6/2 model develops behavior and pathological changes that mimic those occurring in human HD: jerky movements; striatal atrophy; nuclear inclusions of neurons, and neuropil aggregates that are labeled with antibodies directed against ubiquitinated proteins or against mhtt. Although extremely useful, this model only partially mimics the human HD phenotype: the extent and distribution of the aggregates are overwhelming, and neurons are resistant to excitotoxicity compared to the humans.
The N171-82Q mouse, with its shorter polyQ, exhibits a protracted course of symptoms compared to the R6/2. The N171-82Q lines incorporate the prion promoter in contrast to the R6/2 lines, which incorporate the human promoter, as mentioned. The brain of the N171-82Q is “slightly smaller” but “grossly normal”; nuclear inclusions and neuritic aggregates of mhtt are “found in multiple populations of neurons”; light microscopic examination does “not indicate severe loss of neurons in the cortex, hippocampus, striatum, cerebellum, and brainstem” [94].
The HD48Q and HD89Q mice harbor the full length (instead of a fragment) of the human IT15 gene, which is driven by the cytomegalovirus (CMV) promoter. Neuronal loss and reactive astrocytosis involve especially the striatum. Nuclear, ubiquitinated inclusions occur in occasional neurons (<1% in the striatum), thus they are much fewer than in previous models. Furthermore, in contrast to human HD, this model shows Purkinje cells with nuclear inclusions [77].
Another informative transgenic model is the yeast artificial chromosome YAC mouse expressing a full-length human IT15 gene. YAC mice with 48, 72, 89, or 128 CAG repeats are corresponding to adult-onset, or juvenile-onset-causing tracts in humans. YAC 128 is especially interesting because the neuronal loss is confined mainly to the striatum. Nuclear and neuropil aggregates are detectable 10 months after onset of symptoms, and are less abundant in mice carrying the full length mhtt compared to the R6/2 mice [39, 97].
During the development of the full-length YAC128 mouse models, a mouse expressing exons 1 and 2 of 67 of the human IT15 gene occurred spontaneously, yielding a truncated mhtt. The mouse, referred to as “shortstop mouse,” remains asymptomatic although ubiquitinated, nuclear inclusions and neuropil aggregates occur while neuronal density remains within normal limits [96]. This observation supports the hypothesis that htt inclusions may be nontoxic in vivo.
Knockin mice carriers of the HD mutation
The insertion of the human HD mutation into the natural huntingtin gene (Hdh locus) of the mouse allows the endogenous promoter to drive the expression of mhtt. Thus, this model is putatively more liable to replicate the pathogenesis of HD than the transgenic model. Contrary to expectations, symptoms and neuropathologic changes of knockin mice are subtle compared to those exhibited by transgenic mice. Cerebral atrophy is not recorded. Discrete ubiquitinated, nuclear inclusions are rarer and occur later in the brains of knockin mice than in those of transgenic mice [53, 95, 112]. Distinctions of the knockin mouse model are the presence of nuclear staining and microaggregates of mhtt at 2–6 months [54, 111].
Reactive gliosis involving the striatum was reported to occur in at least two distinct knockin models. Lin et al. [55] noticed that these knockin mice suggest a presymptomatic state and reported a new knockin model “with neuropathological features consistent with HD in man”. In this model, “extreme reductions in major brain regions were not observed… Furthermore, cell counts suggest no loss of neurons… There was a marked increase in GFAP immunoreactivity in the striatum of CHL2 mutants… Most CHL2 mutants showed restricted areas of intense staining in the striatum… Presence of ubiquitin- and huntingtin-positive nuclear inclusions throughout the dorsal striatum and nucleus accumbens…less frequent in other brain regions… No evidence of dark neurons.”
In addition to the striatum, reactive astrocytosis is seen within the GP and substantia nigra of a knockin mouse in which the endogenous exon 1 of the Hdh was replaced with the expanded (77 CAG repeats) human HD exon 1 [41]. Furthermore the CAG instability increased with age. Thus, these knockin mice exhibit reactive astrogliosis notably within the striatum, which occurs in human HD.
Further characteristics of mice carriers of human HD mutation are concisely and comprehensively provided by Menalled and Chesselet [64], Rubinsztein [85], Hörsten [106], or by Bates and Gonitel [2].
Conclusions
The neuropathological criteria used to establish a diagnosis rest on a sequence of features including clinical, neuroanatomic, and neuropathologic data. This diagnostic construct is complex, especially in neurodegeneration. The claim that a mouse model carrying the human HD mutation replicates the natural symptoms or pathological changes depends on qualitative and quantitative assessments of the features considered for the comparisons, and on their interpretation. A good command of the versatile expressions of the processes within the human on one hand and mouse on the other hand is crucial to establish a reliable, above all objective, assessment of the validity of the model.
