Abstract
Vibrio cholerae, the bacterium that causes the disease cholera, controls virulence factor production and biofilm development in response to two extracellular quorum-sensing molecules, called autoinducers. The strongest autoinducer, called CAI-1 (for cholera autoinducer-1), was previously identified as (S)-3-hydroxytridecan-4-one. Biosynthesis of CAI-1 requires the enzyme CqsA. Here, we determine the CqsA reaction mechanism, identify the CqsA substrates as (S)-2-aminobutyrate and decanoyl coenzyme A, and demonstrate that the product of the reaction is 3-aminotridecan-4-one, dubbed amino-CAI-1. CqsA produces amino-CAI-1 by a pyridoxal phosphate (PLP)-dependent acyl-CoA transferase reaction. Amino-CAI-1 is converted to CAI-1 in a subsequent step via a CqsA-independent mechanism. Consistent with this, we find cells release ≥100 times more CAI-1 than amino-CAI-1. Nonetheless, V. cholerae responds to amino-CAI-1 as well as CAI-1, whereas other CAI-1 variants do not elicit a quorum-sensing response. Thus, both CAI-1 and amino-CAI-1 have potential as lead molecules in the development of an anti-cholera treatment.
Keywords: Quorum sensing, Vibrio cholerae, CqsA, CAI-1, autoinducer, PLP, virulence
Quorum sensing is a mechanism of bacterial cell-cell communication that relies on the production, release, accumulation, and subsequent detection of signaling molecules called autoinducers1–3. Bacteria use quorum sensing to track changes in cell population density and, in response, to synchronously control community-wide gene expression. Quorum sensing thus enables bacteria to distinguish times when they are solitary from times when they are in a group, and to activate responses tailored to particular population densities. In the human pathogen V. cholerae, quorum sensing regulates activities including biofilm formation and virulence factor expression4–9. V. cholerae is atypical in that accumulation of autoinducers at high cell density results in the repression, rather than the activation, of both biofilm formation and virulence factor production. These traits are generally activated by quorum sensing in pathogens that cause persistent infections. V. cholerae, by contrast, causes an acute disease in which the cessation of virulence factor expression may be important in enabling the bacteria to escape from the host and re-enter the environment4,7. Consistent with this model, a protease proposed to assist in detaching the bacteria from host intestinal epithelial cells is induced in response to autoinducer accumulation5. Both biofilm formation and virulence factor expression are required for V. cholerae to cause infection in humans, and because quorum sensing represses both of these traits, administering autoinducers or other quorum sensing agonists could form the basis of a novel cholera therapeutic6.
V. cholerae employs two autoinducers for quorum sensing, called cholera autoinducer-1 (CAI-1, 1) and autoinducer-2 (AI-2, 2; ref. 6, and see Supplementary Methods). Information from both autoinducers is transduced into the cell via a shared phosphorylation-dephosphorylation cascade that ultimately affects the expression of over 70 target genes8. We have previously identified CAI-1 (1) as (S)-3-hydroxytridecan-4-one and AI-2 (2) as (2S, 4S)-2-methyl-2,3,4-tetrahydroxytetrahydrofuran-borate9–11. The synthesis of CAI-1 (1) and AI-2 (2) requires the enzymes CqsA and LuxS, respectively6,9–11. While the mechanism by which LuxS catalyzes the production of AI-2 (2) is relatively well understood10–14, the specific role of CqsA in the production of CAI-1 (1) has not previously been investigated.
Here, we use a combination of approaches to establish that CqsA is a pyridoxal phosphate (PLP, 3)-dependent acyl-CoA transferase that ligates (S)-2-aminobutyrate (S-4) and decanoylcoenzyme A (5) to form 3-aminotridecan-4-one (amino-CAI-1, 6) with the concomitant release of CoA (7). We find furthermore that cells possess an activity that converts amino-CAI-1 (6) to CAI-1 (1). Although the enzyme(s) responsible for this conversion have not yet been identified, our results establish that amino-CAI-1 (6) is an on-pathway intermediate in the production of the major cholera quorum-sensing signal molecule, CAI-1 (1).