The models available so far do not reproduce the full constellation of changes that compose the characteristic, neuropathologic phenotype seen in the postmortem human HD brains.
The validity of the models probably should be measured by their capacity of reproducing the cellular dysfunction of HD rather than the neuropathologic phenotype of the postmortem human brains.
Nonetheless, two intriguing changes that occur in human HD brains, but not in the models so far, are probably worth investigating. Exploring these changes may provide crucial information relevant to the regional vulnerability not only within the brain, but also notably within the neostriatum itself. These changes are (1) the gradient of neostriatal severity of degeneration (Fig. 3), and (2) the relatively preserved neostriatal islets occurring in a minority of HD brains [107].
Interestingly, the gradient of neostriatal, neuropathologic severity matches the vascular territories of the neostriatum. Indeed, the areas of the neostriatum that are supplied by the most peripheral blood are involved first, and, at death, are the most severely degenerated areas. Despite the mutation, and despite their selective vulnerability, many spiny neurons are still present in the postmortem neostriata of 60-, 70-, 80-year-old, or older HD patients. These neurons, the function of which may or may not be normal, do survive in part thanks to their location within the neostriatum. Thus, these privileged areas harbor the homeostasis that maintains the protective elements against the toxicity of the mutation, which remains to be found and identified. Maybe a subtle difference in the flow or quality of the blood supplying the more distant parts of the neostriatum relative to the heart imposes certain stress that is particularly harmful to spiny neurons. This stress may be sufficient to reduce the life expectancy of spiny neurons by 10–30 years, depending on the length of the CAG repeats, modifier genes, environment, or longevity of the individual. The models may not be ideal for identifying the ingredients protective against the deleterious process, first because their life span is too short; second because a definite gradient is not detected in their striata; third, the models are highly constrained; and fourth because the model might not cause neurodegeneration. However, the models would be useful for testing any candidate agents potentially protecting against or neutralizing the chronic, slow-acting toxicity of the mutation.
The relatively preserved parenchymal islets within the otherwise degenerated human HD neostriatum may reflect a kind of intrinsic mosaicism. Somatic instability had been documented in human HD brains and in heterozygous mice of different ages from line Hhd6/Q72 and Hdh4/Q80 [44, 101]. This suggests that, in spite of the mutation, the human neostriatal milieu remains an environment in which the vulnerable spiny neurons survive. Furthermore, these islets may indicate that an endogenous stopping of the CAG expansion may exist, thus that a therapeutic containment of the expansion may be possible, as nature seems to indicate. The models would be crucial for investigating the achievability of such stopping that would keep the polyQ track within normal range (<36).
Comparative studies on neurogenesis using control or HD brains from humans, and from mouse models clearly illustrate the definite importance of the engineered animals as a reliable research tool. Indeed, the seductive hypothesis that neurogenesis is impaired in neurodegenerative diseases led to several investigations of the subventricular and sub-granular zones of the dentate gyrus of the R6/1 and R6/2 mice [23, 50, 51, 75]. These studies documented that cell proliferation and neurogenesis is impaired especially within the hippocampal dentate gyrus, but not within the subventricular zone compared to the wild type mice. However, using nine, graded human HD brains, Curtis et al. [15] found an increase of proliferative cells in the subventricular zone compared to six control human brains. Of the three types of proliferating cells of the subependymal layer (type A cells resembling neuroblasts; type B cells resembling glial cells; and type C cells resembling progenitor cells), type B cells displayed the largest increase in human HD brains compared to controls. Furthermore, the number of cells labeled with antibodies directed against neuropeptide Y was increased in HD compared to the controls suggesting that neuropeptide Y neurons may participate to the proliferation of progenitor cells [16]. The difference of results of neurogenesis between HD brains and that of the rodent models is intriguing. That the increased thickness of the subependymal layer is proportional to the grade of neuropathological severity suggests that neurogenesis response is in part driven by the extent of the neuronal loss, which is overwhelming in the human compared to the models. Furthermore, the difference of organization of the human neo-striatum and that of the rodents in conjunction with the biological resonance of the artificial mutation may alter the environment in which the cells are located and, therefore, their responses to the occurrence of the mhtt [75].
Combining the observations provided by the painstaking investigations of human postmortem brains and the data through improved models is prerequisite to identify and apply means that will prevent or delay the toxic effects of the HD mutations. Actually, despite their current limitations, the engineered models are essential complements in the laborious attempts to neutralize the harmful gain of function attributed to the pathologic expansion of the polyQ tract of huntingtin.
Acknowledgments
This work was supported by grants from the National Institutes of Health and National Institute on Aging: P01-AG07232, R37-AG15473, and P50-AG08702, the Hereditary Disease Foundation, and the Iseman Fund. The author is grateful to Lisle Merriman for her editorial support, and to Mkeba Cason, Etty Cortes, M.D., and Katerina Mancevska for their help. The New York Brain Bank (NYBB) is especially thankful to the numerous pathologists who referred case material, and to the families of the patients for providing brain tissue for research.
Footnotes
In memory of the Late Dr. Milton Wexler PhD, founder of the Hereditary Disease Foundation.
References
- 1.Albin RL. Selective neurodegeneration in Huntington’s disease. Ann Neurol. 1995;38:835–836. doi: 10.1002/ana.410380602. [DOI] [PubMed] [Google Scholar]
- 2.Bates GP, Gonitel R. Mouse models of triplet repeat diseases. Mol Biotech. 2006;32:147–158. doi: 10.1385/MB:32:2:147. [DOI] [PubMed] [Google Scholar]
- 3.Beal MF. Aging, energy, and oxidative stress in neurodegenerative diseases. Ann Neurol. 1995;38:357–366. doi: 10.1002/ana.410380304. [DOI] [PubMed] [Google Scholar]
- 4.Beal MF, Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington’s disease. Nat Rev Neurosci. 2004;5:373–384. doi: 10.1038/nrn1386. [DOI] [PubMed] [Google Scholar]
- 5.Becher MW, Kotzuk JA, Sharp AH, Davies SW, Bates GP, Price DL, Ross CA. Intranuclear neuronal inclusions in Huntington’s disease and dentatorubropallidoluysian atrophy: correlation between the density of inclusions and IT 15 CAG triplet repeat length. Neurobiol Dis. 1998;4:387–397. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 6.Borrell-Pagès M, Zala D, Humbert S, Saudou F. Huntington’s disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol Life Sci. 2006;63:2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braak H, Braak E. Allocortical involvement in Huntington’s disease. Neuropathol Appl Neurobiol. 1992;18:539–547. doi: 10.1111/j.1365-2990.1992.tb00824.x. [DOI] [PubMed] [Google Scholar]
- 8.Brignull HR, Moore FE, Tang SJ, Morimoto RI. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J Neurosci. 2006:7597–7606. doi: 10.1523/JNEURO-SCI.0990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007 doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 10.Byers RK, Gilles FH, Fung C. Huntington’s disease in children. Neurology. 1973;23:561–569. doi: 10.1212/wnl.23.6.561. [DOI] [PubMed] [Google Scholar]
- 11.Campbell AMG, Corner B, Norman RM, Urich H. The rigid form of Huntington’s disease. J Neurol Neurosurg Psychiatry. 1961;24:71–77. doi: 10.1136/jnnp.24.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carpenter MB, Sutin J, editors. Human neuroanatomy. 8. Williams & Wilkins; Baltimore/London: 1983. pp. 579–586. [Google Scholar]
- 13.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 14.Cha J-H. Transcriptional dysregulation in Huntington’s disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 15.Curtis MA, Penney EB, Pearson AG, van Roon-Mom MC, Butterworth NJ, Dragunow M, Connor B, Faull RLM. Increased cell proliferation and neurogenesis in the adult human Huntington’s disease brain. Proc Natl Acad Sci USA. 2003;100:9023–9027. doi: 10.1073/pnas.1532244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curtis MA, Waldvogel HJ, Synek B, Faull RLM. A histochemical and immunohistochemical analysis of the subependymal layer in the normal and Huntington’s disease brain. J Chem Neutoanat. 2005;30:55–66. doi: 10.1016/j.jchemneu.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 18.de la Monte SM, Vonsattel JP, Richardson EP., Jr Morphometric demonstration of atrophic changes in the cerebral cortex, white matter, and neostriatum in Huntington’s disease. J Neuropathol Exp Neurol. 1988;47:516–525. doi: 10.1097/00005072-198809000-00003. [DOI] [PubMed] [Google Scholar]
- 19.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intra-nuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 20.Ferrante RJ, Kowall NW, Beal MF, Martin JB, Bird ED, Richardson EP. Morphologic and histochemical characteristics of a spared subset of striatal neurons in Huntington’s disease. J Neuropathol Exp Neurol. 1987;46:12–27. doi: 10.1097/00005072-198701000-00002. [DOI] [PubMed] [Google Scholar]