Results
The CqsA enzyme and PLP-dependent acyl-CoA transferases
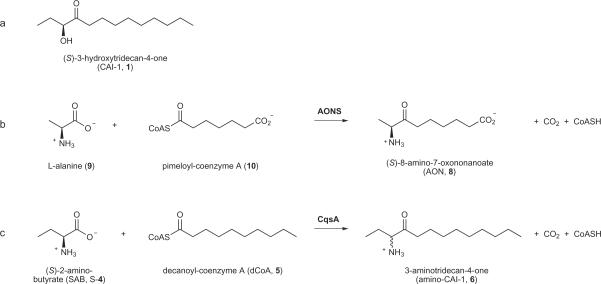
We previously purified V. cholerae CAI-1 (1), determined its chemical structure (Scheme. 1a), and established that its synthesis requires CqsA9. A conundrum was that, based on amino acid sequence analysis, CqsA and its homologues appear likely to be PLP(3)-dependent acyl-CoA transferases. Despite the impressive catalytic potential of PLP(3)-dependent enzymes – they can carry out over 140 different enzymatic reactions – there is as yet no precedent for synthesis of an α-hydroxyketone such as CAI-1 (1)15. The protein most closely related to CqsA by sequence is the metabolic acyl-CoA transferase 8-amino-7-oxononanoate synthase16 (AONS; 29% identical and 49% similar to CqsA). AONS catalyzes the production of the α-aminoketone (S)-8-amino-7-oxononanoate (AON, 8) from L-alanine (9) and pimeloyl-CoA (10)17 (Scheme. 1b). If CqsA were to function via a mechanism analogous to that employed by AONS18, the product most closely resembling CAI-1 (1) would carry an amino group in place of the C3 hydroxyl group (Scheme. 1c). We term this molecule amino-CAI-1 (3-aminotridecan-4-one, 6).
Scheme 1. CAI-1, AON, and amino-CAI-1.
(a) CAI-1; (S)-3-hydroxytridecan-4-one. (b) The AONS reaction. AONS catalyzes the production of the α-aminoketone (S)-8-amino-7-oxononanoate (AON) from L-alanine and pimeloyl-CoA. (c) The proposed CqsA reaction, catalyzing the production of the α-aminoketone amino-CAI-1 from SAB ((S)-2-aminobutyrate) and dCoA (decanoyl-coenzyme A).
CqsA resembles AONS
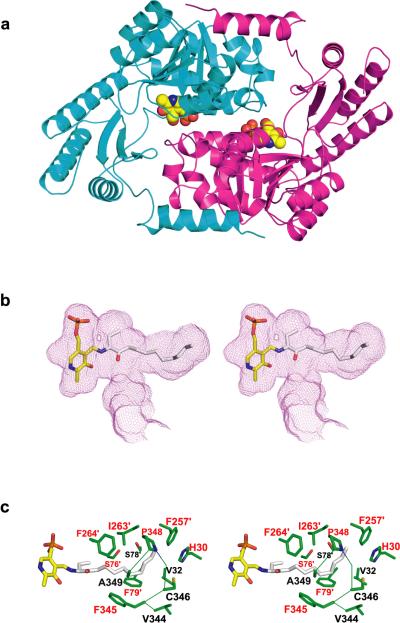
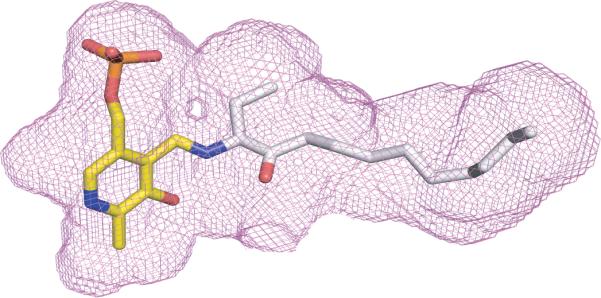
To examine the possibility that CqsA is an acyl-CoA transferase, we determined the CqsA crystal structure using molecular replacement and refined it to 1.8 Å resolution (Fig. 1a and Table S1; see Methods for further details). The crystal structure revealed a homodimer strongly resembling AONS16 (rms deviation = 2.3 Å for 579 equivalent Cα atoms). Key residues surrounding the PLP (3) cofactor and the substrate binding sites are nearly identical to the corresponding residues in AONS16,18. A notable exception is the absence, in CqsA, of a residue analogous to Arg 21 of AONS. Arg 21 coordinates the distal carboxylate in the otherwise aliphatic tail of AON. By contrast, the corresponding region of the CqsA substrate binding pocket is almost entirely lined with hydrophobic residues. Based on the known structure of the substrate AON bound to AONS18, we modeled amino-CAI-1 (6) into the active site of CqsA, bound via a Schiff base linkage to PLP (3, Fig. 1b). Strikingly, amino-CAI-1 (6) fits snugly into the cavity adjacent to the PLP (3) cofactor, without generating steric clashes and without leaving any unfilled volume large enough to accommodate a water molecule. These structural results suggest that the well-characterized mechanism of AONS catalysis18 is an appropriate model for understanding the enzymatic activity of CqsA.