- 21.Folstein SE, editor. Huntington’s disease. A disorder of families. The Johns Hopkins University Press; Baltimore: 1989. The diagnosis of Huntington’s disease; pp. 125–148. [Google Scholar]
- 22.Forno LS, Jose C. Huntington’s chorea: a pathological study. Adv Neurol. 1973;1:453–470. [Google Scholar]
- 23.Gil JMAC, Mohapel P, Araújo IM, Popovic N, Li J-Y, Brundin P, Petersén A. Reduced hippocampal neurogenesis in R6/2 transgenic Huntington’s disease mice. Neurobiol Dis. 2005;20:744–751. doi: 10.1016/j.nbd.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Gómez-Tortosa E, MacDonald ME, Friend JC, Taylor SAM, Weiler LJ, Cupples LA, Srinidhi J, Gusella JF, Bird ED, Vonsattel J-P, Myers RH. Quantitative neuropathological changes in presymptomatic Huntington’s disease. Ann Neurol. 2001;49:29–34. [PubMed] [Google Scholar]
- 25.Gourfinkel-An I, Cancel G, Duyckaerts C, Faucheux B, Hauw J-J, Trottier Y, Brice A, Agid Y, Hirsch EC. Neuronal distribution of intranuclear inclusions in Huntington’s disease with adult onset. Neuroreport. 1998;9:1823–1826. doi: 10.1097/00001756-199806010-00028. [DOI] [PubMed] [Google Scholar]
- 26.Gourfinkel-An I, Cancel G, Trottier Y, Devys D, Tora L, Lutz Y, Imbert G, Saudou F, Stevanin G, Agid Y, Brice A, Mandel J-L, Hirsch EC. Differential distribution of the normal and mutated forms of huntingtin in the human brain. Ann Neurol. 1997;42:712–719. doi: 10.1002/ana.410420507. [DOI] [PubMed] [Google Scholar]
- 27.Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, Warby SC, Doty CN, Roy S, Wellington CL, Leavitt BR, Raymond LA, Nicholson DW, Hayden MR. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 28.Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 29.Greenamyre JT. Huntington’s disease—making connections. N Engl J Med Line. 2007;356:518–520. doi: 10.1056/NEJMcibr067022. [DOI] [PubMed] [Google Scholar]
- 30.Group: The Huntington’s Disease Collaborative Research Group. MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N, Mac-Farlane H, Jenkins B, Anderson MA, Wexler NS, Gusella JF. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 31.Gutekunst C-A, Levey AI, Heilman GJ, Whaley WL, Yi H, Nash NR, Rees HD, Madden JJ, Hersch SM. Identification and localization of huntingtin in brain and human lymphoblastoid cell lines with anti-fusion protein antibodies. Proc Natl Acad Sci USA. 1995;92:8710–8714. doi: 10.1073/pnas.92.19.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutekunst C-A, Li S-H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li X-J. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallervorden J. Handbuch der speziellen pathologischen Anatomie und Histologie (XIII/1 Bandteil A) Springer; Berlin: 1957. Huntingtonsche Chorea (Chorea chronica progressiva hereditaria) pp. 793–822. [Google Scholar]
- 34.Halliday GM, McRitchie DA, Macdonald V, Double KL, Trent RJ, McCusker E. Regional specificity of brain atrophy in Huntington’s disease. Exp Neurol. 1998;154:663–672. doi: 10.1006/exnr.1998.6919. [DOI] [PubMed] [Google Scholar]
- 35.Harper PS, Morris MR, Quarrell OWJ, Shaw DJ, Tyler A, Youngman S. Huntington’s disease. Major problems in neurology. Vol. 22. W.B. Saunders; London: 1991. The clinical neurology of Huntington’s disease; pp. 37–80. [Google Scholar]
- 36.Hedreen JC, Peyser CE, Folstein SE, Ross CA. Neuronal loss in layers V and VI of cerebral cortex in Huntington’s disease. Neurosci Lett. 1991;133:257–261. doi: 10.1016/0304-3940(91)90583-f. [DOI] [PubMed] [Google Scholar]
- 37.Hilditch-Maguire P, Trettel F, Passani LA, Auerbach A, Persichetti F, MacDonald M. Huntingtin: an iron-regulated protein essential for normal nuclear and perinuclear organelles. Hum Mol Genet. 2000;9:2789–2797. doi: 10.1093/hmg/9.19.2789. [DOI] [PubMed] [Google Scholar]
- 38.Hockly E, Cordery PM, Woodman B, Mahal A, van Dellen A, Blakemore C, Lewis CM, Hannan AJ, Bates GP. Environmental enrichment slows disease progression in R6/2 Huntington’s disease mice. Ann Neurol. 2002;51:235–242. doi: 10.1002/ana.10094. [DOI] [PubMed] [Google Scholar]