Figure 1. Structural and functional analysis of CqsA.
(a) The x-ray structure (model B; see Methods) of CqsA, with monomers depicted in cyan and magenta. The two PLP molecules are shown as space-filling models. (b) Stereo-view of the proposed PLP-amino-CAI-1 product aldimine modeled into the large cavity observed within the CqsA homodimer. The product aldimine is shown as a stick representation, with oxygen red, phosphorus orange, nitrogen blue, and carbon atoms yellow (PLP) and gray (amino-CAI-1). The cavity boundary, calculated using the program VOIDOO25, is shown as a purple mesh. (c) Stereo-view of the side chains (green) contacting the modeled product aldimine in panel (b) (except for those side chains in contact with PLP). Residue labels are color-coded according to the strength of defect caused by substitution of each residue with alanine (Table S2), with red labels for those residues whose substitution resulted in <0.05% residual autoinducer production.
Site-directed mutagenesis of the CqsA active site
To probe further the function of CqsA, each residue in the substrate binding site was individually substituted with alanine (Fig. 1c). We evaluated the catalytic potential of the mutant proteins using a V. cholerae reporter strain that produces bioluminescence in response to CAI-16,9 (Table S2). As a control, we confirmed that E. coli cells expressing wild-type CqsA release large amounts of autoinducer activity9. E. coli expressing alanine-substituted proteins were in many cases severely compromised in the production of autoinducer activity. The severity of the defect strongly correlated with the location of the mutation (Fig. 1c). Alanine substitutions in the region of CqsA that binds the PLP (3) cofactor, or in the region modeled to bind the polar head group of amino-CAI-1 (6, Fig. 1b), were not well tolerated (<0.05% residual activity; Table S2). By contrast, alanine substitutions located in the distal region of the substrate binding site predicted to interact with the aliphatic tail of the substrate generally caused little impairment in activity (≥10% residual activity), although several of the alanine substitutions destabilized the enzyme itself (Table S2, Fig. S1). These results are in excellent agreement with the hypothesis that CqsA functions by a mechanism analogous to that used by the acyl-CoA transferase AONS. If so, CqsA would be expected to produce amino-CAI-1 (6) rather than CAI-1 (1, Scheme 1c).