- 39.Hodgson JG, Agopyan N, Gutekunst C-A, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, Jamot L, Li X-J, Stevens ME, Rosemond E, Roder JC, Phillips AG, Rubin EM, Hersch SM, Hayden MR. A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:182–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 40.Huang CC, Faber PW, Persichetti F, Mittal V, Vonsattel J-P, MacDonald ME, Gusella JF. Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins. Somat Cell Mol Genet. 1998;24:217–233. doi: 10.1023/b:scam.0000007124.19463.e5. [DOI] [PubMed] [Google Scholar]
- 41.Ishiguro H, Yamada K, Sawada H, Nishii K, Ichino N, Sawada M, Kurosawa Y, Matsushita N, Kobayashi K, Goto J, Hashida H, Masuda N, Kanazawa I, Nagatsu T. Age-dependent and tissue-specific CAG repeat instability occurs in mouse knock-in for a mutant Huntington’s disease gene. J Neurosci Res. 2001;65:289–297. doi: 10.1002/jnr.1153. [DOI] [PubMed] [Google Scholar]
- 42.Jervis GA. Huntington’s chorea in childhood. Arch Neurol. 1963;9:244–257. doi: 10.1001/archneur.1963.00460090050005. [DOI] [PubMed] [Google Scholar]
- 43.Kegel KB, Meloni AR, Yi Y, Kim YJ, Doyle E, Cuiffo BG, Sapp E, Wang Y, Qin Z-H, Chen JD, Nevins JR, Aronin N, DiFiglia M. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J Biol Chem. 2002;277:7466–7476. doi: 10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- 44.Kennedy L, Shelbourne PF. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Hum Mol Genet. 2000;9:2539–2544. doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- 45.Kremer B, Goldberg P, Andrew SE, Theilmann J, Telenius H, Zeisler J, Squitieri F, Lin B, Bassett A, Almqvist E, Bird TD, Hayden MR. A worldwide study of the Huntington’s disease mutation. The sensitivity and specificity of measuring CAG repeats. N Engl J Med Line. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- 46.Kuemmerle S, Gutekunst C-A, Klein AM, Li X-J, Li S-H, Beal MF, Hersch SM, Ferrante RJ. Huntingtin aggregates may not predict neuronal death in Huntington’s disease. Ann Neurol. 1999;46:842–849. [PubMed] [Google Scholar]
- 47.Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, Becanovic K, Pouladi MA, Sathasivam K, Cha J-HJ, Hannan AJ, Hayden MR, Leavitt BR, Dunnett SB, Ferrante RJ, Albin R, Shelbourne P, Delorenzi M, Augood SJ, Faull RLM, Olson JM, Bates GP, Jones L, Luthi-Carter R. Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 48.Landwehrmeyer GB, McNeil SM, Dure LS, IV, Ge P, Aizawa H, Huang Q, Ambrose CM, Duyao MP, Bird ED, Bonilla E, de Young M, Avila-Gonzales AJ, Wexler NS, DiFiglia M, Gusella JF, MacDonald ME, Penney JB, Young AB, Vonsattel JP. Huntington’s disease gene: regional and cellular expression in brain of normal and affected individuals. Ann Neurol. 1995;37:218–230. doi: 10.1002/ana.410370213. [DOI] [PubMed] [Google Scholar]
- 49.Lange H, Thörner G, Hopf A, Schröder KF. Morphometric studies of the neuropathological changes in choreatic diseases. J Neurol Sci. 1976;28:401–425. doi: 10.1016/0022-510x(76)90114-3. [DOI] [PubMed] [Google Scholar]
- 50.Lazic SE, Grote H, Armstrong JE, Blakemore C, Hannan AJ, van Dellen A, Barker RA. Decreased hippocampal cell proliferation in R6/1 Huntington’s mice. Neuroreport. 2004;15:811–813. doi: 10.1097/01.wnr.0000122486.43641.90. [DOI] [PubMed] [Google Scholar]
- 51.Lazic SE, Grote HE, Blakemore C, Hannan AJ, van Dellen A, Phillips W, Barker RA. Neurogenesis in the R6/1 transgenic mouse model of Huntington’s disease: effects of environmental enrichment. Eur J Neurosci. 2006;23:1829–1838. doi: 10.1111/j.1460-9568.2006.04715.x. [DOI] [PubMed] [Google Scholar]
- 52.Levine MS, Cepeda C, Hickey MA, Fleming SM, Chesselet M-F. Genetic mouse models of Huntington’s and Parkinson’s disease: illuminating but imperfect. Trends Neurosci. 2004;27:691–697. doi: 10.1016/j.tins.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 53.Levine MS, Klapstein GJ, Koppel A, Gruen E, Cepeda C, Vargas ME, Jokel ES, Carpenter EM, Zanjani H, Hurst RS, Efstratiadis A, Zeitlin S, Chesselet M-F. Enhanced sensitivity to N-methyl-D-aspartate receptor activation in transgenic and knockin mouse models of Huntington’s disease. J Neurosci Res. 1999;58:515–532. [PubMed] [Google Scholar]
- 54.Li H, Li S-H, Johnston H, Shelbourne PF, Li S-J. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat Genet. 2000;25:385–389. doi: 10.1038/78054. [DOI] [PubMed] [Google Scholar]