In vitro synthesis of amino-CAI-1 by CqsA
To test directly whether CqsA can catalyze the amino-CAI-1 (6) synthesis reaction shown in Scheme 1c, we combined purified CqsA protein with the presumptive substrates (S)-2-aminobutyrate (SAB, S-4) and decanoyl-CoA (dCoA, 5). Mass spectrometry was used to measure production of amino-CAI-1 (6) and/or CAI-1 (1). We found that amino-CAI-1 (6) was produced while CAI-1 (1) was not (Table S3). As expected (Scheme 1c), the reaction also produced free CoA (7, Fig. S2). Amino-CAI-1 (6) was not produced in control reactions in which an inactive variant of CqsA (K236A; Table S2) was used or in which one of the two substrates was omitted (Table S3). Substitution of (R)-2-aminobutyrate (R-4) for (S)-2-aminobutyrate (S-4) led to production of amino-CAI-1 (6), but with substantially lower yield (and, presumably, with opposite stereochemistry; Table S3). Thus, CqsA is able to convert SAB (S-4) and dCoA (5) to amino-CAI-1 (6). The measured kinetic parameters for the reaction (Fig. S3) are roughly similar to those reported for AONS18. For example, kcat values for CqsA and AONS are 0.024 and 0.06 s−1, respectively. For CqsA, KM = 5.3 mM (SAB, S-4) and 19 μM (dCoA, 5); for AONS, KM = 0.5 mM (alanine, 9) and 25 μM (pimeloyl-CoA, 10). To examine the CqsA reaction in greater detail, we monitored spectral changes induced by the interaction of CqsA with SAB (S-4), dCoA (5), and amino-CAI-1 (6, Supplementary methods and Fig. S4). The results are entirely consistent with the conclusion that CqsA functions via a mechanism closely resembling that of AONS18.
In vivo synthesis of amino-CAI-1 by CqsA
Given that CqsA is capable of producing amino-CAI-1 in vitro, we next tested whether we could detect amino-CAI-1 in bacterial culture fluids. Culture fluids from E. coli overexpressing CqsA and from V. cholerae (cqsA on the chromosome) were collected, spiked with synthetic deuterated CAI-1 (d-1) and amino-CAI-1 (d-6) standards, extracted, and concentrated. CAI-1 (1) and amino-CAI-1 (6), and their deuterated counterparts (d-1 & d-6), were quantified using mass spectrometry (Table S4). When grown in minimal medium, E. coli overexpressing CqsA produced readily detectable amounts of both CAI-1 (1) and amino-CAI-1 (6; Table S4). At least one-hundred-fold more CAI-1 (1) than amino-CAI-1 (6) was present in the E. coli culture fluids at all growth stages (Table S4, Fig. S5a,b), and neither molecule was detected in control experiments using E. coli that lacked CqsA. The lower levels of amino-CAI-1 (6), relative to CAI-1 (1), are not due to instability. Indeed, we found that synthetic amino-CAI-1 (6) is significantly more stable than synthetic CAI-1 (1) in LB medium and in potassium phosphate buffer (Fig. S5c,d). As reported previously9, V. cholerae culture fluids contain much less CAI-1 (1) than do culture fluids from E. coli overexpressing CqsA, presumably because of the difference in enzyme levels (Table S4). Moreover, V. cholerae culture fluids do not contain detectable levels of amino-CAI-1 (6). The failure to detect amino-CAI-1 (6) may be due to the requirement that V. cholerae be grown rich medium, which compromises mass spectral analysis, in order to elicit autoinducer production.
In vivo conversion of amino-CAI-1 to CAI-1
The data presented above show that the presence of CqsA leads to the production of amino-CAI-1 (6), both in vitro and in vivo. We therefore tested whether cells are capable of converting amino-CAI-1 (6) to the quorum-sensing autoinducer CAI-1 (1). We also attempted to detect the reverse reaction, conversion of CAI-1 (1) to amino-CAI-1 (6). For these experiments, we exposed growing E. coli cells lacking CqsA to 40 μM synthetic amino-CAI-1 (6) or synthetic CAI-1 (1). Following incubation, we extracted the samples, spiked them with deuterated amino-CAI-1 (d-6) and CAI-1 standards (d-1), and measured the levels of amino-CAI-1 (6) and CAI-1 (1) using mass spectrometry. Cells provided with amino-CAI-1 (6) converted approximately 23% of the amino-CAI-1 (6) to CAI-1 (1; Table S5). Conversely, when E. coli cells were provided with CAI-1 (1), approximately 6% was converted to amino-CAI-1 (6). Unfortunately, we were unable to demonstrate conversion using V. cholerae cells, apparently for technical reasons (see Methods). Nonetheless, our results establish that E. coli, and presumably V. cholerae, is able to catalyze the interconversion of amino-CAI-1 (6) and CAI-1 (1). Furthermore, it appears that the `forward' reaction, converting amino-CAI-1 (6) to CAI-1 (1), may be thermodynamically favored (Tables S4 and S5). Importantly, conversion occurs via a CqsA-independent mechanism. Taken together, our data suggest that, in vivo, CqsA catalyzes the reaction of SAB (S-4) and dCoA (5) to produce amino-CAI-1 (6; Scheme 1c). Amino-CAI-1 (6) is subsequently converted, via a CqsA-independent reaction, to CAI-1 (1).
Selectivity of the CqsS receptor
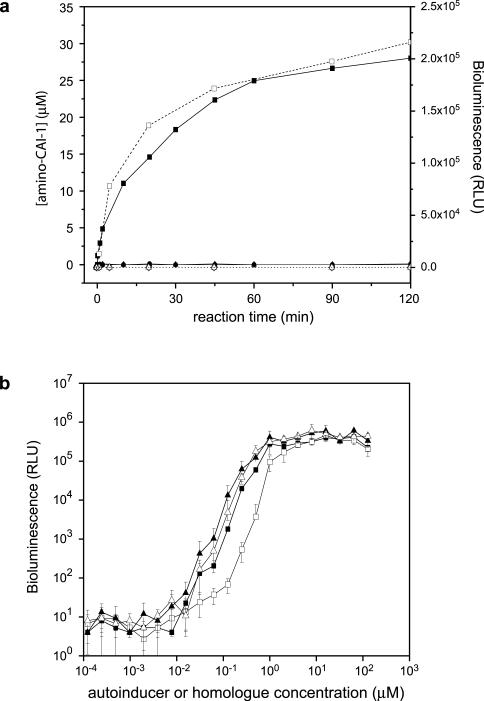
The products of the in vitro CqsA reaction were tested for activity using the V. cholerae bioluminescence bioassay. The signaling activity detected by this method precisely tracked the production of amino-CAI-1 (6) as measured by mass spectrometry (Fig. 2a). To confirm that amino-CAI-1 (6), like CAI-1 (1), is biologically active, we used synthetic amino-CAI-1 (6) in the same assay (Fig. 2b). CAI-1 (1), (S)-amino-CAI-1 (S-6), and (R)-amino-CAI-1 (R-6) displayed nearly indistinguishable activities (EC50's range from 1 μM to 1.5 μM). As reported earlier, (R)-CAI-1 (R-1) is slightly (~2-fold) less active than CAI-1 (i.e., the (S) isomer, 1)9. There are two possible explanations for the finding that amino-CAI-1 (6) is active in the bioassay. First, amino-CAI-1 (6) may bind to and elicit signaling activity from the CqsS receptor directly. Alternatively, amino-CAI-1 (6) may be converted during the course of the bioassay to CAI-1 (1), which then binds CqsS. These possibilities are not mutually exclusive, and our efforts to distinguish them have thus far proved inconclusive.
Figure 2. Activity of in vitro synthesized amino-CAI-1 and related compounds.
(a) Purified CqsA (20 μM) was incubated with 10 mM SAB and 100 μM dCoA and the evolution of amino-CAI-1 was measured using mass spectrometry (filled squares, left Y-axis) and the V. cholerae bioluminescence assay (open squares, right Y-axis). No product is detected by mass spectrometry (filled symbols) or the bioluminescence assay (open symbols) when either SAB (triangles) or dCoA (circles) is omitted, or when catalytically inactive CqsA (K236A, diamonds) is substituted for the wild-type enzyme. Relative light units (RLU) are defined as counts min−1ml−1/OD600. (b) Activity of synthetic (S)- and (R)-amino-CAI-1 (filled and open triangles, respectively) and (S)- and (R)-CAI-1 (filled and open squares, respectively) in the V. cholerae bioluminescence reporter assay.