- 55.Lin C-H, Tallaksen-Greene S, Chien W-M, Cearley JA, Jackson WS, Crouse AB, Ren S, Li X-J, Albin RL, Detloff PJ. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 56.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 57.Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, Chun W, Peters NL, Woods AM, Chan EY, Kooperberg C, Krainc D, Young AB, Tapscott SJ, Olson JM. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum Mol Genet. 2002;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 58.Maat-Schieman MLC, Dorsman JC, Smoor MA, Siesling S, van Duinen SG, Verschuuren JGM, den Dunnen JT, van Ommen G-JB, Roos AC. Distribution of inclusions in neuronal nuclei and dystrophic neurites in Huntington disease brain. J Neuropathol Exp Neurol. 1999;58:129–137. doi: 10.1097/00005072-199902000-00003. [DOI] [PubMed] [Google Scholar]
- 59.Macdonald V, Halliday GM, Trent RJ, McCusker EA. Significant loss of pyramidal neurons in the angular gyrus of patients with Huntington’s disease. Neuropathol Appl Neurobiol. 1997;23:492–495. doi: 10.1111/j.1365-2990.1997.tb01326.x. [DOI] [PubMed] [Google Scholar]
- 60.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 61.Margolis RL, Roos CA. Expansion explosion: new clues to the pathogenesis of repeat expansion neurodegenerative diseases. Trends Mol Med. 2001;7:479–482. doi: 10.1016/S1471-4914(01)02179-7. [DOI] [PubMed] [Google Scholar]
- 62.Markham CH, Knox JW. Observations on Huntington’s chorea in childhood. J Pediatr. 1965;67:46–57. doi: 10.1016/s0022-3476(65)80303-1. [DOI] [PubMed] [Google Scholar]
- 63.McCaughey WTE. The pathologic spectrum of Huntington’s chorea. J Nerv Ment Dis. 1961;133:91–103. [Google Scholar]
- 64.Menalled LB, Chesselet M-F. Mouse models of Huntington’s disease. Trends Pharmacol Sci. 2002;23:32–39. doi: 10.1016/s0165-6147(00)01884-8. [DOI] [PubMed] [Google Scholar]
- 65.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet M-F. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 66.Menalled LB, Sison JD, Wu U, Olivieri M, Li X-J, Li H, Zeitlin S, Chesselet M-F. Early motor dysfunction and striosomal distribution of huntingtin microaggregates in Huntington’s disease knock-in mice. J Neurosci. 2002;22:8266–8276. doi: 10.1523/JNEUROSCI.22-18-08266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Myers RH, Leavitt J, Farrer LA, Jagadeesh J, McFarlane H, Mastromauro CA, Mark RJ, Gusella JF. Homozygote for Huntington disease. Am J Hum Genet. 1989;45:615–618. [PMC free article] [PubMed] [Google Scholar]
- 68.Myers RH, Vonsattel JP, Paskevich PA, Kiely DK, Stevens TJ, Cupples LA, Richardson EP, Jr, Bird ED. Decreased neuronal and increased oligodendroglial densities in Huntington’s disease caudate nucleus. J Neuropathol Exp Neurol. 1991;50:729–742. doi: 10.1097/00005072-199111000-00005. [DOI] [PubMed] [Google Scholar]
- 69.Myers RH, Vonsattel JP, Stevens TJ, Cupples LA, Richardson EP, Martin JB, Bird ED. Clinical and neuropathologic assessment of severity in Huntington’s disease. Neurology. 1988;38:341–347. doi: 10.1212/wnl.38.3.341. [DOI] [PubMed] [Google Scholar]
- 70.Nance MA, Mathias-Hagen V, Breningstall G, Wick MJ, McGlennen RC. Analysis of a very large trinucleotide repeat in a patient with juvenile Huntington’s disease. Neurology. 1999;52:392–394. doi: 10.1212/wnl.52.2.392. [DOI] [PubMed] [Google Scholar]
- 71.Nucifora FC, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, Dawson TM, Ross CA. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 72.Obrietan K, Hoyt KR. CRE-mediated transcription is increased in Huntington’s disease transgenic mice. J Neurosci. 2004;24:791–796. doi: 10.1523/JNEUROSCI.3493-03-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro. 29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 74.Paulson HL. Protein fate in neurodegenerative proteinopathies: polyglutamine diseases join the (mis)fold. Hum Genet. 1999;64:339–345. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Phillips W, Morton AJ, Barker RA. Abnormalities of neurogenesis in the R6/2 mouse model of Huntington’s disease are attributable to the in vivo microenvironment. J Neurosci. 2005;25:11564–11576. doi: 10.1523/JNEUROSCI.3796-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Project: The US—Venezuela Collaborative Research Project. Wexler NS. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc Natl Acad Sci USA. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reddy PH, Williams M, Charles V, Garrett L, Pike-Buchanan L, Whetsell WO, Jr, Miller G, Tagle DA. Behavioural abnormalities and selective neuronal loss in HD transgenic mice expressing mutated full-length HD cDNA. Nat Genet. 1998;20:198–202. doi: 10.1038/2510. [DOI] [PubMed] [Google Scholar]