To further examine CqsS selectivity, we synthesized and examined the bioactivity of a variety of other molecules in which the hydroxyl group at the C3 position was replaced by a proton (11) or by a chloride (12), bromide (13), thiol (14), or carbonyl (15) group. None of these analogs showed significant activity (<1% that of CAI-1) except the molecule containing the carbonyl group (15), which exhibited modest activity (<10%) (data not shown). Thus, the CqsS receptor is highly sensitive to the group at the C3 position.
Finally, since it reports on the presence of both amino-CAI-1 (6) and CAI-1 (1), the V. cholerae bioassay was used to test whether CqsA is capable of producing amino-CAI-1 (6) or CAI-1 (6) from a wide array of potential three-, four-, and ten-carbon substrates. Aside from the reaction described in Scheme 1c, no product was detected with combinations of test compounds containing three carbons (propionaldehyde (16), 1-propanol (17), propionic acid (18), propylamine (19) and pyruvic acid (20)), four carbons (1-butanol (21), 2-ketobutyric acid (22), 1,2-butanediol (23), 2-aminobutyric acid (4) and 2-hydroxybutyric acid (24)), and ten carbons (decanal (25), dCoA (5) and decanoic acid (26)) (structures provided in Fig. S6).
Discussion
Our findings lead to the unexpected conclusion that CqsA, the only enzyme known to be required for synthesis of the major V. cholerae autoinducer CAI-1 (1), does not produce CAI-1 (1) directly. Rather, CqsA produces amino-CAI-1 (6) by a PLP (3) -dependent acyl-CoA transferase reaction. Another enzyme, or set of enzymes, must therefore catalyze the subsequent conversion of amino-CAI-1 (6) to CAI-1 (1). This additional enzyme is present in both E. coli and V. cholerae, suggesting that it is rather ubiquitous.
Our results also raise the question of why, given that amino-CAI-1 (6) is itself a potent autoinducer, cells nonetheless convert most of it to CAI-1 (1). One possible explanation is that conversion of amino-CAI-1 (6) to CAI-1 (1) is accidental; that is, amino-CAI-1 (6) is a fortuitous substrate of a housekeeping enzyme that catalyzes the conversion. This model is consistent with the inferred ubiquity of the enzyme(s) responsible. A second and more interesting possibility is that converting the long-lived amino-CAI-1 (6) molecule to the short-lived CAI-1 (1) molecule is required to allow an immediate response to changes in autoinducer concentration. Decreasing the persistence time of the extracellular signal molecule could force V. cholerae to respond rapidly to changes in cell density, while simultaneously reducing the likelihood that other, potentially competing, bacteria could tune in to the presence of V. cholerae cells. These and other models should become testable once the enzyme required to convert amino-CAI-1 (6) to CAI-1 (1) has been identified and inactivated.
Methods
Cloning, expression and purification of V. cholerae CqsA
The cqsA gene (VCA0523) was amplified by PCR from V. cholerae genomic DNA, cloned into pET-28b (Novagen), and sequenced. N-terminally His6-tagged CqsA was overproduced and purified as described in Supplementary Methods.
Crystallization and data collection
Crystals were grown by vapor diffusion at 23°C from a protein stock (5 mg ml−1) mixed 1:1 (v/v) with well solutions. Diffraction data for two different crystal forms (Table S1) were collected at National Synchrotron Light Source (NSLS) beamline X29 and processed using the HKL Software package19. Crystal A, which diffracted to 2.7 Å resolution, was grown over wells containing 1.25 M ammonium sulfate, 100 mM Bicine (pH 8.0), and 10 mM DTT. Crystal B, which diffracted to 1.8 Å resolution, was grown over wells containing 1.25 M ammonium sulfate, 100 mM bis-Tris propane (BTP, pH 8.5), 10 mM DTT, 50 μM amino-CAI-1 (6), and 0.1% (v/v) dimethyl sulfoxide. Crystals were cryoprotected by brief soaking in mother liquor containing glycerol in concentrations increasing up to 35% (v/v).