- 78.Reiner A, Albin RL, Anderson KD, D’Amato CJ, Penney JB, Young AB. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci USA. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richardson EP., Jr Huntington’s disease: some recent neuropathological studies. Neuropathol Appl Neurobiol. 1990;16:451–460. doi: 10.1111/j.1365-2990.1990.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 80.Richfield EK, Maguire-Zeiss KA, Cox C, Gilmore J, Voorn P. Reduced expression of preproenkephalin in striatal neurons from Huntington’s disease patients. Ann Neurol. 1995;37:335–343. doi: 10.1002/ana.410370309. [DOI] [PubMed] [Google Scholar]
- 81.Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu G-Q, Mucke L. Reducing endogenous tau ameliorates amyloid β-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 82.Robitaille Y, Lopes-Cendes I, Becher M, Rouleau G, Clark AW. The neuropathology of CAG repeat diseases: review and update of genetic and molecular features. Brain Pathol. 1997;7:901–926. doi: 10.1111/j.1750-3639.1997.tb00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roos RAC, Pruyt JFM, de Vries J, Bots GTAM. Neuronal distribution in the putamen in Huntington’s disease. J Neurol Neurosurg Psychiatry. 1985;48:422–425. doi: 10.1136/jnnp.48.5.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rosenberg RN. DNA-triplet repeats and neurologic disease. N Engl J Med Line. 1996;335:1222–1224. doi: 10.1056/NEJM199610173351609. [DOI] [PubMed] [Google Scholar]
- 85.Rubinsztein DC. Lessons from animal models of Huntington’s disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 86.Rubinsztein DC, Carmichael J. Huntington’s disease: molecular basis of neurodegeneration. Expert Rev Mol Med. 2003;5:1–21. doi: 10.1017/S1462399403006549. [DOI] [PubMed] [Google Scholar]
- 87.Rubinsztein DC, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman J-J, Chotai K, Connarty M, Craufurd D, Curtis A, Curtis D, Davidson MJ, Differ A-M, Dode C, Dodge A, Frontali M, Ranen NG, Stine OC, Sherr M, Abbott MH, Franz ML, Graham CA, Harper PS, Hedreen JC, Jackson A, Kaplan J-C, Losekoot M, MacMillan JC, Morrison P, Trottier Y, Novelletto A, Simpson S, Theilmann J, Whittaker JL, Folstein SE, Ross CA, Hayden MR. Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am J Hum Genet. 1996;59:16–22. [PMC free article] [PubMed] [Google Scholar]
- 88.Sapp E, Ge P, Aizawa H, Bird E, Penney J, Young AB, Vonsattel J-P, DiFiglia M. Evidence for a preferential loss of enkephalin immunoreactivity in the external globus pallidus in low grade Huntington’s disease using high resolution image analysis. Neuroscience. 1995;64:397–404. doi: 10.1016/0306-4522(94)00427-7. [DOI] [PubMed] [Google Scholar]
- 89.Sapp E, Kegel KB, Aronin N, Yohyama K, Uchiyama Y, Bhide P, Vonsattel JP, DiFiglia M. Microglia accumulate in the HD striatum and cortex. Soc Neurosci. 1999;25:829. [Google Scholar]
- 90.Sapp E, Penney J, Young AB, Aronin N, Vonsattel J-P, DiFiglia M. Axonal transport of N-terminal huntingtin suggests early pathology of corticostriatal projections in Huntington disease. J Neuropathol Exp Neurol. 1999;58:165–173. doi: 10.1097/00005072-199902000-00006. [DOI] [PubMed] [Google Scholar]
- 91.Sapp E, Schwarz C, Chase K, Bhide PG, Young AB, Penney J, Vonsattel JP, Aronin N, DiFiglia M. Huntingtin localization in brains of normal and Huntington’s disease patients. Ann Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 92.Sathasivam K, Hobbs C, Turmaine M, Mangiarini L, Mahal A, Bertaux F, Wanker EE, Doherty P, Davies SW, Bates GP. Formation of polyglutamine inclusions in non-CNS tissue. Hum Mol Genet. 1999;8:813–822. doi: 10.1093/hmg/8.5.813. [DOI] [PubMed] [Google Scholar]