Structure determination and refinement
Crystals A and B each contained one homodimer in the asymmetric unit (Table S1). An initial model for CqsA was determined for crystal B using the molecular replacement program PHASER20 with an ensemble search model corresponding to the superimposed structures of 8-amino-7-oxononanoate synthase (PDB entries 1BS0 and 1DJ9), 5-aminolevulinate synthase (2BWN and 2BWO), and serine palmitoyl transferase (2JG2). The structure was improved using ARP/wARP21 with data from crystal B to 1.8 Å resolution, and used as a starting point for the refinement of crystal A data at 2.7 Å resolution. In both cases, geometrically restrained individual B factor refinement was used, with weights adjusted on the basis of Rfree. Structures were improved via iterative cycles of building in WINCOOT22 and refinement in REFMAC23. The final model lacked only the first three residues at the N-terminus and the hexahistidine tag. After all water molecules had been incorporated into the model, two magnesium ions, two PLP (3) molecules, and two DTT (27) molecules were built into clear, non-protein electron density. Despite the presence of amino-CAI-1 (6) during crystallization, no corresponding electron density was observed in crystal B. The model derived from crystal B has excellent geometry as determined by MolProbity24 (97.9% in favored regions of the Ramachandran plot and 100% in allowed regions). The model derived from crystal A also has good geometry (95.6% in favored regions of the Ramachandran plot and 100% in allowed regions). The major difference between the two models is the conformation of Lys 236. In AONS and homologous enzymes, this lysine residue is typically engaged in a covalent aldimine linkage between its ε-amino group and the aldehyde of the PLP (3) cofactor15. This aldimine linkage, for which clear electron density was observed, was included in the model derived from the crystal A data. In contrast, convincing electron density for a covalent bond was absent for crystal B; consequently, the interaction was modeled as a hydrogen bond between the ε-amino group of Lys 236 and the free aldehyde of PLP (6). The structure coordinates for both crystals A and B have been deposited in the Brookhaven Protein Data Bank with accession codes of 3HQT and 3HQS, respectively.
Preparation of cell-free culture fluids for bioluminescence activity assays
Site-directed mutagenesis was conducted using PfuUltra high-fidelity polymerase (Stratagene) and complementary PCR primers containing targeted mutations in cqsA. Alanine-substituted or wild-type proteins were overproduced in E. coli BL21 (DE3) cells as described in the Supplementary Methods. After pelleting the cells, the culture fluids were sterilized by filtration through 0.22 μm pore filters (Millipore) and frozen in liquid nitrogen. In 96-well plates, 10 μl of serially-diluted culture fluids were added to 90 μl of a 100-fold diluted overnight culture of reporter strain MM920 (V. cholerae El Tor C6706str ΔcqsA ΔluxQ/pBB1 (luxCDABE from V. harveyi)) grown in LB medium with 5 mg l−1 tetracycline. Bioluminescence was measured as previously reported6,9.
In vitro enzyme activity assay
Enzyme activity was measured by incubating 1 μM CqsA with 25 mM SAB (S-4) and/or 100 μM dCoA (5) at 23°C in 20 mM BTP, pH 8.0, 200 mM NaCl. At each time-point, the reaction was terminated by adding an aliquot of the reaction to acetonitrile (final concentration 30% (v/v)), which caused CqsA to precipitate. Following centrifugation, amino-CAI-1 (6) and CAI-1 (1) present in the supernatant were measured using mass spectrometry as described in Supplementary Methods, while autoinducer activity was measured using the bioluminescence assay.
Mass spectrometry quantification of molecules using deuterated standards
E. coli MG1655 carrying the cqsA gene from V. cholerae on plasmid pTrc99A was grown in M9 minimal medium supplemented with glucose (4 g l−1) and ampicillin (0.1 g l−1) at 30°C with shaking. cqsA expression was induced at an OD600 of 0.12 with 0.1 mM IPTG. Cells were subsequently grown to an OD600 of 2.1. V. cholerae El Tor C6706str cells were grown in tryptone broth at 30°C with shaking to OD600 of 2.5–2.7. Following centrifugation, chemically-synthesized deuterium-labeled CAI-1 (d-1) and amino-CAI-1 (d-2) standards were added to 100-ml culture fluid samples. The samples were extracted with an equal volume of dichloromethane, after which the organic phase was isolated, evaporated, dissolved in 300 μl of 50:50 methanol:water, and analyzed using mass spectrometry (see Supplementary Methods). The concentrations of CAI-1 (1) and amino-CAI-1 (6) were determined by comparing signals of the unlabeled and labeled compounds.
Interconversion of amino-CAI-1 and CAI-1
To optimize uptake and potential interconversion of amino-CAI-1 (6) and CAI-1 (1), membrane-permeable E. coli MC4100 ara+ imp4213 was grown in M9 minimal medium supplemented with glucose (4 g l−1) at 30°C with shaking to an OD600 of 0.7. Two 400-ml cultures were pooled and subsequently divided into six aliquots of 125 ml each. Each aliquot was provided 40 μM CAI-1 (1), amino-CAI-1 (6), or an equivalent volume of DMSO and grown for 4 h at 30°C with shaking (to OD600 ~1.6). Cells were removed by centrifugation and 100 ml of culture fluid was extracted with an equal volume of dichloromethane. The organic phase was isolated, evaporated, dissolved in 300 μl of 50:50 methanol:water and analyzed for CAI-1 (1) or amino-CAI-1 (6) by mass spectrometry (see Supplementary Methods). Multiple attempts to detect interconversion between amino-CAI-1 (6) and CAI-1 (1) in V. cholerae cells failed. Possible explanations include: 1. V. cholerae does not carry out the interconversion reaction, 2. V. cholerae, unlike membrane-permeable E. coli, does not efficiently import amino-CAI-1 (6) or CAI-1 (1), and 3. The rich medium required for growth of V. cholerae interferes with mass spectral detection (see text).
Chemical synthesis
Chemical synthesis of amino-CAI-1 (6), deuterated CAI-1 (d-1) and amino-CAI-1 (d-6) standards, and CAI-1 analogs (12 – 15) is described in the Supplementary Methods. CAI-1 (1) synthesis was described previously9.
Supplementary Material
Figure 4.
Acknowledgments
We thank the staff of the National Synchrotron Light Source beamline X29 for assistance with x-ray data collection; Natacha Ruiz and Anna Arnaudo (Princeton University) for strains; and Jack Kirsch for insightful discussions. This work was supported by HHMI; NIH grants AI054442, GM065859, and AI069326; NSF grant MCB-0639855; and a Dr. Horst Witzel Fellowship from Cephalon Corporation (to M.E.B.).
Literature Cited
- 1.Greenberg EP. Bacterial communication and group behavior. J Clin Invest. 2003;112:1288–90. doi: 10.1172/JCI20099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–46. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 4.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–4. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 5.Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–34. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 6.Miller MB, Skorupski K, Lenz D, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;9:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–56. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U S A. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins DA, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–6. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 11.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum sensing signal molecule. Mol. Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 12.Gopishetty B, et al. Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate analogues. J Am Chem Soc. 2009;131:1243–50. doi: 10.1021/ja808206w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller ST, et al. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–87. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Pei D, Zhu J. Mechanism of action of S-ribosylhomocysteinase (LuxS) Curr Opin Chem Biol. 2004;8:492–7. doi: 10.1016/j.cbpa.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 16.Alexeev D, et al. The crystal structure of 8-amino-7-oxononanoate synthase: a bacterial PLP-dependent, acyl-CoA-condensing enzyme. J Mol Biol. 1998;284:401–19. doi: 10.1006/jmbi.1998.2086. [DOI] [PubMed] [Google Scholar]
- 17.Izumi Y, Morita H, Tani Y, Ogata K. Partial purification and some properties of 7-keto-8-aminopelargonic acid synthetase, an enzyme involved in biotin biosynthesis. Agric Biol Chem. 1973;37:1327–1333. [Google Scholar]
- 18.Webster SP, et al. Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies. Biochemistry. 2000;39:516–28. doi: 10.1021/bi991620j. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1998;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 21.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–63. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.