- 93.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 94.Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, Copeland NG, Price DL, Ross CA, Borchelt DR. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 95.Shelbourne PF, Killeen N, Hevner RF, Johnston HM, Tecott L, Lewandoski M, Ennis M, Ramirez L, Li Z, Iannicola C, Littman DR, Myers RM. A Huntington’s disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Hum Mol Genet. 1999;8:763–774. doi: 10.1093/hmg/8.5.763. [DOI] [PubMed] [Google Scholar]
- 96.Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, Deng Y, Pearson J, Vaid K, Bissada N, Wetzel R, Leavitt BR, Hayden MR. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci USA. 2005;102:11402–11407. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang Y-Z, Li X-J, Simpson EM, Gutekunst C-A, Leavitt BR, Hayden MR. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 98.Sotrel A, Paskevich PA, Kiely DK, Bird ED, Williams RS, Myers RH. Morphometric analysis of the prefrontal cortex in Huntington’s disease. Neurology. 1991;41:1117–1123. doi: 10.1212/wnl.41.7.1117. [DOI] [PubMed] [Google Scholar]
- 99.Squitieri F, Gellera C, Cannella M, Mariotti C, Cislaghi G, Rubinsztein DC, Almqvist EW, Turner D, Bachoud-Lévi A-C, Simpson SA, Delatycki M, Maglione V, Hayden MR, Di Donato S. Homozygosity for CAG mutation in Huntington disease is associated with a more severe clinical course. Brain. 2003;126:946–955. doi: 10.1093/brain/awg077. [DOI] [PubMed] [Google Scholar]
- 100.Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 101.Telenius H, Kremer B, Goldberg YP, Theilmann J, Andrew SE, Zeisler J, Adam S, Greenberg C, Ives EJ, Clarke LA, Hayden MR. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- 102.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007 doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 103.Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, Weber C, David G, Tora L, Agid Y, Brice A, Mandel J-L. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature. 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 104.Turmaine M, Raza A, Mahal A, Mangiarini L, Bates GP, Davies SW. Nonapoptotic neurodegeneration in a transgenic mouse model of Huntington’s disease. Proc Natl Acad Sci USA. 2000;97:8093–8097. doi: 10.1073/pnas.110078997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Dellen A, Blakemore C, Deacon R, York D, Hannan AJ. Delaying he onset of Huntington’s in mice. Nature. 2000;404:721–722. doi: 10.1038/35008142. [DOI] [PubMed] [Google Scholar]
- 106.von Hörsten S, Schmitt I, Nguyen HP, Holzmann C, Schmidt T, Walther T, Bader M, Pabst R, Kobbe P, Krotova J, Stiller D, Kask A, Vaarmann A, Rathke-Hartlieb S, Schulz JB, Grasshoff U, Bauer I, Menezes AM, Vieira-Saecker AMM, Paul M, Jones L, Lindenberg KS, Landwehrmeyer B, Bauer A, Li X-J, Riess O. Transgenic rat model of Huntington’s disease. Hum Mol Genet. 2003:12. doi: 10.1093/hmg/ddg075. [DOI] [PubMed] [Google Scholar]
- 107.Vonsattel J-P, Myers RH, Bird ED, Ge P, Richardson EP., Jr Maladie de Huntington: sept cas avec îlots néostriataux relativement préservés. Rev Neurol. 1992;148:107–116. [PubMed] [Google Scholar]
- 108.Vonsattel J-P, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 109.Vonsattel J-PG, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 110.Wexler NS, Young AB, Tanzi RE, Travers H, Starosta-Rubinstein S, Penney JB, Snodgrass SR, Shoulson I, Gomez F, Arroyo MAR, Penchaszadeh GK, Moreno H, Gibbons K, Faryniarz A, Hobbs W, Anderson MA, Bonilla E, Conneally PM, Gusella JF. Homozygotes for Huntington’s disease. Nature. 1987;326:194–197. doi: 10.1038/326194a0. [DOI] [PubMed] [Google Scholar]
- 111.Wheeler VC, White JK, Gutekunst C-A, Vrbanac V, Weaver M, Li X-J, Li S-H, Yi H, Vonsattel J-P, Gusella JF, Hersch S, Auerbach W, Joyner AL, MacDonald ME. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum Mol Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 112.White JK, Auerbach W, Duyao MP, Vonsattel J-P, Gusella JF, Joyner AL, MacDonald ME. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 113.Yu Z-X, Li S-H, Evans J, Pillarisetti A, Li H, Li X-J. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. J Neurosci. 2003;23:2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zalneraitis EL, Landis DMD, Richardson EP, Jr, Selkoe DJ. A comparison of astrocytic structure in cerebral cortex and striatum in Huntington’s disease. Neurology. 1981;31:151. [Google Scholar]